Abstract
Background
Alterations in pathways including BRAF, CDKN2A, and TERT contribute to the development of melanoma, but the sequence in which the genetic alterations occur and their prognostic significance remain unclear. To clarify the role of these pathways, we analyzed a primary melanoma and its metastasis.
Methods
Immunohistochemistry for BRAF-V600E, Sanger sequencing of BRAF and the TERT promoter, fluorescence in-situ hybridization, and telomere analyses were performed on a primary melanoma and its asynchronous cerebellar metastasis. Using the log-rank test and Cox-proportional model, the cancer genome atlas (TCGA) cohort of melanomas was analyzed for the effect of BRAF mutation and CDKN2A loss on survival.
Results
The primary melanoma expressed mutant BRAF-V600E and possessed a homozygous deletion of CDKN2A. In addition to these early defects, the metastatic lesion also possessed evidence of aneuploidy and an activating mutation of the TERT promoter. In the TCGA melanoma cohort, there was a non-significant trend towards poor prognosis in early stage cutaneous melanoma patients with concomitant BRAF mutation and CDKN2A loss.
Conclusion
BRAF mutation and CDKN2A loss occurred early and TERT promoter mutation later in a case of lethal metastatic melanoma. The effects of these pathways on survival warrant further investigation in early stage cutaneous melanoma patients.
Keywords: melanoma, BRAF, CDKN2A, TERT, FISH
Introduction
The early recognition of clinically significant cutaneous melanoma remains a clinical and pathological challenge. A more thorough understanding of the genetic changes driving melanoma and their order of occurrence could improve both its diagnosis and treatment. Activating mutations in BRAF occur in approximately one-half of malignant melanomas and less frequently in benign and atypical Spitzoid tumors (2–28%). A subset of Spitzoid melanocytic tumors, those with loss of BAP1 expression, more commonly have activating BRAF mutations (1–3). Studies of cutaneous melanomas and nevi have demonstrated that activating mutations in BRAF occur as one of the earliest events in melanoma development; however, they are not sufficient for full transformation (4, 5).
A germinal melanoma with initiating oncogenic mutations, like an activating BRAF mutation, must acquire additional mutations prior to becoming fully transformed. Mutation of tumor suppressors, including CDKN2A, PTEN, TP53 and BAP1, frequently represent potent secondary genetic changes that promote further tumorigenesis (6). Loss, mutation, or methylation of the CDKN2A locus occurs in most melanomas. Specifically, germline mutations in CDKN2A remain the most widely recognized cause of inherited melanoma susceptibility (7), and loss of the 9p21 locus, containing the CDKN2A allele, has been associated with poor prognosis in both cutaneous melanoma and atypical Spitz tumors (8–10).
The attrition of DNA from telomeres, the nucleoprotein complex that protects chromosome ends, and subsequent activation of DNA damage checkpoints function as tumor suppressor mechanism for many cell types including melanoma. Thus, the aberrant activation of telomerase is another molecular alteration frequently found during the development of melanoma. TERT amplification has been identified as a frequent finding in acral lentiginous melanomas (11). More recently, activating mutations in the core promoter of TERT, the catalytic subunit of telomerase, have been found in ~40–70% of melanomas and are independently associated with poor prognosis (12–14). Here, we describe the molecular analysis of a primary melanoma and its asynchronous cerebellar metastasis, which occurred 7 years later. We also analyze the effect of these molecular changes on survival using the TCGA melanoma cohort.
Materials and Methods
Melanoma FISH
Formalin-fixed, paraffin-embedded tissues were cut at 5 µm and mounted on positively-charged glass slides. The selection of tissue and the target areas on the hematoxylin and eosin (H and E)-stained slide was performed by a pathologist. Using the H and E slide as a reference, target areas were etched with a diamond-tipped etcher on the back of the unstained slide to be assayed. Pretreatment, hybridization and post-washes were performed according to the microwave method in the DAKO Histology FISH Accessory kit guide. DNA probes (Cytocell, Ltd.) for RREB1 (6p25), cMYC (8q24), CDKN2A (p16) (9p21) and CCND1 (11q13) were combined and hybridized to the appropriate target areas. 25 interphase nuclei were examined and the numbers of signals per nuclei were enumerated. The FISH signal cut-offs were derived from internal validations and the results were expressed as number of signals per nucleus.
Immunohistochemistry
VE1 staining was completed as previously described (15). Primary antibody incubation (anti-BRAF-V600E clone VE1, diluted 1:20, Spring Bioscience, Pleasanton, CA) was performed for 50 minutes at room temperature. Secondary antibody incubation with anti-mouse horseradish peroxidase-conjugated polymer (PowerVision Poly-HRP anti- Rabbit IgG reference # PV6119, Novocastra / Leica) was performed for 45 minutes at 25C. BAP1 staining was conducted using the Benchmark XT automated stainer (Ventana) as previously described (16). Sections were incubated with BAP1 primary antibody (Santa Cruz, sc-28383, 1:50). Appropriate positive and negative controls were utilized for each run of immunostains.
TERT promoter PCR
PCR for the TERT promoter was performed as previously described (17). Briefly, tissue samples (4µm slide sections) were deparaffinized with xylene (Sigma) and DNA was extracted (QIAamp DNA FFPE Tissue Kit). PCR was performed in a total volume of 25 µl with 50ng of DNA using the AccuPrime GC rich kit using (Life Technologies) and the following primers: Forward 5′-CAGCGCTGCCTGAAACTC and Reverse 5’-GTCCTGCCCCTTCACCTT. Reactions were denatured at 95 °C for 180 s, followed by 35 cycles with denaturation at 95 °C for 30 s, annealing at 62 °C for 25 s, and extension at 72 °C for 30 s. The amplification product was agarose gel purified (NucleoSpin Gel and PCR Clean-up) and sent for bidirectional sequencing using the amplification primers (GeneWiz).
Telomere dysfunction induced foci (TIF) assay
Melanoma tissue sections were processed for TIF analysis by fluorescence in situ hybridization (FISH) as described previously (18). Tissue sections were hybridized with a FITC-conjugated telomere sequence (TTAGGG)3-specific peptide nucleic acid (PNA) probe (PNA Bio, Thousand Oaks, CA) in formamide followed by incubation with a polyclonal anti-53BP1 antibody (1:500) (Novus Bio, Littleton, CO) and then with Alexaflour 568 conjugated goat anti Rabbit (1:500) (Invitrogen). Images were captured with a Deltavision microscope at 100x and then deconvoluted using Autoquant X3. TIFs were quantified using Imaris software.
Survival analysis
The survival analysis was carried out using the TCGA dataset on skin cutaneous melanoma (SKCM, https://confluence.broadinstitute.org/display/GDAC/Home). This dataset included 204 patients with 108 deaths during the study. We utilized the clinical information (time of diagnosis, time of death, tumor stage, gender, age), data on somatic mutations in BRAF (V600E or K601E) and copy number changes. To assess deletion of CDKN2A we analyzed the copy number changes of the focal deletion 9p21.3. Deletion was defined to be present if the Log2 of copy N change was lower than -0.415 (corresponding to a ratio of 0.75 copy N between tumor and normal). The survival time and the vital status during the study were used to obtain Kaplan-Meier curves and the log-rank test was used to determine the difference between the survival distributions of two groups of samples. The Cox-proportional model was used to determine the effect of single parameters on survival and the corresponding HR and Wald test p-value are reported. We analyzed the survival distribution among stage I, stage II, stage III, early stage (stage I/II), late stage (stage III/IV), and all stage cutaneous melanoma (6 groups total). The limited sample size precluded a subgroup analysis of stage IV patients. Among these groups, we studied the effect of CDKN2A deletion among the patients concomitantly bearing (or not bearing) a BRAF mutation using a significance of α= 0.0083 after employing the Bonferroni correction for multiple comparisons.
Results
Case Report
On December 24, 2005, a 19 year-old white female with no family history of melanoma was diagnosed with a 1.6mm melanoma on her left antecubital fossa (anterior elbow). The patient promptly had the lesion excised with clear margins and a concurrent negative sentinel lymph node biopsy. At the time of diagnosis, the patient had Stage Ib (T2aN0M0) disease. The patient had 7 years of unremarkable followup. However, in December 2012, she began experiencing neurological symptoms including neck paresthesia, headaches, and nausea that ultimately prompted her to seek medical attention. A computed tomography scan revealed a ~3.0cm mass in the posterior fossa, which was excised, and a diagnosis of metastatic melanoma was rendered. Soon after the excision of the initial metastasis, the patient developed widespread metastatic brain lesions including leptomeningeal involvement. Despite whole brain irradiation, the patient ultimately succumbed to metastatic disease in 2013.
Histopathologic findings
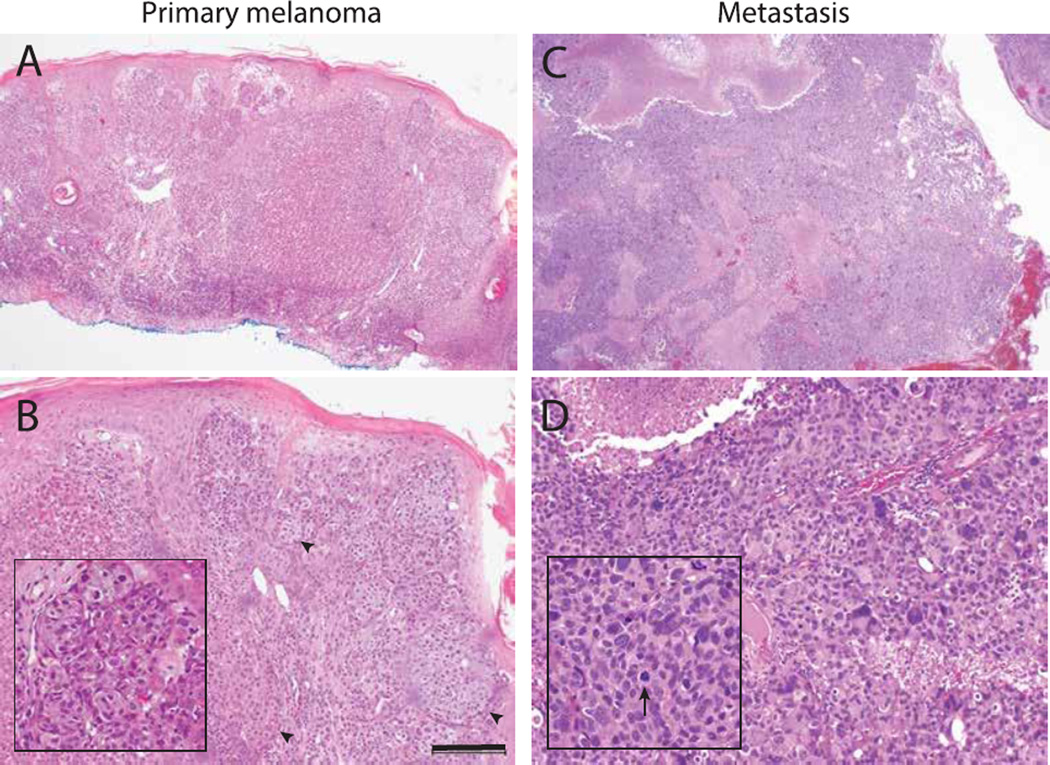
With consent from the patient’s family, the remaining tissue from the patient’s original biopsy was requested. The histopathology sections of the original lesion biopsied in 2005 show an atypical epithelioid neoplasm invading into the dermis with asymmetry, a disorganized growth pattern, and several mitotic figures consistent with a diagnosis of nodular melanoma (Figure 1A). Some areas of the primary melanoma showed Spitzoid features including epithelial hyperplasia and nests of large epitheliod cells with abundant cytoplasm (Figure 1B). The sections of the metastatic posterior cranial fossa lesion showed brain tissue with large areas of involvement by a pleomorphic and epithelioid large cell malignant neoplasm (Figure 1C, D). Tumor cells were negative for low molecular weight cytokeratin (CAM5.2) and focally positive for MART-1 and HMB-45 consistent with the diagnosis of metastatic melanoma.
Figure 1.
Histology of primary and metastatic melanoma. (A) The primary cutaneous lesion is an asymmetric cellular tumor filling the dermis with lack of maturation at the base of the lesion. (B) The primary lesion also shows epithelial hyperplasia and some nests comprised of large and epithelioid cells with abundant cytoplasm (arrowheads). Inset highlights a nest of epitheliod melanocytes. (C) The metastatic cerebellar lesion shows a densely cellular neoplasm with zones of necrosis and hemorrhage. (D) There is marked pleomorphism at higher magnification. Arrows indicates an aberrant mitotic figure. Scale bar=200µm.
Immunohistochemistry
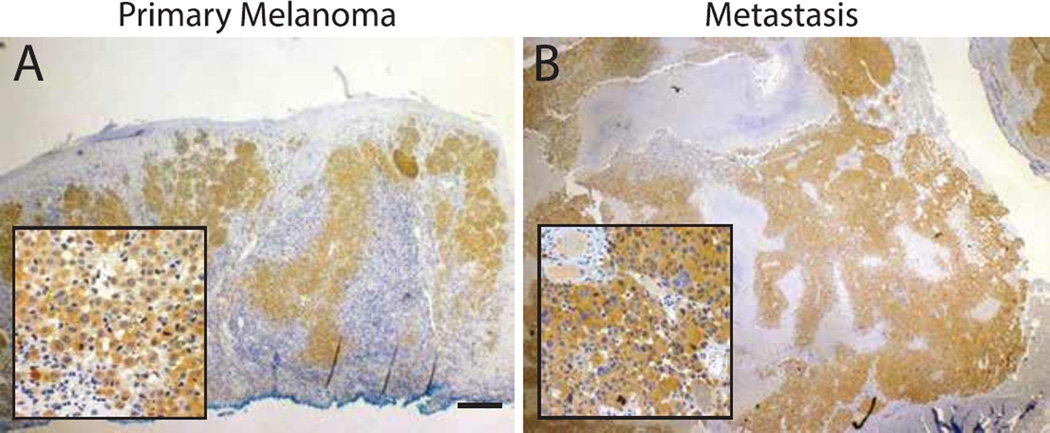
Postmortem immunohistochemistry of the primary lesion with a BRAF-V600E mutation specific antibody (VE1) revealed diffuse cytoplasmic staining of melanocytic nests and single epithelioid melanocytes (Figure 2A). The widespread and uniform staining of the melanocytes of the primary lesion suggests that the BRAF-V600E mutation was an early event in the development of the melanoma. After the initial diagnosis of metastatic melanoma, the metastatic lesion was also sent for VE1 staining. Similar to the primary lesion, the cytoplasm of the malignant cells stained strongly and diffusely, consistent with an underlying BRAF-V600E mutation (Figure 2B). Sanger sequencing confirmed the presence of a mutation (c.1799T>A, p.600V>E) in the metastasis. The concordance of the BRAF mutation of the primary and metastatic lesion is consistent with that of another recent study (19). Given early reports of significant tissue toxicity with concurrent use of B-RAF inhibitors and radiation therapy (20), treatment with B-RAF inhibition had been scheduled upon the completion of radiation therapy.
Figure 2.
BRAF immunohistochemistry. (A) Melanoma biopsy shows diffuse cytoplasmic staining by the BRAF-V600E specific antibody (VE1) supporting the presence of underlying BRAF-V600E mutation. Inset shows individual B-Raf-mutant melanocytes invading the dermis. (B) Cerebellar lesion shows strong BRAF-V600E reactivity by the infiltrating tumor cells. Scale bar=200µm.
BAP1 is a recently identified tumor suppressor implicated in the development of uveal melanoma and melanocytic tumors (21). A subset of atypical Spitz tumors characterized by BRAF mutations and loss of BAP1 expression has been described (3). Because some discrete regions of the primary lesion had Spitzoid features, we sought to determine the mutational status of BAP1 using immunohistochemistry (21, 22). Both the primary melanoma and the metastatic lesion demonstrated strong nuclear staining by the BAP1 antibody throughout the lesions (data not shown), suggesting that homozygous loss or truncating mutations of BAP1 were absent in both lesions and did not contribute to the pathogenesis of this tumor.
Fluorescence in situ Hybridization Studies
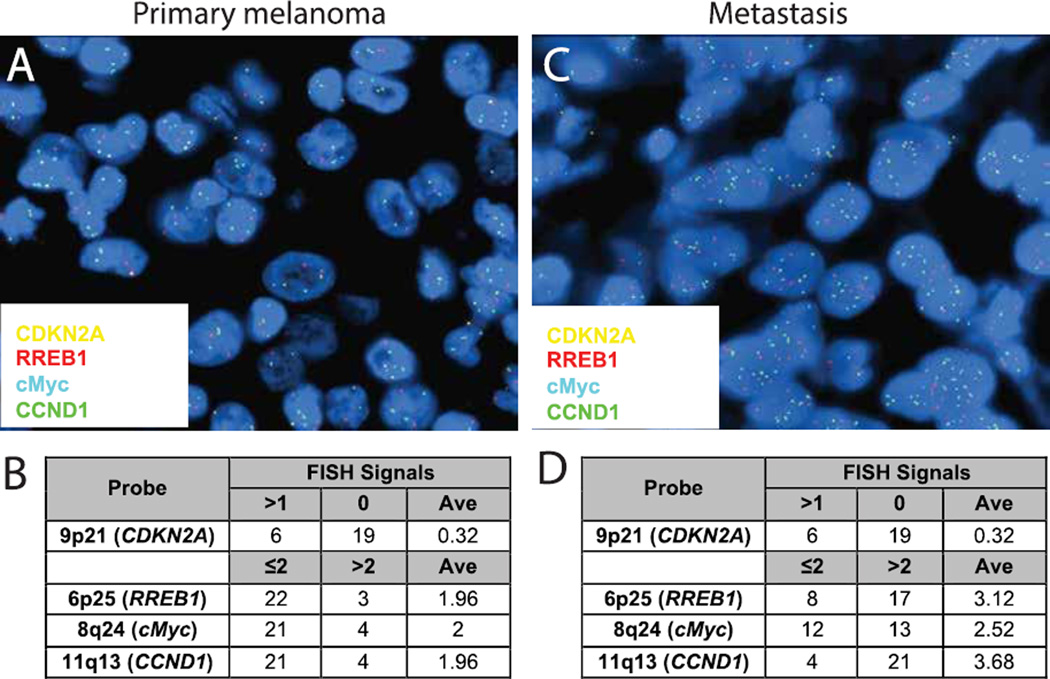
A 4-probe FISH test targeting 6p25 (RREB1), 9p21 (CDKN2A), 11q13 (CCND1), and 8q24 (MYC) was used to assess both lesions (23). The primary lesion showed homozygous loss of the CDKN2A locus (Figure 3A). In the primary lesion, 76% of cells showed homozygous loss of 9p21 with an average of 0.32 signals per cell (Figure 3B). The other loci had normal copy numbers with an average of approximately 2 signals per cell. In the metastatic lesion, in addition to homozygous loss of 9p21, more than 50% of the cells showed greater than 2 copies of 6p25, 11q13, and 8q24 with an average of 3.12, 3.68, and 2.52 signals, respectively, per cell (Figure 3C, D). The amplification of these loci demonstrates the presence of aneuploidy in the metastatic lesion.
Figure 3.
FISH analysis of tumors. (A) Representative image of 4 probe analysis of primary melanoma shows homozygous loss of CDKN2A. (B) Table summarizing the scoring of signal number from 25 cells. Average (Ave) indicates the mean number of foci per nuclei. (C) FISH from metastatic melanoma shows homozygous loss of CDKN2A but increased signals from all remaining probes suggested genomic instability and possible aneuploidy. (D) Table summarizing the scoring of the brain lesion.
TERT promoter and telomere analysis
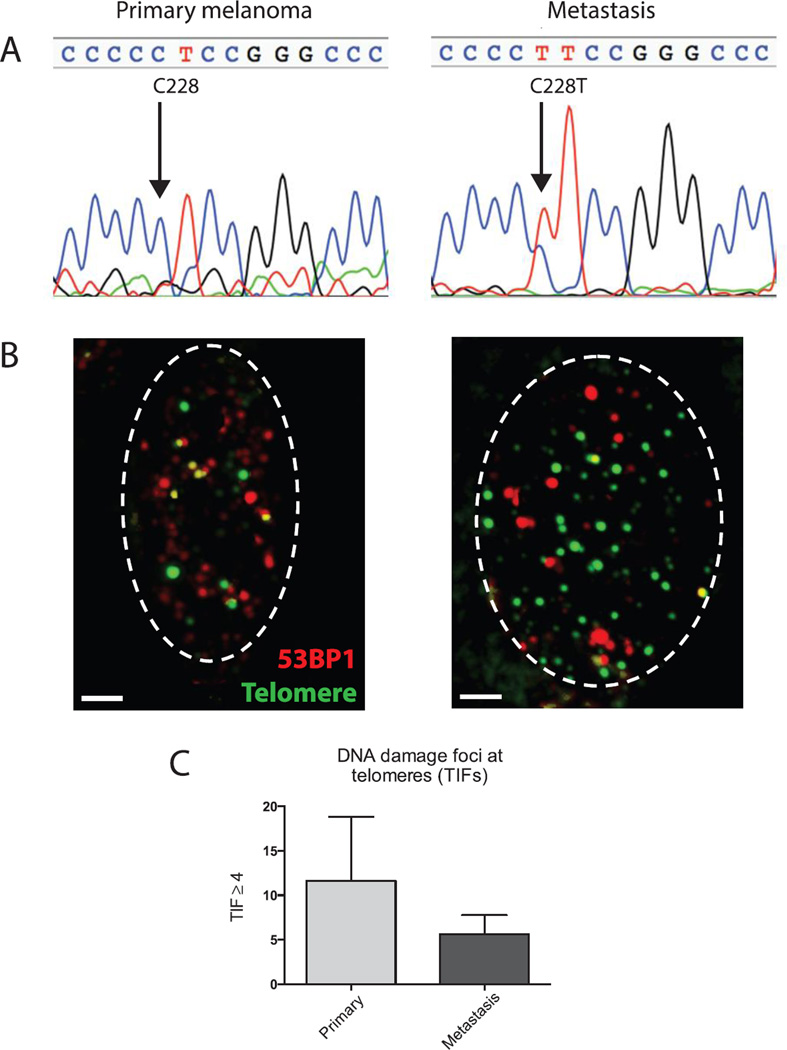
The mutational status of the TERT promoter was assessed in both lesions. Highly recurrent mutations of nucleotide 1,295,228 and 1,295,250 on chromosome 5 generate binding sites for ETS transcription factors in sporadic melanomas (12). While both sites were wild type in the primary melanoma, the metastatic melanoma demonstrated a C228T mutation by Sanger sequencing (Figure 4A). Due to the limitations of tissue availability and Sanger sequencing, we cannot definitively exclude the presence of the TERT promoter mutations in a small proportion of the cells of the primary lesion. To further assess the possible role of telomeres in the development of the tumor, both lesions were analyzed for telomere dysfunction-induced foci (TIF) (24). The TIF assay identifies telomere-specific sites of DNA damage by co-localizing them with telomeres. DNA damage foci were localized with 53BP1 (red label) and telomeres were labeled with a telomere specific (TTAGGG) FITC (green) labeled peptide nucleic acid (PNA)-probe. Consistent with a study from melanoma cell lines (25), abundant DNA damage foci were noted in both the original melanoma and the metastatic lesion (data not shown). Co-localization of DNA damage foci and telomeres revealed more cells with ≥4 telomere dysfunction induced foci (TIFs) per cell in the primary melanoma compared to the metastatic brain lesion (Figure 4B, C). This was true in spite of the fact that the metastatic melanoma cells had dramatically larger nuclei and increased numbers of telomere foci.
Figure 4.
TERT promoter and telomere analysis. (A) Sanger sequencing of the TERT promoter sequencing of the primary melanoma reveals a wild type promoter. The metastatic cerebellar lesion carried a C228T mutation in the TERT promoter. (B) Representative nuclei stained for TIF foci using 53BP1 to identify sites of DNA damage (red) and TTAGGG FISH to identify telomeres. Yellow signals indicate co-localization. Dashed lines outline the cell nucleus. Scale bar=2µm. (C) Quantitation of the percentage of cells with ≥4 TIFs per nucleus. Greater than 100 cells were scored for each replicate.
Survival analyses
Because of the aggressive nature of this patient’s tumor, albeit delayed, we examined the mutations identified in the primary tumor to determine whether they might predict an aggressive clinical course. Loss of CDKN2A has been associated with poor prognosis in cutaneous melanoma (8–10, 26). While BRAF mutations predict poor prognosis in patients with metastatic disease, the effects of BRAF mutations on early stage disease is not clear (27). Using TCGA survival cohorts, we have analyzed CDKN2A deletion and BRAF mutation in relationship to age, stage and gender. Consistent with the literature (28), there was an association between younger age and BRAF mutations. The median age of patients carrying a BRAF mutation was 47 years compared to 58 years for those without mutations (Wilcoxon test p=2.98e-06).
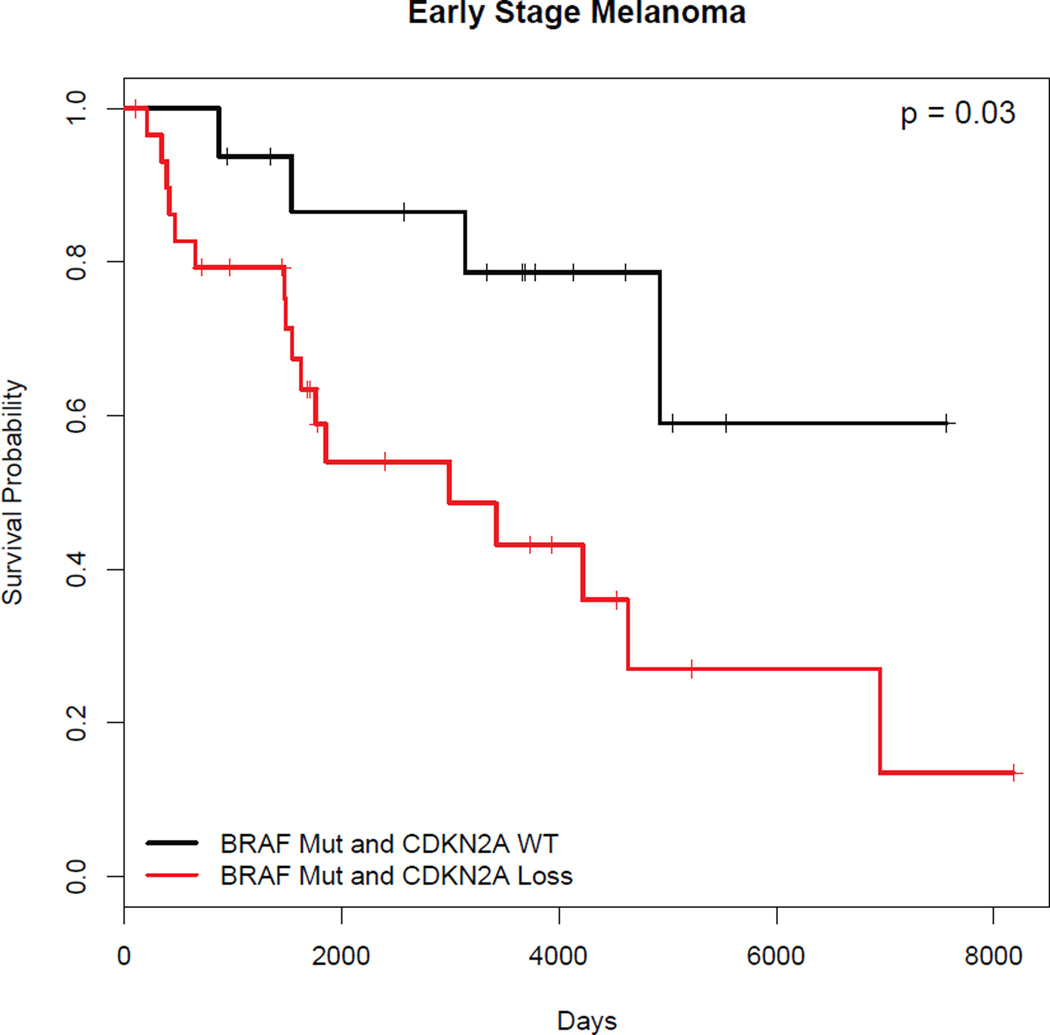
We next investigated the effects of BRAF and CDKN2A on survival. In contrast to existing data suggesting that BRAF mutation was associated with worse disease-free survival (29), we found that the presence of BRAF mutation correlated with better survival without reaching significance (HR=0.73, p=0.11, N=204). Existing studies suggest that CDKN2A locus loss predicts poor survival (21–23), but it is not clear how this effect relates to other genetic events such as an activating BRAF mutation and to the tumor stage. In our dataset, CDKN2A locus deletion alone did not predict a worse prognosis, even though it trended in this direction (HR=1.25, p=0.26, N=231). Among stage I cutaneous melanoma patients bearing BRAF mutations, the concomitant loss of CDKN2A did not significantly predict poor survival but also trended in this direction (HR=7.02, p=0.026, N=18). Similarly, in early stage cutaneous melanoma (stages I and II), the concomitant presence of BRAF mutation and loss of CDKN2A trended towards poor survival without reaching significance (HR=3.32, p=0.032, N=46) (Figure 5). CDKN2A deletion plus BRAF mutation did not predict poor survival among stage III and IV patients where it actually trended towards better survival (HR=0.60, p=0.31, N=32). Thus, in the TCGA dataset, the loss of CDKN2A did not significantly affect survival regardless of BRAF mutation status.
Figure 5.
Kaplan-Meier survival curve demonstrates that early stage (Stage I and II) cutaneous melanoma patients with BRAF mutation and CDKN2A loss do not have a significantly worse prognosis compared to those without CDKN2A loss. Wald test: p = 0.032.
Discussion
BRAF and CDKN2A are the most frequently mutated oncogene and tumor suppressor, respectively, in cutaneous melanoma. We found that this patient’s stage I melanoma harbored a mutation in BRAF and a loss of CDKN2A. By immunohistochemistry and FISH, respectively, these events were present in a majority of the melanocytes of the primary lesion, suggesting that those changes occurred early during this cancer’s evolution. Homozygous loss of CDKN2A loss has been validated as an important diagnostic marker in melanoma. However, using copy number variation data from the TCGA, we found that the loss of CDKN2A did not predict poor prognosis in cutaneous melanoma. The differences between our study and others may be explained by the different methods used to detect loss of the CDKN2A locus, different disease cohorts, and the relatively small sample size in this study. We do note that the loss of CDKN2A did trend towards poor prognosis, especially in the presence of an activating BRAF mutation.
A number of recent studies suggested that gene expression profiles of early stage cutaneous melanoma may be able to predict which patients are at higher risk of aggressive clinical disease. While many of these emerging diagnostics appear promising, they frequently use techniques that are not widely available in clinical pathology laboratories. In contrast, BRAF mutation status and CDKN2A allele status are routinely assessed in pathology laboratories. Thus, determining the precise effects of activating BRAF mutations and CDKN2A loss on survival may reveal important prognostic factors in early stage cutaneous melanoma.
While the role of BRAF as an oncogenic driver and CDKN2A as a tumor suppressor are indisputable, the contribution of telomeres to the development of melanoma appears to be more nuanced. Because TERT promoter mutations are found more frequently in sun-exposed sites and show mutations that could be consistent with UV-induced cytidine-to-thymidine transitions (13, 14), it has been suggested that these mutations might occur early in the development of cutaneous melanoma. As the primary melanoma occurred on sun-exposed skin, it is somewhat surprising that a UV-induced TERT promoter was not detected in the primary lesion. However, as C228T mutations are also frequently found in UV-protected internal malignancies (14), it is possible that the melanoma acquired its TERT promoter mutation after metastasizing. Alternatively, the C228T mutant cells could have been present as a small subset of the primary lesion, which was not detected by Sanger sequencing, but then became the dominant cell type by the time the melanoma was metastatic. In either case, because both a BRAF mutation and loss of CDKN2A were present in the majority of the primary lesion by immunohistochemistry and FISH, respectively, it is likely that these changes preceded any TERT promoter activating mutation.
In addition to the identification of recurrent mutations in the TERT promoter in sporadic melanomas, recent studies have identified mutations in the POT1 telomere binding protein in families with inherited cutaneous melanoma (30, 31). These mutations in POT1 appear to be loss-of-function mutants that impair telomere capping, which are also associated with increased telomere length. Separate epidemiologic studies have suggested that increased telomere length might be a risk factor for the development of nevi and melanoma (32, 33). There remain several possible explanations for the contribution of telomeres to the development of melanoma. Increased telomere length may promote melanoma formation by delaying telomere-driven senescence in melanoma precursor lesions, such as ‘dysplastic’ nevi. Increased telomere length may itself drive telomere genomic instability by promoting chromosome mis-segregation and aneuploidy through effects on mitosis (34–36). Finally, heterogeneous or increased telomere length might simply be a marker of telomere uncapping (37), which would be the primary driver of genomic instability and aneuploidy (38, 39). Both experimental and patient tissue-based studies will be critical to resolve this important issue in the pathogenesis of melanoma.
Aneuploidy has been implicated in the pathogenesis of a variety of malignancies (40). Although aneuploidy has been recognized as a feature of melanocytic neoplasms, there is limited evidence on its role in pathogenesis of melanoma (41–45). It has been reported that increased levels of aneuploidy correlate with tumor stage and predict poor survival in melanoma (46) as well as most solid tumors. The degree of aneuploidy correlates with the degree of focal aneuploidy (copy number changes of arm regions). Analysis of the recurrent patterns of copy number changes indicates that these frequent events represent potent drivers selected during tumorigenesis (47, 48). Through the analysis of large genomic datasets, it will be important to determine how the different copy number changes interact with each other and with other genetic events to influence tumor behavior and clinical outcome.
Telomere dysfunction has been implicated as a major source of aneuploidy in tumors. Although telomere shortening is initially a tumor-suppressor mechanism (49–51), most advanced tumors reactivate telomerase. In the early stages of tumorigenesis, telomere dysfunction, which usually occurs in the absences of telomerase (52), in combination with other genetic changes drives genomic instability, aneuploidization, and the development of cancerous cellular clones. Late in tumor development, the reactivation of telomerase re-establishes telomere protection and promotes the growth of the aneuploid tumor. The genomic characterization of this tumor is consistent with this model of tumor formation that allows us to provide a speculative model on its evolution. Unchecked cell division driven by BRAF and the loss of cell cycle checkpoints (CDKN2A) coupled with the absence of TERT expression ultimately caused the primary lesion to experience telomere attrition and damage. Despite the presence of persistent DNA damage signals due to critically shortened telomeres (TIFs) in the primary lesion, the cancerous cells ultimately bypassed cell cycle arrest and became increasingly aneuploid. Subsequently, the upregulation of telomerase expression through mutations of the TERT promoter then stabilized the telomeres of the metastatic lesion resulting in less DNA damage at telomeres in the metastatic lesion. In this model, the C228T TERT promoter mutation was acquired after aneuploidization to stabilize the genome of the metastatic melanoma. While it not clear whether aneuploidy and the TERT mutation contributed to the brain metastasis, both events provided those cells with a growth advantage by the time the lesion became metastatic.
Acknowledgements
We thank the patient’s family and Quest Diagnostics for their participation in the study. We thank A. von Deimling for the early gift of VE1 monoclonal antibody and B. Chong and G. Xiao for their help with statistical analyses. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number K08CA164047 a Burroughs Wellcome Fund CAMS to R.W. J.W.S. is the Southland Foundation Distinguished Chair in Geriatric Research and this work was supported in part by the Simmons Cancer Center Support Grant 5P30CA142543. Molecular studies were supported in part by ProPath. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
References
- 1.Yazdan P, Cooper C, Sholl LM, et al. Comparative Analysis of Atypical Spitz Tumors With Heterozygous Versus Homozygous 9p21 Deletions for Clinical Outcomes, Histomorphology, BRAF Mutation, and p16 Expression. Am J Surg Pathol. 2014;38(5):638. doi: 10.1097/PAS.0000000000000160. [DOI] [PubMed] [Google Scholar]
- 2.Fullen DR, Poynter JN, Lowe L, et al. BRAF and NRAS mutations in spitzoid melanocytic lesions. Mod Pathol. 2006;19(10):1324. doi: 10.1038/modpathol.3800653. [DOI] [PubMed] [Google Scholar]
- 3.Wiesner T, Murali R, Fried I, et al. A distinct subset of atypical Spitz tumors is characterized by BRAF mutation and loss of BAP1 expression. Am J Surg Pathol. 2012;36(6):818. doi: 10.1097/PAS.0b013e3182498be5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33(1):19. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 5.Yeh I, von Deimling A, Bastian BC. Clonal BRAF mutations in melanocytic nevi and initiating role of BRAF in melanocytic neoplasia. Journal of the National Cancer Institute. 2013;105(12):917. doi: 10.1093/jnci/djt119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastian BC. The molecular pathology of melanoma: an integrated taxonomy of melanocytic neoplasia. Annual review of pathology. 2014;9:239. doi: 10.1146/annurev-pathol-012513-104658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill VK, Gartner JJ, Samuels Y, Goldstein AM. The genetics of melanoma: recent advances. Annual review of genomics and human genetics. 2013;14:257. doi: 10.1146/annurev-genom-091212-153429. [DOI] [PubMed] [Google Scholar]
- 8.Conway C, Beswick S, Elliott F, et al. Deletion at chromosome arm 9p in relation to BRAF/NRAS mutations and prognostic significance for primary melanoma. Genes, chromosomes & cancer. 2010;49(5):425. doi: 10.1002/gcc.20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerami P, Scolyer RA, Xu X, et al. Risk assessment for atypical spitzoid melanocytic neoplasms using FISH to identify chromosomal copy number aberrations. Am J Surg Pathol. 2013;37(5):676. doi: 10.1097/PAS.0b013e3182753de6. [DOI] [PubMed] [Google Scholar]
- 10.Grafstrom E, Egyhazi S, Ringborg U, Hansson J, Platz A. Biallelic deletions in INK4 in cutaneous melanoma are common and associated with decreased survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(8):2991. doi: 10.1158/1078-0432.CCR-04-1731. [DOI] [PubMed] [Google Scholar]
- 11.Bastian BC, Kashani-Sabet M, Hamm H, et al. Gene amplifications characterize acral melanoma and permit the detection of occult tumor cells in the surrounding skin. Cancer Res. 2000;60(7):1968. [PubMed] [Google Scholar]
- 12.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidenreich B, Nagore E, Rachakonda PS, et al. Telomerase reverse transcriptase promoter mutations in primary cutaneous melanoma. Nature communications. 2014;5:3401. doi: 10.1038/ncomms4401. [DOI] [PubMed] [Google Scholar]
- 14.Griewank KG, Murali R, Puig-Butille JA, et al. TERT Promoter Mutation Status as an Independent Prognostic Factor in Cutaneous Melanoma. Journal of the National Cancer Institute. 2014;106(9) doi: 10.1093/jnci/dju246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buresh CJ, Oliai BR, Miller RT. Reactivity with TdT in Merkel cell carcinoma: a potential diagnostic pitfall. American journal of clinical pathology. 2008;129(6):894. doi: 10.1309/R494HQ9VRDJWDY30. [DOI] [PubMed] [Google Scholar]
- 16.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44(7):751. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koelsche C, Sahm F, Capper D, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta neuropathologica. 2013;126(6):907. doi: 10.1007/s00401-013-1195-5. [DOI] [PubMed] [Google Scholar]
- 18.Suram A, Kaplunov J, Patel PL, et al. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 2012;31(13):2839. doi: 10.1038/emboj.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colombino M, Capone M, Lissia A, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30(20):2522. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 20.Satzger I, Degen A, Asper H, Kapp A, Hauschild A, Gutzmer R. Serious skin toxicity with the combination of BRAF inhibitors and radiotherapy. J Clin Oncol. 2013;31(13):e220. doi: 10.1200/JCO.2012.44.4265. [DOI] [PubMed] [Google Scholar]
- 21.Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43(10):1018. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gammon B, Traczyk TN, Gerami P. Clumped perinuclear BAP1 expression is a frequent finding in sporadic epithelioid Spitz tumors. J Cutan Pathol. 2013;40(6):538. doi: 10.1111/cup.12133. [DOI] [PubMed] [Google Scholar]
- 23.Gerami P, Li G, Pouryazdanparast P, et al. A highly specific and discriminatory FISH assay for distinguishing between benign and malignant melanocytic neoplasms. Am J Surg Pathol. 2012;36(6):808. doi: 10.1097/PAS.0b013e31824b1efd. [DOI] [PubMed] [Google Scholar]
- 24.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13(17):1549. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 25.Warters RL, Adamson PJ, Pond CD, Leachman SA. Melanoma cells express elevated levels of phosphorylated histone H2AX foci. J Invest Dermatol. 2005;124(4):807. doi: 10.1111/j.0022-202X.2005.23674.x. [DOI] [PubMed] [Google Scholar]
- 26.Straume O, Sviland L, Akslen LA. Loss of nuclear p16 protein expression correlates with increased tumor cell proliferation (Ki-67) and poor prognosis in patients with vertical growth phase melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2000;6(5):1845. [PubMed] [Google Scholar]
- 27.Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29(10):1239. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 28.Bauer J, Buttner P, Murali R, et al. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment cell & melanoma research. 2011;24(2):345. doi: 10.1111/j.1755-148X.2011.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagore E, Requena C, Traves V, et al. Prognostic value of BRAF mutations in localized cutaneous melanoma. J Am Acad Dermatol. 2014;70(5):858. doi: 10.1016/j.jaad.2013.10.064. [DOI] [PubMed] [Google Scholar]
- 30.Robles-Espinoza CD, Harland M, Ramsay AJ, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet. 2014;46(5):478. doi: 10.1038/ng.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi J, Yang XR, Ballew B, et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet. 2014;46(5):482. doi: 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han J, Qureshi AA, Prescott J, et al. A prospective study of telomere length and the risk of skin cancer. J Invest Dermatol. 2009;129(2):415. doi: 10.1038/jid.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nan H, Du M, De Vivo I, et al. Shorter telomeres associate with a reduced risk of melanoma development. Cancer Res. 2011;71(21):6758. doi: 10.1158/0008-5472.CAN-11-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lisaingo K, Uringa EJ, Lansdorp PM. Resolution of telomere associations by TRF1 cleavage in mouse embryonic stem cells. Mol Biol Cell. 2014 doi: 10.1091/mbc.E13-10-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim MK, Smith S. Persistent telomere cohesion triggers a prolonged anaphase. Mol Biol Cell. 2014;25(1):30. doi: 10.1091/mbc.E13-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKerlie M, Walker JR, Mitchell TR, Wilson FR, Zhu XD. Phosphorylated (pT371)TRF1 is recruited to sites of DNA damage to facilitate homologous recombination and checkpoint activation. Nucleic Acids Res. 2013;41(22):10268. doi: 10.1093/nar/gkt775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126(1):63. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi MT, Cesare AJ, Fitzpatrick JA, Lazzerini-Denchi E, Karlseder J. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nature structural & molecular biology. 2012;19(4):387. doi: 10.1038/nsmb.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21(6):765. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 41.Isaac AK, Lertsburapa T, Pathria Mundi J, Martini M, Guitart J, Gerami P. Polyploidy in spitz nevi: a not uncommon karyotypic abnormality identifiable by fluorescence in situ hybridization. The American Journal of dermatopathology. 2010;32(2):144. doi: 10.1097/DAD.0b013e3181b72d6f. [DOI] [PubMed] [Google Scholar]
- 42.Satoh S, Hashimoto-Tamaoki T, Furuyama J, Mihara K, Namba M, Kitano Y. High frequency of tetraploidy detected in malignant melanoma of Japanese patients by fluorescence in situ hybridization. Int J Oncol. 2000;17(4):707. doi: 10.3892/ijo.17.4.707. [DOI] [PubMed] [Google Scholar]
- 43.Gattuso P, Reddy V, Solans E, et al. Is DNA ploidy of prognostic significance in stage I cutaneous melanoma? Surgery. 1990;108(4):702. [PubMed] [Google Scholar]
- 44.Vogt T, Stolz W, Glassl A, et al. Multivariate DNA cytometry discriminates between Spitz nevi and malignant melanomas because large polymorphic nuclei in Spitz nevi are not aneuploid. The American Journal of dermatopathology. 1996;18(2):142. doi: 10.1097/00000372-199604000-00006. [DOI] [PubMed] [Google Scholar]
- 45.LeBoit PE, Van Fletcher H. A comparative study of Spitz nevus and nodular malignant melanoma using image analysis cytometry. J Invest Dermatol. 1987;88(6):753. doi: 10.1111/1523-1747.ep12470449. [DOI] [PubMed] [Google Scholar]
- 46.Kheir SM, Bines SD, Vonroenn JH, Soong SJ, Urist MM, Coon JS. Prognostic significance of DNA aneuploidy in stage I cutaneous melanoma. Annals of surgery. 1988;207(4):455. doi: 10.1097/00000658-198804000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solimini NL, Xu Q, Mermel CH, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337(6090):104. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davoli T, Xu AW, Mengwasser KE, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Seminars in cancer biology. 2011;21(6):349. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shay JW, Wright WE. Telomeres and telomerase: implications for cancer and aging. Radiation research. 2001;155(1 Pt 2):188. doi: 10.1667/0033-7587(2001)155[0188:tatifc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 51.Shay JW. Are short telomeres predictive of advanced cancer? Cancer discovery. 2013;3(10):1096. doi: 10.1158/2159-8290.CD-13-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Artandi SE, DePinho RA. A critical role for telomeres in suppressing and facilitating carcinogenesis. Current opinion in genetics & development. 2000;10(1):39. doi: 10.1016/s0959-437x(99)00047-7. [DOI] [PubMed] [Google Scholar]