Abstract
BACKGROUND
The genetic basis of nonobstructive azoospermia is unknown in the majority of infertile men.
METHODS
We performed array comparative genomic hybridization testing in blood samples obtained from 15 patients with azoospermia, and we performed mutation screening by means of direct Sanger sequencing of the testis-expressed 11 gene (TEX11) open reading frame in blood and semen samples obtained from 289 patients with azoospermia and 384 controls.
RESULTS
We identified a 99-kb hemizygous loss on chromosome Xq13.2 that involved three TEX11 exons. This loss, which was identical in 2 patients with azoospermia, predicts a deletion of 79 amino acids within the meiosis-specific sporulation domain SPO22. Our subsequent mutation screening showed five novel TEX11 mutations: three splicing mutations and two missense mutations. These mutations, which occurred in 7 of 289 men with azoospermia (2.4%), were absent in 384 controls with normal sperm concentrations (P = 0.003). Notably, five of those TEX11 mutations were detected in 33 patients (15%) with azoospermia who received a diagnosis of azoospermia with meiotic arrest. Meiotic arrest in these patients resembled the phenotype of Tex11-deficient male mice. Immunohistochemical analysis showed specific cytoplasmic TEX11 expression in late spermatocytes, as well as in round and elongated spermatids, in normal human testes. In contrast, testes of patients who had azoospermia with TEX11 mutations had meiotic arrest and lacked TEX11 expression.
CONCLUSIONS
In our study, hemizygous TEX11 mutations were a common cause of meiotic arrest and azoospermia in infertile men. (Funded by the National Institutes of Health and others.)
Nearly half of all cases of male infertility are thought to be associated with genetic defects.1-3 Up to 20% of infertile men receive a diagnosis of azoospermia.3 Nonobstructive azoospermia is spermatogenic failure that is defined by the absence of spermatozoa in the seminal fluid.1,4 Azoospermia is a heterogeneous condition with several histologic pheno-types.5 The most severe form of azoospermia is the Sertoli-cell–only syndrome, which is defined as a complete absence of germ cells.6,7
Azoospermia with meiotic arrest is a milder form of infertility with a cessation at the spermatocyte stage of germ-cell formation.7 Both the Sertoli-cell–only syndrome and meiotic arrest affect all seminiferous tubules. Mixed testicular atrophy is yet a milder form of azoospermia with a variable degree of germ-cell loss and spermatozoa detected in at least some tubules.1,7
Up to 20% of men with nonobstructive azoospermia have a detectable chromosomal abnormality; these abnormalities include sex-chromosome anomalies (e.g., Klinefelter's syndrome), structural aberrations (e.g., translocations and inversions), and Y-chromosome microdeletions of azoospermia factors.8 However, the remaining 80% of men with nonobstructive azoospermia have negative results on genetic testing and receive a diagnosis of “idiopathic” azoospermia.3
Numerous mouse models that have linked hundreds of genes with azoospermia and infertility provide insight into the molecular mechanisms responsible for this condition in mice. The loss of function of these genes causes infertility.2 Yet, only a few studies involving humans with azoospermia have identified mutations in genes that are associated with infertility in mice (e.g., HSF2, SYCP3, PRM1, PRM2, SOHLH1, and NR5A1).9-13 This could be explained by the low discriminative power of a single candidate gene; hundreds of genes may contribute to the diagnosis of histologically diverse azoospermia.
We performed array comparative genomic hybridization (aCGH) in blood samples obtained from patients with azoospermia and mutation screening by means of direct Sanger sequencing of the testis-expressed 11 (TEX11) gene open reading frame in patients with azoospermia and controls.
METHODS
PATIENTS
The initial study cohort was composed of 49 men recruited from the Center for Fertility and Reproductive Endocrinology at Magee–Womens Hospital of the University of Pittsburgh Medical Center and the Institute of Human Genetics of the Polish Academy of Sciences in Poznań, Poland. The diagnoses in men with azoospermia were further classified, on the basis of histologic examination of testicular biopsy specimens, in one of three categories: the Sertoli-cell–only syndrome (no germ cells), meiotic arrest (no maturation beyond spermatocytes), and mixed testicular atrophy (few elongated spermatids). The cohort included 9 men with the Sertoli-cell–only syndrome, 19 with meiotic arrest, and 21 with mixed testicular atrophy.
The follow-up study was composed of a cohort of 240 patients, including 54 with the Sertoli-cell–only syndrome, 14 with meiotic arrest, and 172 with heterogeneous mixed testicular atrophy. These patients were recruited from the Center of Reproductive Medicine and Andrology, University Clinic, Münster, Germany.7
In all patients, a diagnosis of nonobstructive azoospermia was confirmed by means of semen analysis performed according to the guidelines of the World Health Organization.4 Men in whom the cause of infertility (such as chromosomal abnormalities and Y-chromosome microdeletions) was known were not included in the study.
ISOLATION OF GENOMIC DNA
Genomic DNA was isolated from whole blood with the use of the Gentra Puregene kit (Qiagen). A total of 384 samples obtained from men with normal sperm concentrations (>39×106 sperm per milliliter) were used as normal controls. Genomic DNA from 192 semen samples was isolated with the use of the TRIzol reagent (Life Technologies), and 192 matched DNA samples were obtained from peripheral blood.
MICROARRAY TESTING
We performed aCGH with the use of a whole-genome 400K oligonucleotide microarray (Agilent Technologies) on DNA samples obtained from 15 American patients of European descent who had azoospermia. Male DNA purified from blood samples donated by men and provided by Promega was used as a reference. A targeted X-chromosome high-resolution microarray designed at the Pittsburgh Cytogenetics Laboratory (180K oligonucleotide aCGH with the use of the Agilent platform) was used for delineation of X-chromosome deletions. The aCGH data were analyzed with the use of Agilent CytoGenomics software, version 3.0. Genomic copy-number variants, also known as losses (deletions) and gains (duplications), were analyzed with the use of the following bioinformatic databases: Online Mendelian Inheritance in Man, UniGene, the Conserved Domain Database, and BioGPS. Polymorphic copy-number variants reported in the Database of Genomic Variants14 were removed from further consideration.
POLYMERASE-CHAIN-REACTION TESTING
Polymerase-chain-reaction (PCR) testing was performed with 15 ng of DNA, with the use of HiFi HotStart DNA Polymerase (Kapa Biosystems). Each coding exon of TEX11 and at least 50 bp of flanking introns (ranging from 274 to 632 bp) were amplified, including exon 2 (isoform 2) and exons 3 to 31 (isoform 1) (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Long-range PCR was performed with 200 ng of DNA, with the use of the Takara LA PCR kit, version 2.1 (Clontech), and the forward primer TCTGTCCGAAAAGTCACATATCTCTGTTTCTG and reverse primer TATACAGTTGCTATGGACCGAATGTTTGTGTC. PCR products were run on an ABI Prism 3130xl sequencer with the use of BigDye Terminator (version 3.1) Cycle Sequencing Kit (Applied Biosystems).
Data from direct Sanger sequencing were analyzed with the use of Sequencher software (Gene Codes). An association between TEX11 mutations and azoospermia was evaluated by means of Fisher's exact test (with P values of <0.05 considered to indicate statistical significance). Quantitative PCR was performed with 20 ng of DNA, with the use of iQ SYBR Green Supermix (Bio-Rad Laboratories). Primers were used exclusively within TEX11 exons 10 to 12 and beta-actin (ACTB) was used as a control (Table S2 in the Supplementary Appendix).
We calculated the relative quantification number with the use of the ΔΔCt method. ΔCt for each sample was calculated as ΔCt = CtTEX11exon –CtACTB, (with Ct denoting cycling threshold and ACTB, beta-actin). ΔΔCt for each experimental sample was calculated as ΔΔCt = ΔCtexperimental–Ctcontrol. Relative quantification (RQ) was calculated as RQ = 2(–ΔΔCt).
ASSESSMENT OF TEX11 EXPRESSION IN TESTICULAR TISSUE
Sections of testicular tissue from healthy and fertile mice, macaques (Macaca fascicularis), and humans were stained with a primary anti-TEX11 antibody (1:100 dilution of goat polyclonal antibody) (ab99461, Abcam). The primary antibody was detected with the use of a secondary antibody (a 1:250 dilution of horseradish peroxidase–conjugated chicken antigoat secondary antibody; sc-2984, Santa Cruz Biotechnology). Staining was visualized with the use of 3,3′-diaminobenzidine (D4168, Sigma-Aldrich) and hematoxylin as the counterstain.
MODELING TEX11
The protein structure prediction server (PSIPRED),15 homology, detection, and structure prediction server (HHpred),16 and protein structure and function prediction server (I-TASSER)17 algorithms were used to predict the secondary structure of protein. We used the Pfam database18 to search for protein domains in TEX11. The TPRpred method19 was used to identify tetratricopeptide repeat (TPR) regions in the protein. To visualize the three-dimensional structure of TEX11, we used the Phyre215 and I-TASSER algorithms.17,20 Structural models were generated with the use of the PyMOL Molecular Graphics System (Schrödinger) (see the Methods section in the Supplementary Appendix).
RESULTS
GENOMIC ACGH STUDY
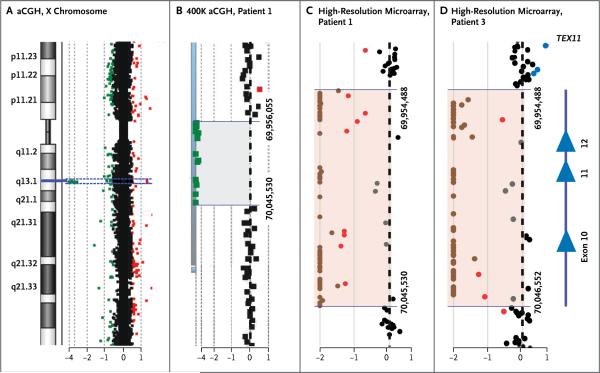
We initially performed a genomewide 400K oligonucleotide aCGH microarray analysis of blood samples obtained from 15 unrelated patients of European descent who had azoospermia in order to identify genomic losses and gains associated with this condition. We detected 181 copy-number variants, with an average of 38 per sample (Table S3 in the Supplementary Appendix). A total of 161 of the 181 copy-number variants (89%) that were reported as being polymorphic in the Database of Genomic Variants were excluded from further analysis. We identified a 90-kb hemizygous loss encompassing a part of TEX11 on chromosome Xq13.1 in Patient 1, who had azoospermia with mixed testicular atrophy (Table 1, and Fig. 1A and 1B).
Table 1.
Mutations in TEX11 Detected in Three Samples Obtained from the Initial Study Cohort of 49 Men of European Descent Who Had Azoospermia.*
| Patient No. | Change in Coding DNA | Protein/RNA Change | Phenotype | Study Controls | Controls from dbSNP Database | Race or Ethnic Group† | |
|---|---|---|---|---|---|---|---|
| SNP | Frequency | ||||||
| % | no./total no. (%) | ||||||
| 1 | c.652del237bp | p.218del79aa | Mixed testicular atrophy | 0 | NA | NA | White |
| 2 | c.511A→G | p.M171V | Meiotic arrest | 0 | rs143246552 | het 2/2270 (0.1) | White |
| 3 | c.652del237bp | p.218del79aa | Meiotic arrest | 0 | NA | NA | White |
The term aa denotes amino acid substitution, bp base pairs, dbSNP Single Nucleotide Polymorphism Database, het heterozygous, NA not available, rs reference number, and SNP single-nucleotide polymorphism.
Race and ethnic group were self-reported.
Figure 1. Hemizygous Deletion of TEX11 Exons 10 to 12 and Flanking Intronic Regions in Two Men with Azoospermia.
Panel A shows the array comparative genomic hybridization (aCGH) plot of the X chromosome. On the left, an idiogram of the X chromosome shows a region of interest (blue line) at the Xq13.2 band. On the right, dots represent oligonucleotide DNA probes, arranged according to their physical map locations from the distal p arm (top) to the distal q arm (bottom) of the X chromosome. For each probe, the fluorescence intensity of the test signal relative to the reference signal is converted to a logarithmic (log2) value and shown along the x axis. Probes with a log2 ratio clustered around zero (black dots) indicate DNA segments with a normal copy number. A positive log2 ratio (above +0.3 [red dots]) indicates a gain (extra copy) of the chromosomal region, whereas intervals with a negative log2 ratio (below −0.5 [green dots]) represent a loss (deletion) in DNA copy number. Panel B shows a magnified view of the deleted region detected by 12 probes with the use of the 400K whole-genome array (gray shaded area). Genomic coordinates (GRCh37/hg19 assembly) of the deletion start at 69,956,055 and stop at 70,045,530 nucleotides. Panels C and D show that the aCGH plot of the 180K oligonucleotide X-chromosome high-resolution microarray more precisely delineates the deletion intervals in Patients 1 and 3. Chromosomal alterations suggestive of a homozygous or heterozygous deletion are indicated by brown and red dots, respectively. In a sample obtained from Patient 1 (Panel C), the deletion spans chrX:69,954,488 to 70,045,530. In a sample obtained from Patient 3 (Panel D), the deletion spans chrX:69,954,488 to 70,046,552 regions (pink shaded areas). On the right side of Panel D, Xq13.2 deletions in both patients encompass exons 10 to 12 (blue triangles) of TEX11.
With the use of an X-chromosome high-resolution microarray, we mapped the deletion to a 91,042-bp segment from chrX:69,954,448 to 70,045,530 (GRCh37/hg19) (Fig. 1C). This region spans exons 10 to 12 (isoform 1, coding exons 8 to 10) with the breakpoints located in introns 9 and 12 (Fig. 1C and 2A). This in-frame genomic deletion predicts a protein lacking 79 amino acids in the highly conserved sporulation domain SPO22 (ZIP4) (Fig. 2C).
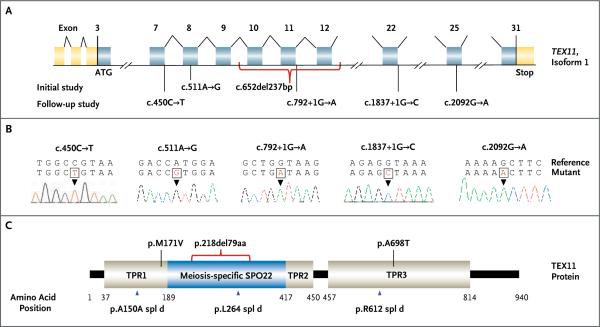
Figure 2. TEX11 Mutations Detected in Men with Azoospermia.
Panel A shows the genomic structure of TEX11, with mutations mapped to isoform 1 (GenBank accession number, NM 001003811.1). Gold rectangles represent noncoding exons, and blue rectangles represent coding exons. The coding sequence of the gene begins at nucleotides that encode a start codon in exon 3 and ends in exon 31 at a stop codon. Point mutations detected in men with azoospermia in the initial and follow-up studies are shown. The red bracket below exons 10 to 12 shows the approximately 99-kb deletion detected in two men with azoospermia. Panel B shows sequence chromatograms of five point mutations detected in TEX11. Reference and mutant DNA sequences are shown, with an arrowhead identifying the mutation. Panel C shows predicted TEX11 domains with multiple tetratricopeptide repeat (TPR)–containing regions (amino acid positions 37–188, 418–450, and 457–814) and a sporulation domain (SPO22) meiosis-specific motif (amino acid positions 189–417). Mutations in the coding region (black lines) and splicing changes (blue arrowheads) are located in predicted domains. The red bracket shows the deletion that encompasses 79 amino acids of the SPO22 domain (35% of the entire length of the SPO22 domain) and includes 52 conserved positions.
SEARCH FOR TEX11 MUTATIONS
Because the deletion has been shown to cause TEX11 loss of function, meiotic arrest at the pachytene stage, and sterility in male mice,21,22 we performed PCR assays and sequenced coding regions of TEX11 in 48 patients of European descent who had azoospermia (49 patients total, including Patient 1). We identified TEX11 mutations in two additional patients, both with meiotic arrest (Table 1 and Fig. 2). We identified a missense mutation, c.511A→G, predicting the amino acid substitution p.M171V in Patient 2. Amino acid position 171 is located in the TPR1-predicted protein–protein interaction region.
We also observed the absence of PCR products spanning coding exons 10 to 12 (c.652del237bp), which suggested a deletion, in Patient 3. The high-resolution microarray analysis of the sample obtained from Patient 3 revealed that this loss was identical to the deletion of coding exons 10 to 12 detected in Patient 1 (Fig. 1D). TEX11 intragenic deletions of exons 10 to 12 in Patients 1 and 3 were confirmed by means of quantitative PCR analysis (Fig. S1 and Table S4 in the Supplementary Appendix). Using long-range PCR, we sequenced the deletion junctions in both patients and mapped the deletions to a 99,329-bp segment (chrX:69,948,411 to 70,047,740). The breakpoints were located inside of two homologous L1 repeats (L1MA9, 2862 bp; and L1PA4, 6135 bp) that are oriented in opposite directions (Fig. S2 and S3 in the Supplementary Appendix) and are susceptible to DNA breaks. Also, two nearly identical L1PA4 repeats and multiple highly homologous Alu repeats (AluSx and AluSz, >85% homology) were detected near the breakpoints (Fig. S3 in the Supplementary Appendix). Sequencing analysis of the junction regions revealed two noncontiguous deletions separated by a 268-bp inverted sequence retained from intron 9 (chrX:70,047,357 to 70,047,625) and flanked by seven identical short (10-bp to 12-bp) sequences at each junction site (Fig. S3 in the Supplementary Appendix).
REPLICATION STUDY OF TEX11 IN MÜNSTER POPULATION
We sequenced the constitutive DNA of 240 men with azoospermia from Germany to determine the prevalence of TEX11 mutations in a larger, well-phenotyped group. Among these 240 men, we identified 4 (1.7%) who had azoospermia with TEX11 mutations: one missense mutation and three mutations that are predicted to result in splicing alterations (Table 2).
Table 2.
Mutations in TEX11 Detected in Four Samples Obtained from the Follow-up Study Cohort of 240 Patients with Azoospermia from Munster, Germany.*
| Patient No. |
Change in Coding DNA |
Protein/RNA Change |
Phenotype | Study Controls |
Controls from dbSNP Database | Race or Ethnic Group† |
Testicular Sperm Extraction |
Follicle- Stimulating Hormone |
Total Testosterone |
|
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Frequency | |||||||||
| % | no./total no. (%) | mIU/ml | nmol/liter | |||||||
| 4 | C.1837+1G→C | p.R612spl d | Meiotic arrest | 0 | NA | NA | White | No sperm | 6 | 15.1 |
| 5 | C.792+1G→A | p.L264spl d | Meiotic arrest | 0 | NA | NA | Arabic | No sperm | 2.9 | 12.8 |
| 6 | c.450C→T | p.A150Aspl d | Mixed testicular atrophy | 0 | rs147088100 | het2/2267 (0.1) | White | Few sperm | 8.4 | 17.2 |
| 7 | c.2092G→A | p.A698T | Partial meiotic arrest‡ | 0 | rs140984555 | het4/2268 (0.2) | Black | Few sperm | 28 | 18.4 |
The term spl d denotes splicing donor site.
Race and ethnic group were self-reported.
Histologic analysis showed complete arrest, and testicular sperm extraction showed very few postmeiotic cells.
Patient 6, who had mixed testicular atrophy, had a splicing mutation in the last base of exon 7, c.450C→T (p.A150A, coding exon 5) that interrupted the exonic donor splice site (Fig. 2). The mutation was expected to affect a TPR-containing region (TPR1). Patient 5, who had meiotic arrest, had a splicing mutation in the donor splice site of intron 11, c.792+1G→A (Fig. 2). This splicing mutation was expected to interrupt the highly conserved meiosis-specific domain SPO22 (Fig. 2C). Patient 4, who had meiotic arrest, had a donor splice-site mutation in intron 22, c.1837+1G→C, that was predicted to affect a TPR-containing region (TPR3) (Fig. 2). Patient 7, who also had meiotic arrest, had a missense mutation in exon 25, c.2092G→A (p.A698T), that was predicted to result in the substitution of an alanine residue to a larger and polar threonine residue in a TPR-containing region (TPR3) (Fig. 2). ClustalW software and Polymorphism Phenotyping, version 2 (PolyPhen-2) software were used to predict that this mutation, which affects a highly conserved amino acid, was deleterious (Fig. S4 in the Supplementary Appendix). Overall, TEX11 mutations were present in 5 of 33 patients with meiotic arrest (15%) and in 2 of 193 men with mixed testicular atrophy (1%) (Table 3). No TEX11 mutations were identified in the 63 men with the Sertoli-cell–only syndrome (Table 3). Our sequencing analysis of coding exons did not identify any of the TEX11 mutations in 384 controls with normal sperm concentrations (P=0.003 by Fisher's exact test) (Table S5 in the Supplementary Appendix). Moreover, none of the mutations were detected in men in the Single Nucleotide Polymorphism Database (dbSNP); only female carriers of three heterozygous mutations, c.511A→G, c.450C→T, and c.2092G→A, were identified, at a frequency of 0.1% (2 of 2270 controls), 0.1% (2 of 2267), and 0.2% (4 of 2268), respectively (Table 1).
Table 3.
Incidence and Frequency of the TEX11 Mutation, According to Cohort and Phenotype of the Study Population.
| Cohort | Phenotype | All Men with Azoospermia | All Men with Normozoospermia | ||
|---|---|---|---|---|---|
| Meiotic Arrest | Mixed Testicular Atrophy | Sertoli Cell–Only | |||
| no. of patients/total no. (%) | no./total no. (%) | ||||
| Initial study cohort | 2/19 (11) | 1/21 (4.8) | 0/9 | 3/49 (6.1) | 0/192 |
| Follow-up study cohort | 3/14 (21) | 1/172 (0.6) | 0/54 | 4/240 (1.7) | 0/192 |
| Total | 5/33 (15) | 2/193 (1.0) | 0/63 | 7/289 (2.4) | 0/384 |
TEX11 PROTEIN STRUCTURE MODELING
We modeled the TEX11 tertiary structure and estimated the potential effects of the identified mutations on TEX11 conformation (Fig. S5 and S6 in the Supplementary Appendix). More than 70% of TEX11 is predicted to be composed of alpha helixes. Our structure analysis predicted one SPO22 or ZIP4 domain and multiple TPR motifs in the protein. These results are consistent with initial TEX11 modeling that identified TPRs in the SPO22 domain.23
Using the algorithms Phyre215 and I-TASSER,17 we built three-dimensional homology models for the SPO22 domain, N-terminal two-thirds fragment of TEX11, and full-length TEX11. These models predicted tandem pairs of antiparallel alpha helixes that form a superstructure with curvatures ranging from fully closed to fully extended (Fig. S6 in the Supplementary Appendix). The deletion of exons 10 to 12 removes nearly three alpha helixes in the SPO22 motif; mutation p.M171V is likely to destabilize inter-helix packing, substituting a smaller side chain for methionine24; and mutation p.A698T is expected to affect the loop geometry and packing of the two surrounding alpha helixes (Fig. S5 and S6 in the Supplementary Appendix).
TEX11 PROTEIN IN HEALTHY CONTROLS AND IN PATIENTS WITH AZOOSPERMIA
We investigated the cellular localization of TEX11 in seminiferous tubules by immunostaining normal testicular tissue sections obtained from mice, macaques, and men with anti-TEX11 antibodies (Fig. 3A and 3B, and Fig. S7 in the Supplementary Appendix). In mice, TEX11 was prominent in preleptotene to pachytene spermatocytes (Fig. S7A in the Supplementary Appendix). In macaques, TEX11 signal was detected in zygotene spermatocytes and spermatids (Fig. S7C in the Supplementary Appendix). Similarly, in humans, TEX11 was detected in late-pachytene spermatocytes (stage V) and in round and elongated spermatids (Fig. S7E in the Supplementary Appendix).
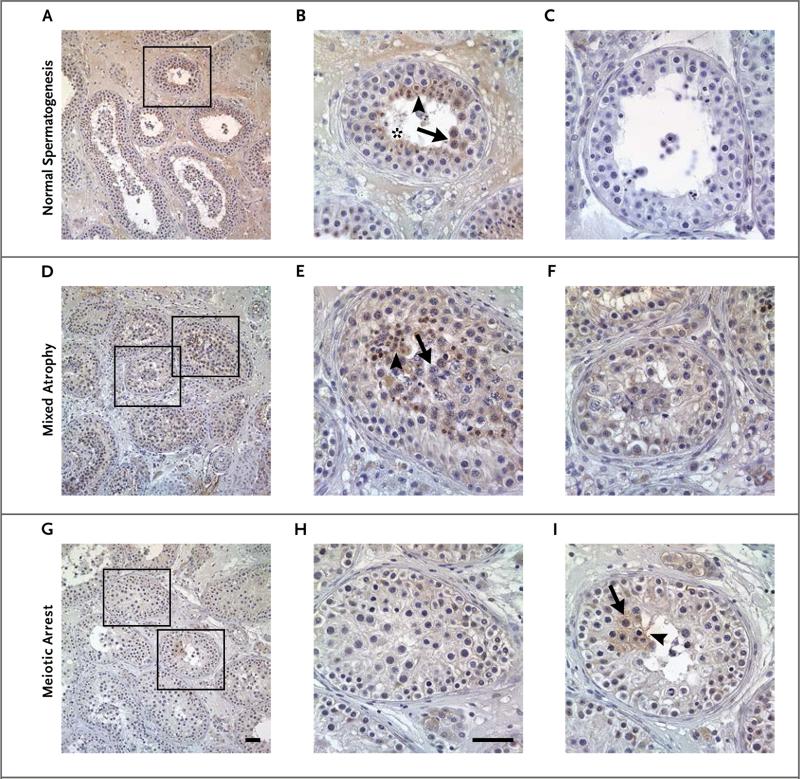
Figure 3. Immunohistochemical Detection of TEX11 in Testicular Tissue Sections Obtained from a Man with Normal Spermatogenesis, a Man with Mixed Testicular Atrophy, and a Man with Meiotic Arrest.
In a man with normal spermatogenesis (Panels A and B), TEX11 was highly expressed in late spermatocytes (arrow), round spermatids (arrowhead), and elongated spermatids (asterisk). Testicular tissue stained with IgG antibody was used as a negative control (Panel C). TEX11 was also present in tubules with remaining spermatogenesis in a patient with mixed testicular atrophy. TEX11 expression was observed in late spermatocytes (arrow) and round spermatids (arrowhead) (Patient 6) (Panels D and E). Seminiferous tubules containing only less-differentiated germ cells and Sertoli cells show no TEX11 expression (Panel F). In a man with complete meiotic arrest (Patient 4), germ cells did not have TEX11 expression in the majority of tubules (Panels F, G, and H). However, TEX11 staining was observed in rare seminiferous tubules with remaining late spermatocytes (arrow) and round spermatids (arrowhead) (Panel I). Scale bars indicate 50 μm.
TEX11 expression was completely absent in surrounding somatic cells such as Sertoli cells and interstitial cells; this suggests that human TEX11 protein is a germ cell–specific factor. To establish a genotype–phenotype correlation, we examined the histologic characteristics and performed immunohistochemical analysis of testes from patients who had azoospermia and deleterious TEX11 mutations (Fig. 3). Two distinct patterns of TEX11 expression were observed in these specimens. In testicular biopsy specimens that showed mixed testicular atrophy, TEX11 staining was present in spermatocytes and in round and elongated spermatids in the majority of tubules (Fig. 3D, 3E, and 3F). However, in biopsy specimens obtained from patients with complete meiotic arrest, spermatids were not seen, and TEX11 expression was not detected in most seminiferous tubules; only a few tubules with late spermatocytes and round spermatids had TEX11 expression (Fig. 3G, 3H, and 3I).
DISCUSSION
In most metazoans, meiosis has a role in generating biologic diversity and supporting species survival. It is not surprising that many meiotic proteins that are involved in chromosomal crossing over and DNA recombination are highly conserved in evolution. Therefore, alterations in meiotic proteins are not typically tolerated in nature, and disrupted gametogenesis leads to partial or complete sterility.
Despite many animal models implicating meiotic arrest in infertility, little is known about meiotic defects in human gametogenesis.2 One example in animal models is the infertility in male Tex11-knockout mice that is due to meiotic arrest and azoospermia.22 TEX11 contains a meiosis-specific domain (SPO22) and numerous TPRs of unknown function. TPRs are protein–protein interaction modules composed of helix-turn-helix repeats that typically appear in tandem and pack with each other to form superhelical structures with various curvatures that can provide docking surfaces for other molecules. TEX11 is present in the lateral synaptonemal complex in prophase I and interacts with components of the double-strand DNA break-repair MRN complex (MRE11 [meiotic recombination 11]–RAD50–NBS1 [Nijmegen breakage syndrome protein 1]).21 It regulates homologous chromosome synapsis and double-strand DNA break repair. Thus, it is critical for synaptonemal complex formation and the chiasma in chromosomal crossover.21,22,25,26 The protein is highly conserved and functionally uniform across species.21
To overcome inconsistencies of genomewide association studies27,28 and to include rare variants, we performed a whole-genome aCGH screening study involving 15 patients with azoospermia. We identified a recurrent 99-kb TEX11 intragenic deletion in 2 unrelated patients. The deletion genomic region is rich in transposable elements and prone to genomic rearrangements, as is evident by a clustering of breakpoints of multiple duplications around TEX11 introns 9 and 12 (Fig. S2 in the Supplementary Appendix).29-32
The molecular structure of the deletion junction fragments suggests a complex mechanism involving an initial inversion of the region, leading to X-chromosome deletions, duplications, or both29,30,33 (Fig. S2 in the Supplementary Appendix). Although inversions and deletions involving TEX11 were not observed in a fertile male population, they may occur frequently in the general population and could be associated with azoospermia. Therefore, we hypothesize that the TEX11 inversion recurs in asymptomatic female carriers, as was shown in the case of exon 22 of F8 (encoding coagulation factor VIII) and multiple exonic inversions in the Duchenne's muscular dystrophy gene that encodes the protein dystrophin.29,30,34,35 This suggests that TEX11-associated male infertility is underestimated because of a potential, undetected high-carrier frequency of inversions in the female population and a high risk of new inversion formation in germ cells of fathers of advanced age.36,37
Overall, TEX11 mutations were identified in 7 of 289 patients (2.4%). Histologic examination of the patients with azoospermia and TEX11 mutations indicated that most of these mutations were detected in patients with complete meiotic arrest (5 of 33 patients, 15%); only two mutations (in 2 of 193 patients, 1%) were detected in patients with azoospermia and mixed testicular atrophy, and no mutations were detected in patients with azoospermia and the Sertoli-cell–only syndrome.
The likely explanation for such phenotype variability in these two patients with mixed testicular atrophy is histologic heterogeneity that was captured among the analyzed sections owing to random section selection. Another possible explanation is that TEX11 mutations are not fully penetrant because of the genetic backgrounds of these persons, as has been shown in mice38 and in patients with Klinefelter's syndrome, in whom foci of spermatogenesis are occasionally detected, whereas testicular degeneration starts early in infancy and accelerates during puberty. Moreover, in some instances, histologic forms of azoospermia (mixed testicular atrophy, meiotic arrest, and the Sertoli-cell–only syndrome) may reflect the stage of disease progression.
We propose that the deletion and splicing mutations severely affect the tertiary structure of the SPO22 domain and the entire protein, disrupting its function or stability. We hypothesize that TEX11 mutations disturb the formation and function of the synaptonemal complex, causing major disruption of pachytene synapsis and anaphase spindle checkpoints; the disruption, in turn, triggers meiotic arrest, spermatocyte apoptosis, and azoospermia. This model is supported by computational structure analysis and immunostaining results in patients with azoospermia, meiotic arrest, and TEX11 mutations; in these patients, we did not detect TEX11 expression and noted massive loss of late spermatocytes and round and elongated spermatids. Notably, wild-type TEX11 expression differs substantially among various mammalian species with normal sperm concentrations (e.g., in humans, macaques, mice, and pigs). However, RNA-based studies in mice39 and antibody-based studies in porcine testes40 have clearly shown that TEX11 expression is reactivated in spermatids. We therefore conclude that the difference in protein expression between primates and mice is probably due to antibody sensitivity in detecting mouse protein in postmeiotic (spermatid) stages.
Our finding that TEX11 mutations occurred in infertile men with meiotic arrest is important for the diagnosis of azoospermia and meiotic arrest. It is also important for preconception testing in men who are partners of women undergoing in vitro fertilization, intracytoplasmic sperm injection, or both.
Supplementary Material
Acknowledgments
Supported by a Mentored Clinical Scientist Development Award from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (K08HD058073, to Dr. A. Yatsenko), awards from the Pennsylvania Department of Health (67-16, to Dr. A. Yatsenko), the Magee–Womens Research Institute (to Dr. A. Yatsenko), the University of Pittsburgh, Kenneth P. Dietrich School of Arts and Sciences (to Dr. Berman), the National Science Center, Poland (2011/01/B/NZ2/04819, to Dr. Kurpisz), and the German Research Foundation (TU298/1-2, to Drs. Tüttelmann and Röpke; and SCHL394/11, to Dr. Schlatt).
We thank the patients for their participation; Dr. Andrew Althouse for his advice on the statistical analysis of our data; Rose Ann King, Bethany Jones, Christina Kubiak, Irene Laffoon, Melissa Lombardozzi, Pamela Print, and Etta Volk for their help in andrology testing and semen sample collection; Jutta Salzig, Anne-Lena Bröcher, Mandy Hoffmann, and Steffi Burkhardt from the University of Münster for their technical support; Dr. Archana Kishore and Randall Beadling from the University of Pittsburgh for their assistance with bench experiments; and Bruce Campbell, scientific editor at the Magee–Womens Research Institute, for his careful editing and useful comments on an earlier version of the manuscript.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.American Urological Association The evaluation of the azoospermic male: AUA best practice statement. 2011 ( https://www.auanet.org/common/pdf/education/clinical-guidance/Male-Infertility-b.pdf)
- 2.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JY, Dada R, Sabanegh E, Carpi A, Agarwal A. Role of genetics in azoospermia. Urology. 2011;77:598–601. doi: 10.1016/j.urology.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization . WHO laboratory manual for examination and processing of human semen. 5th ed. WHO Press; Geneva: 2010. pp. 56–102. [Google Scholar]
- 5.McLachlan RI, Rajpert-De Meyts E, Hoei-Hansen CE, de Kretser DM, Skakkebaek NE. Histological evaluation of the human testis — approaches to optimizing the clinical value of the assessment: mini review. Hum Reprod. 2007;22:2–16. doi: 10.1093/humrep/del279. [DOI] [PubMed] [Google Scholar]
- 6.Nieschlag E, Behre HM, Nieschlag S. Andrology: male reproductive health and dysfunction. 3rd ed. Springer; New York: 2010. [Google Scholar]
- 7.Tüttelmann F, Werny F, Cooper TG, Kliesch S, Simoni M, Nieschlag E. Clinical experience with azoospermia: aetiology and chances for spermatozoa detection upon biopsy. Int J Androl. 2011;34:291–8. doi: 10.1111/j.1365-2605.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- 8.O'Flynn O'Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: a review. Fertil Steril. 2010;93:1–12. doi: 10.1016/j.fertnstert.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 9.Choi Y, Jeon S, Choi M, et al. Mutations in SOHLH1 gene associate with nonobstructive azoospermia. Hum Mutat. 2010;31:788–93. doi: 10.1002/humu.21264. [DOI] [PubMed] [Google Scholar]
- 10.Miyamoto T, Hasuike S, Yogev L, et al. Azoospermia in patients heterozygous for a mutation in SYCP3. Lancet. 2003;362:1714–9. doi: 10.1016/S0140-6736(03)14845-3. [DOI] [PubMed] [Google Scholar]
- 11.Röpke A, Tewes AC, Gromoll J, Kliesch S, Wieacker P, Tüttelmann F. Comprehensive sequence analysis of the NR5A1 gene encoding steroidogenic factor 1 in a large group of infertile males. Eur J Hum Genet. 2013;21:1012–5. doi: 10.1038/ejhg.2012.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mou L, Wang Y, Li H, et al. A dominant-negative mutation of HSF2 associated with idiopathic azoospermia. Hum Genet. 2013;132:159–65. doi: 10.1007/s00439-012-1234-7. [DOI] [PubMed] [Google Scholar]
- 13.Imken L, Rouba H, El Houate B, et al. Mutations in the protamine locus: association with spermatogenic failure? Mol Hum Reprod. 2009;15:733–8. doi: 10.1093/molehr/gap056. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–5. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 16.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biegert A, Mayer C, Remmert M, Söding J, Lupas AN. The MPI bioinformatics toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adelman CA, Petrini JH. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 2008;4(3):e1000042. doi: 10.1371/journal.pgen.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Gell K, van der Heijden GW, et al. Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 2008;22:682–91. doi: 10.1101/gad.1613608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry J, Kleckner N, Börner GV. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc Natl Acad Sci U S A. 2005;102:17594–9. doi: 10.1073/pnas.0508581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaber M, Zhang XJ, Matthews BW. Structural basis of amino acid alpha helix propensity. Science. 1993;260:1637–40. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- 25.Tsubouchi T, Zhao H, Roeder GS. The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev Cell. 2006;10:809–19. doi: 10.1016/j.devcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Primig M, Williams RM, Winzeler EA, et al. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–23. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 27.Hu Z, Xia Y, Guo X, et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat Genet. 2012;44:183–6. doi: 10.1038/ng.1040. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H, Xu J, Zhang H, et al. A genome-wide association study reveals that variants within the HLA region are associated with risk for nonobstructive azoospermia. Am J Hum Genet. 2012;90:900–6. doi: 10.1016/j.ajhg.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mühle C, Zenker M, Chuzhanova N, Schneider H. Recurrent inversion with concomitant deletion and insertion events in the coagulation factor VIII gene suggests a new mechanism for X-chromosomal rearrangements causing hemophilia A. Hum Mutat. 2007;28:1045. doi: 10.1002/humu.9506. [DOI] [PubMed] [Google Scholar]
- 30.Lakich D, Kazazian HH, Jr, Antonarakis SE, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet. 1993;5:236–41. doi: 10.1038/ng1193-236. [DOI] [PubMed] [Google Scholar]
- 31.Huang CR, Schneider AM, Lu Y, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–82. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing J, Zhang Y, Han K, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–26. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aston KI, Carrell DT. Emerging evidence for the role of genomic instability in male factor infertility. Syst Biol Reprod Med. 2012;58:71–80. doi: 10.3109/19396368.2011.635751. [DOI] [PubMed] [Google Scholar]
- 34.Toffolatti L, Cardazzo B, Nobile C, et al. Investigating the mechanism of chromosomal deletion: characterization of 39 deletion breakpoints in introns 47 and 48 of the human dystrophin gene. Genomics. 2002;80:523–30. [PubMed] [Google Scholar]
- 35.Madden HR, Fletcher S, Davis MR, Wilton SD. Characterization of a complex Duchenne muscular dystrophy-causing dystrophin gene inversion and restoration of the reading frame by induced exon skipping. Hum Mutat. 2009;30:22–8. doi: 10.1002/humu.20806. [DOI] [PubMed] [Google Scholar]
- 36.Archer NP, Langlois PH, Suarez L, Brender J, Shanmugam R. Association of paternal age with prevalence of selected birth defects. Birth Defects Res A Clin Mol Teratol. 2007;79:27–34. doi: 10.1002/bdra.20316. [DOI] [PubMed] [Google Scholar]
- 37.Wyrobek AJ, Eskenazi B, Young S, et al. Advancing age has differential effects on DNA damage, chromatin integrity, gene mutations, and aneuploidies in sperm. Proc Natl Acad Sci U S A. 2006;103:9601–6. doi: 10.1073/pnas.0506468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan J, Eckardt S, Leu NA, et al. Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev Biol. 2009;330:167–74. doi: 10.1016/j.ydbio.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet. 2005;14:2911–8. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang L, Zeng W, Clark RK, Dobrinski I. Characterization of the porcine testis-expressed gene 11 (Tex11). Spermatogenesis. 2011;1:147–51. doi: 10.4161/spmg.1.2.16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.