Abstract
Circadian rhythms are the approximate 24-h biological cycles that function to prepare an organism for daily environmental changes. They are driven by the molecular clock, a transcriptional:translational feedback mechanism that in mammals involves the core clock genes Bmal1, Clock, Per1/2, and Cry1/2. The molecular clock is present in virtually all cells of an organism. The central clock in the suprachiasmatic nucleus (SCN) has been well studied, but the clocks in the peripheral tissues, such as heart and skeletal muscle, have just begun to be investigated. Skeletal muscle is one of the largest organs in the body, comprising approximately 45% of total body mass. More than 2300 genes in skeletal muscle are expressed in a circadian pattern, and these genes participate in a wide range of functions, including myogenesis, transcription, and metabolism. The circadian rhythms of skeletal muscle can be entrained both indirectly through light input to the SCN and directly through time of feeding and activity. It is critical for the skeletal muscle molecular clock not only to be entrained to the environment but also to be in synchrony with rhythms of other tissues. When circadian rhythms are disrupted, the observed effects on skeletal muscle include fiber-type shifts, altered sarcomeric structure, reduced mitochondrial respiration, and impaired muscle function. Furthermore, there are detrimental effects on metabolic health, including impaired glucose tolerance and insulin sensitivity, which skeletal muscle likely contributes to considering it is a key metabolic tissue. These data indicate a critical role for skeletal muscle circadian rhythms for both muscle and systems health. Future research is needed to determine the mechanisms of molecular clock function in skeletal muscle, identify the means by which skeletal muscle entrainment occurs, and provide a stringent comparison of circadian gene expression across the diverse tissue system of skeletal muscle.
Keywords: Bmal1, MyoD1, metabolism, Clock, sarcomere, insulin signaling
The importance of skeletal muscle to health is often underestimated, yet skeletal muscle is the most abundant tissue in the human body, comprising approximately 45% of total body mass (Goodpaster et al., 2000; Hoppeler and Fluck, 2002). Skeletal muscle tissue is an intricate network of more than 600 individual muscles that have different fiber-type compositions, metabolic capacities, and mechanical functions (Poole, 1986). As a whole, skeletal muscle tissue is critical for systemic health and quality of life. It is well understood that skeletal muscle functions to produce force and enable locomotion, but skeletal muscle is involved in a number of integral processes that are frequently forgotten. Skeletal muscle serves as a principal reservoir for amino acids in the absence of nutrient intake, thereby allowing the maintenance of protein synthesis in other tissues. This supply of amino acids also serves as a pool of precursors for hepatic gluconeogenesis, which is significant in sustaining blood glucose levels in the fasting state (Ripperger et al., 1995). Not only does skeletal muscle serve as a reserve of amino acids, but it also acts as a depot for glucose in the postprandial state. As much as 80% of postprandial glucose is taken up by skeletal muscle (Defronzo et al., 1981; Ferrannini et al., 1988). Consequently, skeletal muscle is vital for systemic-level glucose homeostasis. Furthermore, it has been demonstrated in recent years that skeletal muscle is an endocrine tissue. Factors (known as myokines) are secreted from the muscle and work both locally on the muscle tissue and remotely on other tissues. For example, interleukin (IL)–6 is a myokine that is secreted from muscle following exercise. Contracting muscle releases IL-6 into the bloodstream, and it acts locally on muscle to promote glucose uptake and fat oxidation and remotely on liver and adipose tissue to increase glucose production and lipolysis, respectively (Febbraio and Pedersen, 2002). Other identified myokines include IL-8, IL-15, and brain-derived neurotrophic factor and Irisin (Febbraio and Pedersen, 2002; Nielsen and Pedersen, 2007; Matthews et al., 2009).
Changes in muscle composition and function are also strongly correlated with disease development. Two of the most common diseases in the United States, cardiovascular disease and cancer, are associated with loss of muscle mass, diminished strength, and impaired muscle metabolism. Notably, severe loss of muscle mass is a significant risk factor for mortality in these disease states (Kadar et al., 2000; Akashi et al., 2005). Another common health concern, diabetes, is intricately tied to skeletal muscle function. Decreased insulin sensitivity in skeletal muscle is involved in the onset of type 2 diabetes (DeFronzo et al., 1992; Reaven, 2005). It is clear that changes in skeletal muscle mass and metabolism affect human health, but recently a new skeletal muscle property that may affect disease development has come to light. Skeletal muscle, like virtually every cell in the body, has circadian rhythms, and recent studies have begun to demonstrate that disruptions in circadian rhythms can be detrimental to skeletal muscle health (Yoo et al., 2004; Andrews et al., 2010). The goal of this review is to present the concept of circadian rhythms in skeletal muscle, provide insight into the role of circadian rhythms in skeletal muscle function and metabolism, and highlight skeletal muscle as a potential therapeutic target in circadian rhythms disruption and associated diseases.
Circadian Rhythms in Skeletal Muscle
As previously stated, skeletal muscle, similar to almost every cell of the body, has circadian rhythms. These rhythms are generated by a transcriptional-translational feedback loop known as the molecular clock, and this mechanism has been reviewed in more complete detail elsewhere (Shearman et al., 2000; Buhr and Takahashi, 2013; Robinson and Reddy, 2014). At a basic level, Clock, Bmal1, Cry1/2, and Per1/2 are all core molecular clock components (Kume et al., 1999; Shearman et al., 2000). Bmal1 and Clock make up the positive limb of the molecular clock. They are bHLH transcription factors, which heterodimerize in the nucleus and transactivate Per and Cry family genes by binding E-box elements in their regulatory regions. PER and CRY then accumulate, multimerize, and translocate to the nucleus, where they inhibit BMAL1:CLOCK activity, thereby repressing their own expression. The accumulation of PER and CRY protein is also tightly controlled through phosphorylation and degradation via E3 ubiquitin ligases and the proteasome system, so the inhibition of BMAL1:CLOCK activity is lifted (Gallego and Virshup, 2007; Yoo et al., 2013). Proper timing of the molecular clock mechanism requires transcription, translation, and important rate-modifying posttranslational steps, thus presenting many sites through which information from environmental cues and physiological function can support or modify the clock.
Aside from a timekeeping role, the clock modulates the transcription of a large number of genes within the cell (clock-controlled genes [CCGs]); some of these are regulated directly by the binding of the core clock transcription factors Bmal1 and/or Clock to their promoters. To date, the identities of the direct CCGs in a specific tissue, such as skeletal muscle, have not been defined, but circadian transcriptome results suggest that they often encode transcription factors (e.g., MyoD1, Pgc1α) or proteins that control rate-limiting steps in cell physiology (e.g., Pdk4, Dbp; Fig. 1). Tissue-specific detailed reviews of the molecular clock mechanism and CCGs within individual tissues are available in several recent reviews by other groups (Panda et al., 2002; Storch et al., 2002; Kornmann et al., 2007; Nakahata et al., 2008; Lee et al., 2013; Shostak et al., 2013). The most direct evidence for the contribution of BMAL1:CLOCK regulation of gene expression outside of the timekeeping function comes from chromatin immunoprecipitation studies followed by DNA sequencing. These studies, performed in liver, determined that the BMAL1: CLOCK transcription factors bind to more than 2000 sites across the chromatin, with up to 85% of the sites being associated with actively expressed genes. Gene ontology analysis showed that the BMAL1:CLOCK–targeted expressed genes are highly enriched for metabolic, cancer, and insulin signaling pathways in liver (Rey et al., 2011; Koike et al., 2012). Chromatin immunoprecipitation studies have not yet been performed in skeletal muscle, but the results from liver highlight the breadth of transcriptional regulation by the molecular clock factors BMAL1 and CLOCK.
Figure 1.
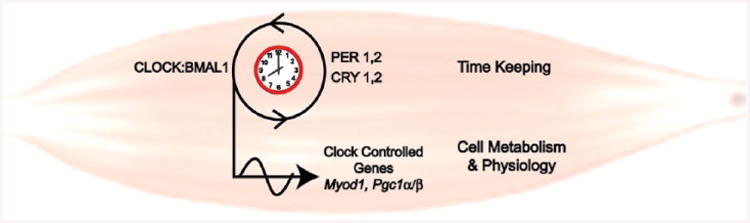
Simplified cartoon of molecular clock components in muscle. This cartoon highlights the role of CLOCK:BMAL1 as part of the core molecular clock and also illustrates their role directly targeting transcription of genes important for muscle-specific function.
Although more thorough investigations of molecular clock targets are needed in skeletal muscle, there have been studies identifying circadian-expressed genes using expression profiling. The first article to define circadian gene expression in skeletal muscle was published in 2007 and identified 215 circadian genes (McCarthy et al., 2007). However, increased sampling frequency, with tissues collected every 2 h for 48 h, and continued development of analysis tools for circadian gene expression studies have expanded this list to more than 2300 genes (Pizarro et al., 2013). A large portion of these genes are involved in metabolism, transcription, and signaling in muscle (McCarthy et al., 2007). The function of the molecular clock in skeletal muscle is only starting to be uncovered. Our lab has demonstrated that MyoD1, which is well characterized for its role as a master regulator of muscle gene expression, is under direct BMAL1:CLOCK control (Andrews et al., 2010; Zhang et al., 2012). In work looking at myogenesis, Chatterjee et al. (2013) has demonstrated that the core molecular clock gene, Bmal1, through transcriptional regulation of Wnt signaling, temporally regulates myogenic differentiation. Myogenin, a transcription factor that induces myogenesis, has also been suggested to be circadian in expression (Shavlakadze et al., 2013). While there is still much to learn, the results of these studies suggest important links between the molecular clock mechanism and the highly conserved muscle lineage transcription factors, MyoD1 and Myogenin, consistent with a daily role in muscle maintenance (Andrews et al., 2010).
Entrainment of the Skeletal Muscle Molecular Clock
Circadian rhythms are oscillations over a period of approximately 24 h, and in skeletal muscle, these rhythms include oscillations in transcription, metabolism, and myogenic capacity (Andrews et al., 2010; Chatterjee et al., 2011; Zhang et al., 2012). This oscillation of physiological processes is believed to be beneficial as it allows an organism to anticipate changes in environmental cues. These rhythms run in the absence of any external environmental cues, but a fundamental property of circadian rhythms is the ability to be entrained. The molecular clock has a phase, defined as the time relative to a specific point of the circadian cycle. For example, phase of the molecular clock may be determined by the time of peak Bmal1 expression. Entrainment occurs when the phase of the molecular clock is reset or modulated to be aligned with the timing of an environmental cue, such as light (Roenneberg et al., 2003). The skeletal muscle molecular clock can be entrained by cues such a light, time of feeding, and activity. In the case of light, the skeletal muscle clock is entrained in an indirect manner through the central clock in the suprachiasmatic nucleus (SCN). Light is transmitted via the retinohypothalamic tract from the retina to the SCN. Light evokes signaling in the SCN, through elements such as cyclic-AMP, that then modulate the molecular clock to affect the peak/phase of molecular clock oscillations (Gooley et al., 2001; Panda et al., 2002; Lee et al., 2010a; An et al., 2011). The SCN molecular clock communicates with other tissues, such as the skeletal muscle, using neurohumoral and temperature signals (Balsalobre et al., 2000; Brown et al., 2002; Abraham et al., 2010; Saini et al., 2012). It is in this indirect manner that the skeletal muscle clock is modulated by light cues.
Time of feeding also serves as an entrainment cue. Studies of time-restricted feeding in mice have demonstrated a phase shift in core molecular clock genes in liver and adipose tissue (Hara et al., 2001; Stokkan et al., 2001; Zvonic et al., 2006). In liver, this was shown to be independent of SCN input, since time-restricted feeding prevented the shift of the clock genes when the mice were exposed to a 7-h light:dark cycle advance (Hara et al., 2001). Although research on time of feeding and skeletal muscle is limited, time of feeding has been shown to entrain skeletal muscle circadian rhythms. A study from our lab demonstrated this using PER2:LUC mice. These mice were developed by Yoo et al. (2004) and have luciferase cDNA knocked into the Per2 coding region to generate a chimeric protein. Tissues from these mice may be explanted and placed in culture with luciferin to observe real-time light emission as an indicator of PER2 and thus molecular clock oscillations (Yoo et al., 2004). To evaluate the effect of time of feeding on skeletal muscle rhythms, our lab limited access to food to only 4 h/d for 2 wk. The restricted feeding resulted in a shift in gene expression (PER2:LUC bio-luminescence) in the skeletal muscle of the mice. In addition to studying the effect of time-restricted feeding, our lab also demonstrated the ability of scheduled activity to entrain the skeletal muscle molecular clock (Wolff and Esser, 2012). Scheduled bouts of either voluntary or involuntary endurance exercise resulted in a significant shift in clock gene expression (PER2:LUC bioluminescence) in 3 different muscle types as well as the lung. The shift in gene expression was observed in these tissues but not in the SCN, supporting a role for exercise as a non-SCN–associated entrainment cue for skeletal muscle. Notably, the phase of the each of the 3 muscles, soleus, extensor digitorum longus (EDL), and flexor digitorum brevis (FDB), for pre-exercise was distinct, highlighting the complexity of skeletal muscle and skeletal muscle circadian rhythms. This may be due to the differences in composition and function of the 3 muscles. The soleus is a postural muscle that is composed of mostly type I slow, oxidative fibers, whereas the EDL and FDB are intermittently recruited muscles (for either extension or flexion of the toes) and composed of more type II fast oxidative-glycolytic and glycolytic fibers. Despite differences in the pre-exercise phase of the muscles, scheduled activity shifted the phases to about the same magnitude, 2 to 3 h (Wolff and Esser, 2012). In addition, studies with the ClockΔ19 mice have demonstrated decreases in protein levels of proliferator-activated receptor-γ coactivator-1α (PGC1α) and mitochondrial transcription factor A with a concomitant decrease in mitochondrial content in skeletal muscle. Access to a running wheel under conditions of 12-h:12-h dark/light cycle and ad libitum access to food resulted in daily exercise (during the dark/active phase) in the clock mutant mice, which partially rescued the metabolic phenotype of the skeletal muscle (Pastore and Hood, 2013). This illustrates the potential impact physical activity may have as an entrainment cue for skeletal muscle and also suggests that entrainment cues for skeletal muscle may be useful as therapies in conditions of circadian disruption.
Synchrony Between Skeletal Muscle and Other Peripheral Tissues
Another critical concept is the idea of synchrony and the coordination of rhythms between clocks among different tissues. A study by Yoo et al. (2004) using the PER2:LUC mice demonstrated that peripheral (non-SCN) tissues had oscillators and that these oscillators had very distinct phases. Furthermore, they illustrated the importance of coordination of the tissue clocks by lesioning the SCN. In the presence of a lesioned SCN, the mice behave arrhythmically, and the clocks in other tissues no longer express their normal phase relationships (Yoo et al., 2004).
As mentioned earlier, expression of genes involved in metabolism have been shown to oscillate in skeletal muscle. In fact, in the study by McCarthy et al. (2007), one of the largest groups of oscillatory genes in skeletal muscle consisted of genes involved in substrate metabolism. Skeletal muscle and liver are considered key metabolic tissues, and as such, it follows that coordination between these tissues would be critical for normal metabolic function at the systems level. A complete investigation of the coordination of the metabolic functions of these tissues with regard to circadian rhythms has not been done. However, data on the expression of circadian genes in skeletal muscle and liver have been collected and are available on the database CircaDB (Pizarro et al., 2013). Muscle and liver serve as tissues that participate in the regulation of blood glucose. From CircaDB, one can see that genes involved in insulin signaling (Irs1, Irs2, Akt2, and Tbc1d1) and genes important for hepatic glucose production (Pck1, G6pc, Pep, and Pyg) are circadian in both of these tissues, and the phases are coordinated in a manner to facilitate blood glucose regulation (Table 1). Although protein and activity data are essential to confirm the gene data, it is apparent in Table 1 that during the light phase (or fasting phase for mice), genes involved in insulin signaling (Irs1, Tbc1d1, Akt, and Insr) have reduced expression in both skeletal muscle and liver. During this time, genes involved in hepatic glucose production (Pck1, G6pc, Pep, and Pyg) have increased expression. Furthermore, it has been shown in rodents that hepatic glucose production peaks during the fasting phase, at which time insulin sensitivity is low (la Fleur et al., 2001; Matsumoto et al., 2007; Zhang et al., 2010). In contrast to the fasting phase, it is necessary for greater insulin sensitivity in the metabolic tissues during the feeding phase in order to take up and store postprandial glucose (Oakes et al., 1997; Radziuk and Pye, 2001). Furthermore, the feeding phase is also often the active phase, during which skeletal muscle requires more substrate. Therefore, during the feeding/active phase, insulin sensitivity in liver and skeletal muscle needs to be higher (Fig. 2). In this regard, synchrony between the skeletal muscle and other tissues is critical for normal physiological function.
Table 1.
Circadian genes involved in carbohydrate homeostasis.
| Skeletal Muscle | Liver Expression | |
|---|---|---|
|
|
|
|
| Peak | Peak | |
| Akt2 | ∼48 | |
| Irs1 | ∼42 | ∼24 |
| Irs2 | ∼30 | ∼23 |
| Tbc1d1 | ∼42 | ∼42 |
| Insr | ∼30 | |
| Pck1 | ∼37 | |
| Pep | ∼48 | |
| Pyg | ∼42 | |
| G6pc | ∼42 |
Approximate time of peak expression of metabolic genes in skeletal muscle and liver based on CircaDB (Pizarro et al., 2013). Dark gray squares represent the dark phase, and light gray cells represent the light phase. Uncolored cells reflect borderline light/dark times.
Figure 2.
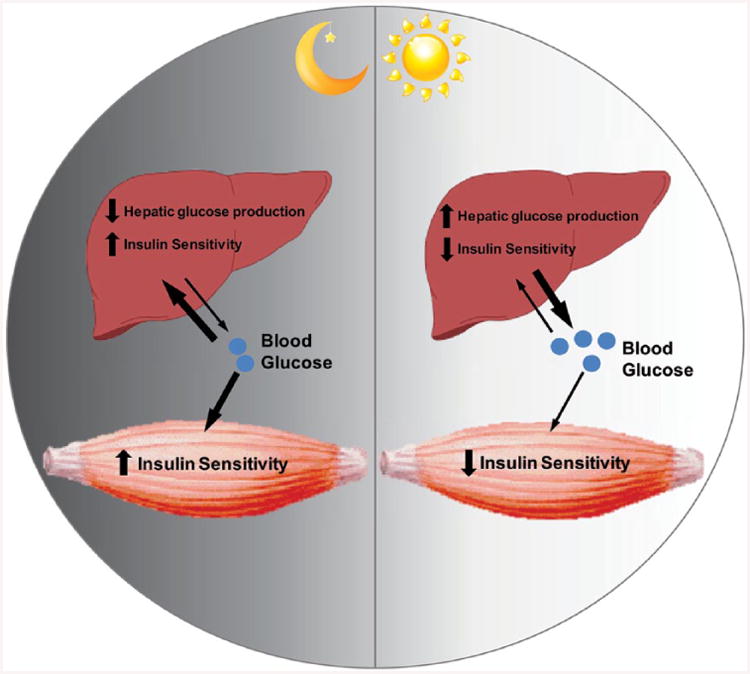
Cartoon depicting time-of-day coordination between skeletal muscle and liver metabolism for the regulation of systems glucose homeostasis. The dark phase/fed phase for mice is characterized by higher insulin sensitivity in skeletal muscle/liver and decreased hepatic glucose production. The light phase/fasting phase for mice is characterized by reduced insulin sensitivity in skeletal muscle/liver and increased hepatic glucose production.
Skeletal Muscle in Circadian Mutant Models
The role of circadian clocks in regulating physiological processes has been largely studied through the use of genetic mouse models of clock disruption. These models include the Per1/Per2 deficient mice, Cry1/Cry2 double knockouts, and Clock-/-, ClockΔ19, and Bmal1-/- mice. The first mouse model of clock disruption, the ClockΔ19, was identified through a forward genetics screen and is a mouse model in which exon 19 (an exon critical for the DNA binding of the Clock gene) is mutated. These mice have a free running period of 28 h, becoming behaviorally arrhythmic after 1 to 2 wk in constant darkness (Vitaterna et al., 1994). ClockΔ19 mice are moderately more susceptible to cancer and display a marked metabolic phenotype involving obesity, dyslipidemia, hepatic steatosis, and hyperglycemia (Rudic et al., 2004; Turek et al., 2005; Lee, Li, et al., 2010a). Interestingly, the Clock-/- mice (mice deficient of the Clock gene due to targeted gene knock down) do not exhibit the same phenotype as the ClockΔ19 mice. Clock-/- display a slightly shorter period length with preserved behavioral rhythms. They have reduced life span, age-related cataract development, and increased risk for dermatitis (Debruyne et al., 2006, 2007; Dubrovsky et al., 2010). Differences between ClockΔ19 and Clock-/- mouse models likely arise from the fact that the ClockΔ19 mice have a dominant negative mutation in Clock whereas Clock-/-are deficient of Clock altogether and maybe compensation from NPAS2. Models involving genes of the negative limb of the molecular clock, Period and Cryptochrome, appear to have less severe consequences in comparison to the Clock-/-and ClockΔ19 mice. Period and Cryptochrome knockout mice do not display an altered life span and the serious physiological phenotype observed in ClockΔ19 and Bmal1-/- mice. However, they do reveal a significant role for the negative limb clock genes in tumorigenesis. In addition, activity rhythms are disrupted in these mice. Cry1-/- mice have a shorter period length, and Cry2-/- mice have a significantly longer period length, but both maintain rhythmic behavior. Only the double knockouts, Per1-/-/Per2m/m and Cry1/Cry2-/-, result in arrhythmic behavior (Vitaterna et al., 1994; van der Horst et al., 1999; Zheng et al., 2001; Liu et al., 2007).
Of the studied circadian mouse models, Bmal1-/-mice exhibit the most severe pathology. These mice display significantly reduced life span, age-associated weight decline, behavioral arrhythmicity ectopic calcification, and sterility (Bunger et al., 2005; Kondratov et al., 2006; Yu and Weaver, 2011). Furthermore, Bmal1-/- mice have impaired insulin sensitivity and glucose tolerance, they are weak, and they develop dilated cardiomyopathy (Rudic et al., 2004; Andrews et al., 2010; Lefta et al., 2012). These models all illustrate an essential role for the molecular clock in whole-body physiology and tissue-specific processes. Although there has been limited research on skeletal muscle circadian rhythms, skeletal muscle is a major site of pathology in clock-disrupted models. Skeletal muscle structure and function have been shown to be greatly affected by clock disruption (Andrews et al., 2010). Bmal1-/- and ClockΔ19 mice show significantly reduced force production at both the whole-muscle and single-fiber level. This may in part be due to disrupted organization of thick and thin filaments in the sarcomere. The sarcomere is the basic functional unit of skeletal muscle and is defined by the presence of a conserved group of proteins in stoichiometric ratios that comprise the thick myofilaments containing myosin heavy and light chains and thin myofilaments containing actin, tropomyosin, and troponins as well as important critical accessory proteins such as titin (Huxley and Hanson, 1959; Eisenberg and Cohen, 1983; Squire, 1997). It has been well established that selective decreases in the content of proteins within the sarcomere or disruptions in the spatial organization of the thick and thin myofilaments are causal for diminished force production by skeletal muscle (Wang et al., 1979; Riley et al., 2000; Laing and Nowak, 2005; Trappe, 2009; Tardiff, 2011). This highly conserved hexagonal arrangement of thick and thin filaments, with 6 thin filaments surrounding every thick filament, is lost in the Bmal1-/- and ClockΔ19 mice. Using 2 skeletal muscle–specific models of Bmal1 disruption, Dyar and colleagues (2014) showed a reduction in force production in one of their models without a concomitant change in myofilament architecture. They suggested the difference in force production between the models was probably the result of developmental differences in one of their mouse strains (Dyar et al., 2014).
Disruption of circadian rhythms is associated with an increased risk for development of metabolic disruption in both animals and humans (Karlsson et al., 2001; Rudic et al., 2004; Turek et al., 2005; Kroenke et al., 2007; Morikawa et al., 2007; Scott et al., 2008; Scheer et al., 2009; Marcheva et al., 2010). As stated earlier, skeletal muscle comprises ∼45% of the body mass of most mammals and is a critical component of normal metabolic health. Skeletal muscle is responsible for approximately 80% of postprandial insulin-mediated glucose disposal (DeFronzo et al., 1981; Ferrannini et al., 1988). In addition, altered muscle function can contribute to insulin resistance and metabolic syndrome (Kelley et al., 1999; Petersen et al., 2007). Our previous work demonstrated that Bmal1-/- and ClockΔ19 mice display an ∼40% decrease in skeletal muscle mitochondrial volume. The mitochondria present in these mutant mice displayed aberrant morphology and increased respiratory uncoupling. Specifically, a significant reduction in state III respiration (ADP-stimulated, mmol O2/min/mg protein) in mitochondria isolated from gastrocnemius muscle was observed (Andrews et al., 2010). Muscle-specific loss of Bmal1 resulted in decreased skeletal muscle glucose uptake, reduced glucose oxidation, and increased PDK4 expression (Dyar et al., 2014). The increase in PDK4 expression coupled with an observed shift toward more oxidative fibers in the soleus muscle may indicate greater use of lipids for energy metabolism in the muscle of skeletal muscle–specific Bmal1 knockout mice. These data demonstrate the importance of the skeletal muscle molecular clock in the maintenance of systemic metabolic health.
The contribution of the molecular clock in skeletal muscle to muscle function and muscle metabolism is currently being established. In 2006, a study investigating the effects of Bmal1 rescue in the germline Bmal1-/- mouse demonstrated a critical role for the skeletal muscle clock in skeletal muscle maintenance and systemic health (McDearmon et al., 2006). McDearmon et al. rescued Bmal1 in the brain or muscle of Bmal1-/- mice using the tetracycline transactivator system with the brain-specific promoter Scg2 or the muscle-specific promoter Acta1 upstream of the Bmal1 transgene. As stated earlier, whole-body Bmal1 knockout mice are behaviorally arrhythmic in constant darkness and have decreased overall activity levels, reduced body weight, and decreased longevity. When Bmal1-/- was rescued in the brain, the mice regained rhythmic behavior but daily activity and body weight remained low. Interestingly, the muscle-specific rescue did not restore rhythmic behavior but did return daily activity levels back to normal and was associated with maintenance of body weight. Even more surprisingly, only 75% of brain-specific rescued mice survived to the end of the experiment, while 100% of the muscle-specific rescued mice survived, suggesting that muscle-specific rescue of Bmal1-/- may be sufficient to restore longevity. These data demonstrate the importance of the muscle clock in overall systems health.
Summary
Skeletal muscle, similar to practically every organ/tissue in the body, has circadian rhythms (Yoo et al., 2004). Circadian rhythms and the molecular clock mechanism modulate the expression of a substantial number of genes in skeletal muscle, many of which participate in transcription, myogenesis, and metabolism (McCarthy et al., 2007). It is important for processes such as these to be entrained to the environment to allow an organism to anticipate environmental changes and respond accordingly. For instance, during the fasting period, glucose availability is low and the skeletal muscle must alter substrate metabolism accordingly (Pelley, 2012). Furthermore, the skeletal muscle must work in coordination with other tissues and so must have rhythms in synchrony with other tissue rhythms. In the example of fasting, other metabolic tissues, such as liver and adipose tissue, must also alter their metabolism to work collectively with the skeletal muscle to maintain glucose homeostasis and spare glucose for the brain. Circadian rhythms may be entrained by cues such as light, time of feeding, and time of activity (Gooley et al., 2001; Hara et al., 2001; Panda et al., 2002; Roenneberg et al., 2003; Lee, Li, et al., 2010a; An et al., 2011; Wolff and Esser, 2012). If skeletal muscle rhythms are disrupted or asynchronous, entrainment cues may be a useful strategy in resetting the skeletal muscle clock so that it is both entrained to the environment and in synchrony with other tissue rhythms.
The importance of circadian rhythms for skeletal muscle is made evident by the skeletal muscle phenotype observed in models of molecular clock disruption, such as the Bmal1-/- mice. Loss of the core clock gene Bmal1 results in severe muscle pathology, including fiber-type shifts, decreased mitochondria, impaired mitochondrial respiration, altered sarcomeric structure, and debilitated function (Andrews et al., 2010; Dyar et al., 2014). This model highlights a role for circadian rhythms in muscle structure, function, and metabolism. Many common diseases are associated with altered muscle mass, function, and metabolism, including cancer and heart disease. In fact, the prognosis for these diseases is strongly linked to the magnitude of skeletal muscle changes (Kadar et al., 2000; Akashi et al., 2005). Metabolic diseases are also becoming an epidemic in the United States, and the skeletal muscle is a primary metabolic site disrupted in the development of these diseases (Defronzo et al., 1981; DeFronzo et al., 1992). The molecular clock significantly affects skeletal muscle metabolism, as demonstrated in the Bmal1-/- mice. Perhaps there is a disruption or weakening of circadian rhythms in these diseased states that contributes to the pathogenesis of skeletal muscle. In this case, entrainment by environmental cues or pharmacological agents may synchronize or strengthen circadian rhythms and therefore aid in the treatment of these diseases. In addition to alleviating the skeletal muscle pathology, entrainment of skeletal muscle circadian rhythms may also contribute to systemic health. This is quite feasible considering that skeletal muscle–specific rescue of the Bmal1-/- mice was able to protect mice from the decrease in activity, body weight, and longevity observed (McDearmon et al., 2006). These data all suggest a vital role for skeletal muscle circadian rhythms in both skeletal muscle and systems health and support skeletal muscle circadian rhythms as a possible target of intervention in prevalent diseases. Further research is needed to elucidate skeletal muscle molecular clock function across all muscles and to investigate the role of skeletal muscle circadian rhythms in both skeletal muscle and systemic health.
Acknowledgments
Support for this study was provided by the National Institutes of Health, grant AR066082 and University of Kentucky Center for Muscle Biology to K.A.E.
References
- Abraham U, Granada AE, Westermark PO, Heine M, Kramer A, Herzel H. Coupling governs entrainment range of circadian clocks. Mol Syst Biol. 2010;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi YJ, Springer J, Anker SD. Cachexia in chronic heart failure: prognostic implications and novel therapeutic approaches. Curr Heart Fail Rep. 2005;2:198–203. doi: 10.1007/BF02696650. [DOI] [PubMed] [Google Scholar]
- An S, Irwin RP, Allen CN, Tsai C, Herzog ED. Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol. 2011;105:2289–2296. doi: 10.1152/jn.00966.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci USA. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- Buhr ED, Takahashi JS. Molecular components of the mammalian circadian clock. Handb Exp Pharmacol. 2013;217:3–27. doi: 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Walisser JA, Sullivan R, Manley PA, Moran SM, Kalscheur VL, Colman RJ, Bradfield CA. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Barquero NP, Li L, Ma K. Circadian clock gene, Bmal1, regulates skeletal muscle metabolism and development. Endocr Rev. 2011;32:3–434. [Google Scholar]
- Chatterjee S, Nam D, Guo B, Kim JM, Winnier GE, Lee J, Berdeaux R, Yechoor VK, Ma K. Brain and muscle Arnt-like 1 is a key regulator of myogenesis. Journal of cell science. 2013;126:2213–2224. doi: 10.1242/jcs.120519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFronzo RA, Bonadonna RC, Ferrannini E. Pathogenesis of NIDDM: a balanced overview. Diabetes Care. 1992;15:318–368. doi: 10.2337/diacare.15.3.318. [DOI] [PubMed] [Google Scholar]
- Defronzo RA, Simonson D, Ferrannini E, Barrett E. Insulin resistance: a universal finding in diabetic states. Bull Schweiz Akad Med Wiss. 1981:223–238. [PubMed] [Google Scholar]
- Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging. 2010;2:936–944. doi: 10.18632/aging.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyar KA, Ciciliot S, Wright LE, Bienso RS, Tagliazucchi GM, Patel VR, Forcato M, Paz MI, Gudiksen A, Solagna F, et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab. 2014;3:29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg BR, Cohen IS. The ultrastructure of the cardiac Purkinje strand in the dog: a morphometric analysis. Proc R Soc Lon B Biol Sci. 1983;217:191–213. doi: 10.1098/rspb.1983.0006. [DOI] [PubMed] [Google Scholar]
- Febbraio MA, Pedersen BK. Muscle-derived interleukin-6: mechanisms for activation and possible biological roles. FASEB J. 2002;16:1335–1347. doi: 10.1096/fj.01-0876rev. [DOI] [PubMed] [Google Scholar]
- Ferrannini E, Simonson DC, Katz LD, Reichard G, Jr, Bevilacqua S, Barrett EJ, Olsson M, DeFronzo RA. The disposal of an oral glucose load in patients with non-insulin-dependent diabetes. Metabolism. 1988;37:79–85. doi: 10.1016/0026-0495(88)90033-9. [DOI] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Goodpaster BH, Kelley DE, Thaete FL, He J, Ross R. Skeletal muscle attenuation determined by computed tomography is associated with skeletal muscle lipid content. J Appl Physiol. 2000;89:104–110. doi: 10.1152/jappl.2000.89.1.104. [DOI] [PubMed] [Google Scholar]
- Gooley JJ, Lu J, Chou TC, Scammell TE, Saper CB. Melanopsin in cells of origin of the retinohypothalamic tract. Nature neuroscience. 2001;4:1165. doi: 10.1038/nn768. [DOI] [PubMed] [Google Scholar]
- Hara R, Wan K, Wakamatsu H, Aida R, Moriya T, Akiyama M, Shibata S. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- Hoppeler H, Fluck M. Normal mammalian skeletal muscle and its phenotypic plasticity. J Exp Biol. 2002;205:2143–2152. doi: 10.1242/jeb.205.15.2143. [DOI] [PubMed] [Google Scholar]
- Huxley HE, Hanson J. The structural basis of the contraction mechanism in striated muscle. Ann N Y Acad Sci. 1959;81:403–408. doi: 10.1111/j.1749-6632.1959.tb49323.x. [DOI] [PubMed] [Google Scholar]
- Kadar L, Albertsson M, Areberg J, Landberg T, Mattsson S. The prognostic value of body protein in patients with lung cancer. Ann N Y Acad Sci. 2000;904:584–591. doi: 10.1111/j.1749-6632.2000.tb06520.x. [DOI] [PubMed] [Google Scholar]
- Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup Environ Med. 2001;58:747–752. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DE, Goodpaster B, Wing RR, Simoneau JA. Skeletal muscle fatty acid metabolism in association with insulin resistance, obesity, and weight loss. Am J Physiol. 1999;277:E1130–E1141. doi: 10.1152/ajpendo.1999.277.6.E1130. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Reinke H, Saini C, Schibler U. Regulation of circadian gene expression in liver by systemic signals and hepatocyte oscillators. Cold Spring Harb Symp Quant Biol. 2007;72:319–330. doi: 10.1101/sqb.2007.72.041. [DOI] [PubMed] [Google Scholar]
- Kroenke CH, Spiegelman D, Manson J, Schernhammer ES, Colditz GA, Kawachi I. Work characteristics and incidence of type 2 diabetes in women. Am J Epidemiol. 2007;165:175–183. doi: 10.1093/aje/kwj355. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, Buijs RM. A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes. 2001;50:1237–1243. doi: 10.2337/diabetes.50.6.1237. [DOI] [PubMed] [Google Scholar]
- Laing NG, Nowak KJ. When contractile proteins go bad: the sarcomere and skeletal muscle disease. BioEssays. 2005;27:809–822. doi: 10.1002/bies.20269. [DOI] [PubMed] [Google Scholar]
- Lee B, Li A, Hansen KF, Cao R, Yoon JH, Obrietan K. CREB influences timing and entrainment of the SCN circadian clock. J Biol Rhythms. 2010a;25:410–420. doi: 10.1177/0748730410381229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Moulik M, Fang Z, Saha P, Zou F, Xu Y, Nelson DL, Ma K, Moore DD, Yechoor VK. Bmal1 and beta-cell clock are required for adaptation to circadian disruption, and their loss of function leads to oxidative stress-induced beta-cell failure in mice. Mol Cell Biol. 2013;33:2327–2338. doi: 10.1128/MCB.01421-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefta M, Campbell KS, Feng HZ, Jin JP, Esser KA. Development of dilated cardiomyopathy in Bmal1-deficient mice. Am J Physiol Heart Circ Physiol. 2012;303:H475–H485. doi: 10.1152/ajpheart.00238.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Matthews VB, Astrom MB, Chan MH, Bruce CR, Krabbe KS, Prelovsek O, Akerstrom T, Yfanti C, Broholm C, Mortensen OH, et al. Brain-derived neurotrophic factor is produced by skeletal muscle cells in response to contraction and enhances fat oxidation via activation of AMP-activated protein kinase. Diabetologia. 2009;52:1409–1418. doi: 10.1007/s00125-009-1364-1. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genom. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Nakagawa H, Miura K, Soyama Y, Ishizaki M, Kido T, Naruse Y, Suwazono Y, Nogawa K. Effect of shift work on body mass index and metabolic parameters. Scand J Work Environ Health. 2007;33:45–50. doi: 10.5271/sjweh.1063. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circa-dian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AR, Pedersen BK. The biological roles of exercise-induced cytokines: IL-6, IL-8, and IL-15. Appl Physiol Nutr Metab. 2007;32:833–839. doi: 10.1139/H07-054. [DOI] [PubMed] [Google Scholar]
- Oakes ND, Cooney GJ, Camilleri S, Chisholm DJ, Kraegen EW. Mechanisms of liver and muscle insulin resistance induced by chronic high-fat feeding. Diabetes. 1997;46:1768–1774. doi: 10.2337/diab.46.11.1768. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Pastore S, Hood DA. Endurance training ameliorates the metabolic and performance characteristics of circadian Clock mutant mice. J Appl Physiol. 2013;114:1076–1084. doi: 10.1152/japplphysiol.01505.2012. [DOI] [PubMed] [Google Scholar]
- Pelley JW. Elsevier's Integrated Review Biochemistry. New York: Elsevier; 2012. [Google Scholar]
- Petersen KF, Dufour S, Savage DB, Bilz S, Solomon G, Yonemitsu S, Cline GW, Befroy D, Zemany L, Kahn BB, et al. The role of skeletal muscle insulin resistance in the pathogenesis of the metabolic syndrome. Proc Natl Acad Sci USA. 2007;104:12587–12594. doi: 10.1073/pnas.0705408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucl Acids Res. 2013;41:D1009–D1013. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole DC. Measurements of the anaerobic work capacity in a group of highly trained runners. Med Sci Sports Exerc. 1986;18:703–705. [PubMed] [Google Scholar]
- Radziuk J, Pye S. Hepatic glucose uptake, gluconeogenesis and the regulation of glycogen synthesis. Diabetes Metab Res Rev. 2001;17:250–272. doi: 10.1002/dmrr.217. [DOI] [PubMed] [Google Scholar]
- Reaven GM. Insulin resistance, the insulin resistance syndrome, and cardiovascular disease. Panminerva Medica. 2005;47:201–210. [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley DA, Bain JL, Thompson JL, Fitts RH, Widrick JJ, Trappe SW, Trappe TA, Costill DL. Decreased thin filament density and length in human atrophic soleus muscle fibers after spaceflight. J Appl Physiol. 2000;88:567–572. doi: 10.1152/jappl.2000.88.2.567. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Fritz S, Richter K, Hocke GM, Lottspeich F, Fey GH. Transcription factors Stat3 and Stat5b are present in rat liver nuclei late in an acute phase response and bind interleukin-6 response elements. J Biol Chem. 1995;270:29998–30006. doi: 10.1074/jbc.270.50.29998. [DOI] [PubMed] [Google Scholar]
- Robinson I, Reddy AB. Molecular mechanisms of the circadian clockwork in mammals. FEBS Lett. 2014;588:2477–2483. doi: 10.1016/j.febslet.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Daan S, Merrow M. The art of entrainment. J Biol Rhythms. 2003;18:183–194. doi: 10.1177/0748730403018003001. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Morf J, Stratmann M, Gos P, Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circa-dian oscillators. Genes Dev. 2012;26:567–580. doi: 10.1101/gad.183251.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci USA. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes. 2008;32:658–662. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- Shavlakadze T, Anwari T, Soffe Z, Cozens G, Mark PJ, Gondro C, Grounds MD. Impact of fasting on the rhythmic expression of myogenic and metabolic factors in skeletal muscle of adult mice. Am J Physiol Cell Physiol. 2013;305:C26–C35. doi: 10.1152/ajpcell.00027.2013. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shostak A, Husse J, Oster H. Circadian regulation of adipose function. Adipocyte. 2013;2:201–206. doi: 10.4161/adip.26007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire JM. Architecture and function in the muscle sarcomere. Curr Opin Struct Biol. 1997;7:247–257. doi: 10.1016/s0959-440x(97)80033-4. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Tardiff JC. Thin filament mutations: developing an integrative approach to a complex disorder. Circ Res. 2011;108:765–782. doi: 10.1161/CIRCRESAHA.110.224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappe T. Influence of aging and long-term unloading on the structure and function of human skeletal muscle. Appl Physiol Nutr Metab. 2009;34:459–464. doi: 10.1139/h09-041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, McClure J, Tu A. Titin: major myofibrillar components of striated muscle. Proc Natl Acad Sci USA. 1979;76:3698–3702. doi: 10.1073/pnas.76.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G, Esser KA. Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med Sci Sports Exerc. 2012;44:1663–1670. doi: 10.1249/MSS.0b013e318255cf4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2∷LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EA, Weaver DR. Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging. 2011;3:479–493. doi: 10.18632/aging.100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Patel SP, McCarthy JJ, Rabchevsky AG, Goldhamer DJ, Esser KA. A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle. Nucl Acids Res. 2012;40:3419–3430. doi: 10.1093/nar/gkr1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]


