Abstract
There are significant challenges in developing in vitro human tissue and tumor models that can be used to support new drug development and evaluate personalized therapeutics. The challenges include: (1) working with primary cells which are often difficult to maintain ex vivo, (2) mimicking native microenvironments from which primary cells are harvested, and (3) lack of culture devices that can support these microenvironments to evaluate drug responses in a high-throughput manner. Here we report a versatile well plate-based perfusion culture device that was designed, fabricated and used to: (1) ascertain the role of perfusion in facilitating the expansion of human multiple myeloma cells and evaluate drug response of the cells, (2) preserve the physiological phenotype of primary murine osteocytes by reconstructing the 3D cellular network of osteocytes, and (3) circulate primary murine T cells through a layer of primary murine intestine epithelial cells to recapitulate the interaction of the immune cells with the epithelial cells. Through these diverse case studies, we demonstrate the device’s design features to support: (1) the convenient and spatiotemporal placement of cells and biomaterials into the culture wells of the device; (2) the replication of tissues and tumor microenvironments using perfusion, stromal cells, and/or biomaterials; (3) the circulation of non-adherent cells through the culture chambers; and (4) conventional tissue and cell characterization by plate reading, histology, and flow cytometry. Future challenges are identified and discussed from the perspective of manufacturing the device and making its operation for routine and wide use.
Introduction
There is a rapidly growing recognition for critical importance of developing physiologically relevant human tissue and tumor models, as a new means for: (1) preclinical drug evaluation to reduce our reliance on animal models that correlate poorly with clinical outcomes and (2) patient-specific diagnostic screening of therapeutic options, for example, for optimum care of cancer patients.1 Although in vitro tissue models would never be able to fully reproduce the biological complexity associated with homeostasis and disease progression, the models are expected to provide “snapshot” replications of authentic phenotypic cell functions of specific persons and their response to drug treatments. The development and realization of this exciting new technology will certainly require significant advances on three major research fronts: (1) ability to work with primary human cells which are often difficult to maintain ex vivo; (2) mimicking native tissue microenvironments from which primary cells are harvested; and (3) manufacturing of devices that can be easily used by laboratory technicians to replicate the microenvironments and evaluate cell response to drugs in a high-throughput manner.
The use of primary cells is important since immortalizing human cells into cell-lines by gene transfection perturbs the cells’ gene expression profiles and cellular physiology as well as physical integrity of their genome.2–4 Even if primary cells can be grown and maintained, resulting gene expression and cellular physiology can be rather different in conventional versus microenvironment-mimicking culture environments, as shown over two decades ago by the pioneering work of Bissel.5 Since then, research by many investigators has demonstrated the value of using microenvironment-reconstructed cell culture, often with the enabling use of biomaterials, for reproducing authentic cell phenotypes and functions.6
Microfluidic-based perfusion culture has also been increasingly used to mimic mechanical forces and mass transfer conditions associated with in vivo microenvironments.7–9 However, for practical and wide use, there are considerable engineering challenges. Ideally, perfusion culture devices should: (1) be constructed with previously proven and well accepted biocompatible materials, (2) be able to support the replication of various 3D tissue and tumor types, (3) allow the convenient and spatiotemporal placement of cells and biomaterials in wells, (4) be able to support several weeks of multicellular culture which may be required for functional 3D tissue replication as well as monitoring long-term cell response to drugs, and (5) be compatible for use with conventional tissue and cell characterization techniques such as well plate readers, histology, polymerase chain reaction (PCR), and flow cytometry. Ultimately, devices should be commercially available at reasonable costs relative to those utilized in traditional in vitro cultures. Most importantly, devices must be easy to use for routine and reproducible laboratory work by technician-level workforce.
We have approached these challenges by developing a 96-well plate-modified perfusion culture device for reconstructing several tissue and tumor microenvironments with primary human and murine cells. The idea of producing microfluidic culture devices to the format of well plates has been recently pursued by several companies such as Millipore (CellASIC ONIX) and Fluxion Bioscience (Bioflux) as well as academic researchers.10–13 Advantages of using the industry standard well plate format include high-throughput use and compatibility with plate readers. However, the commercial and research devices are currently not capable of: (1) placing cells and/or biomaterials conveniently into culture wells to assemble and modify tissue microenvironments at various time points during culture and (2) circulating non-adherent cells through a multiple number of tissue culture wells. The circulation of non-adherent cells is important for: (1) “organs-on-a-chip” applications6 where different tissues interact physiologically through circulating immune cells in the body and (2) emulating the development of metastatic cancers via circulating tumor cells. These unmet design attributes were incorporated during our device development in addition to providing perfusion and plate reading capabilities, as the main design features of our prototype device.
In this paper, these design features of our prototype device are evaluated and demonstrated by the exploratory use of the device for three specific cases: (1) evaluation of perfusion effect on interactions between human primary multiple myeloma cells and osteoblasts, (2) reconstruction of the 3D cellular network of osteocytes with primary murine osteocytes and microbeads, and (3) circulation of primary murine T cells through a layer of primary murine intestine epithelial cells cultured on nanofibrous mesh. Also, through the prism of these case studies, remaining device design and fabrication challenges are discussed from the perspective of making devices that can be manufactured and will be easy to use.
Device Design and Fabrication
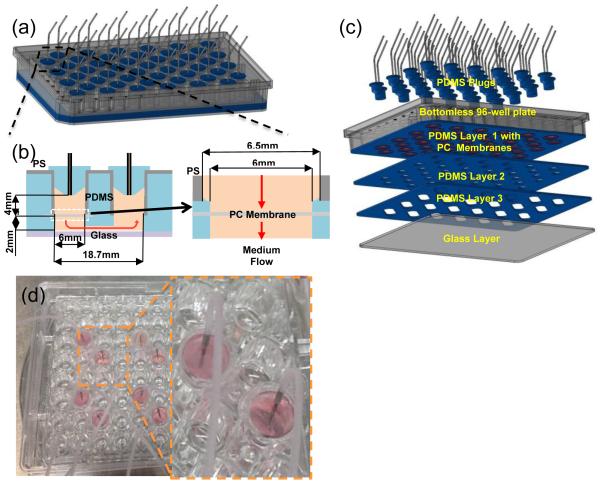
As illustrated in Fig. 1a, the prototype device was assembled on a commercial polystyrene (PS) bottomless 96-well plate. In this design, 2 wells were used to produce 1 culture chamber and 1 outlet chamber (i.e., 48 chambers available for cell culture). The outlet chamber was used to direct the effluent stream to exit through the top of the device so that the device bottom could be accessible for plate reading and microscopy. As shown in Figs. 1b and 1c, three polydimethylsiloxane (PDMS) layers were used to: (1) provide a fluidic channel of 2 mm thick and 5 mm wide between the inlet and outlet chambers and (2) anchor the placement of transparent polycarbonate (PC) membrane discs in culture wells during the device assembly. Tissue and tumor microenvironments were reconstructed on the membrane surface. The bottom of the device was sealed with a glass layer. The device was designed to have an overall thickness of 13.5 mm (without plugs) to be fit into a plate reader. Details associated with device fabrication procedures are provided in Supplementary Information 1 and Supplementary Figs. 1 through 3. An example of the actual device is shown in Fig. 1d.
Figure 1.
Well plate-based perfusion device: (a)–(c) schematic illustrations of device and fabrication and (d) actual device.
The use of PC membranes enabled the culture medium to perfuse through tissue and tumor samples while providing optical access through the bottom of the culture chamber. Also, PDMS plugs were used at the top of the culture wells to enable: (1) the convenient placement of cells and biomaterials into the culture wells at various time points during culture when necessary and (2) the external fluidic connection between culture wells to circulate non-adherent cells such as T cells. For the culture cases described in the next sections, Table I summarizes specific cell types and seeding numbers, flow rates, and characterization techniques utilized. Also, materials and methods used for all the culture experiments are provided in Supplementary Information 2 through 4. Also, Table II compares the design features of our prototype device demonstrated through these culture cases in comparison to those of other well plate-based commercial and research culture devices.
Table 1.
Basic features of the prototype device specifically utilized for three case studies.
| MMC Expansion | Osteocyte Network | T Cell Circulation | |
|---|---|---|---|
|
Cell Types &
Seeding Number |
104 Patient BM cells 104 OSB cell line |
105 Primary murine osteocytes |
105 Primary murine T cells 104 Primary murine IECs |
| Biomaterials | - | Microbeads | Nanofiber mesh |
|
Membrane
Pore Size (μm) |
1 | 3 | 10 |
| Flow Rate (μL/min) | 0.25-2.5 | 1 | 7.6 |
| Readouts | Flow cytometry Plate reader |
Histology Immunostaining |
Cell count Optical microscopy SEM |
Table 2.
Major design features of the prototype device demonstrated through culture case studies in comparison to those of other commercial and research devices
| Features | A 11 | B | C 10 | Prototype |
|---|---|---|---|---|
| Well plate based | Y | Y | Y | Y |
| 3D tissue architecture | Y | N | Y | Y |
| >105 cells | Y | N | Y | Y |
| Perfusion | N | N | Y | Y |
| Non-adhering cells | N | N | Y | Y |
| Biomaterials inclusion | N | N | Y | Y |
| Recirculating cells | N | N | Y | Y |
| Interactive culture chambers | N | N | N | Y |
| Real-time imaging | Y | Y | Y | Y |
| Plate reader | N | N | N | Y |
B: CellASIC™ ONIX Microfluidic Platform, Y (Yes), N (No)
Perfusion Effect on Interactions between Human Multiple Myeloma Cells and Osteoblasts
Multiple myeloma (MM), an incurable B cell malignancy, is the second most common blood cancer in the U.S. with a typical survival of 5 to 7 years.14 Although it has been stipulated that the stromal compartment of bone marrow offers a supportive milieu for the survival and proliferation of multiple myeloma cells (MMCs), the study of mechanistic interactions between stromal cells and MMCs has been previously hindered by the inability of conventional culture techniques to maintain primary MMCs ex vivo. We recently found15 that co-culture of patient bone marrow mononucleate cells (BMMCs) with an ossified tissue pre-grown from osteoblasts (OSB) in microfluidic culture wells can enable the ex vivo preservation and expansion of the MMCs present in the BMMCs population.
This finding is consistent with in vivo observations that have implicated the preferential adhesive interactions of MMCs with a layer of OSBs residing at the endosteum niche (i.e., interface region between bone marrow and bone) while conferring drug resistance and facilitating the survival of MMCs.16,17 We also observed that MMCs slowly killed OSB in this platform over a period of five weeks, which in turn causes their own death. The result suggested that maintaining OSB viability is critical for the long-term maintenance and expansion of primary MMCs ex vivo.18 Our approach enables mechanistic studies of adhesive interactions between primary MMCs and OSBs, which have not been previously possible due to the inability to maintain the viability of primary MMCs during conventional culture.
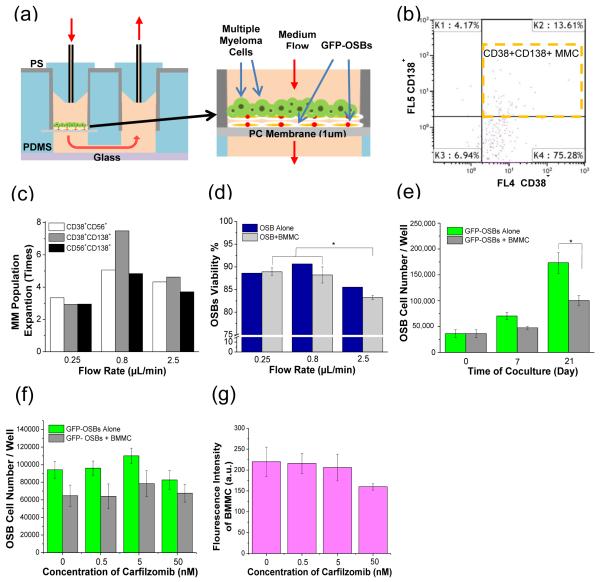
As illustrated in Fig. 2a, the prototype device was used in this investigation to study the potential role of perfusion on influencing MMC-OSB interactions. BMMCs isolated from 3 MM patients’ bone marrow aspirate were used. For the BMMC sample used for the results presented in Figs. 2c, 2d, and 2e, the percentages of MMC subpopulations in BMMC before culture were: (1) 2.6% of CD38+CD138+, (2) 9.3% of CD38+CD56+ and (3) 5.2% of CD56+CD138+. Human OSB cell line cells (hFOB 1.19) were first cultured for 4 days to form a confluent layer in the culture chambers, and then BMMCs were seeded and co-cultured for 7 days. 3 different flow rates (0.25, 0.8 and 2.5 μL/min) were used to evaluate the perfusion effect on interactions between MMC and OSB. Cells in the culture chambers were trypsinized, removed from the chambers, and characterized by flow cytometry to analyze: (1) the proliferation of the MMC subpopulations and (2) the viability of OSB.
Figure 2.
Expansion and drug response of primary human MMCs: (a) schematic illustration of the device configuration and use; (b) flow cytometry dot plot of CD38+ CD138+ MMCs cultured at 0.8 μL/min flow rate; (c) flow rate effect on the expansion of CD38+ CD56+, CD38+ 138+, and CD56+ CD138+ MMC populations, as determined by flow cytometry; (d) flow rate effect on OSB viability, as determined by flow cytometry; and (e) effect of BMMCs on the cell number of GFP-OSBs, as determined by plate reading. Response to drug (CFZ) of (f) OSBs and (g) BMMCs, as determined by plate reading. *p<0.05
As an example of the data obtained, Fig. 2b shows a dot plot of detected CD38+CD138+ MMC subpopulation of the tissue sample cultured at the flow rate of 0.8 μL/min. Fig. 2c summarizes the effect of flow rate on the expansion of CD38+CD56+, CD38+138+, and CD56+CD138+ subpopulations. 0.8 μL/min flow rate resulted in the most expansion of all the MMC subpopulations. For example, CD38+CD138+ MMCs were expanded about 7.5 times at this flow rate. Also, as shown in Fig. 2d, OSB viability was the highest at 0.8 μL/min. Since it is known that flow-induced shear stress can stimulate OSB proliferation and differentiation,20,21 the increased OSB viability at the optimum flow rate of 0.8 μL/min may be responsible for the highest expansion of MMC subpopulations (Fig. 2c). The result suggested that appropriate flow-induced shear stress can stimulate the proliferation of OSB and thus help maintain the viability and expansion of MMCs.
The effect of BMMCs on the viability of GFP-OSBs was evaluated by calculating the number of viable OSBs after 0, 7 and 21 days using the fluorescence data obtained from a plate reader (Fig. 2e). The results showed that OSBs, which were modified to express green fluorescent protein (GFP-OSBs), proliferated by about 2.8 times in 21 days during co-culture. However, in comparison to OSB-only culture control, the OSB proliferation was significantly inhibited by the presence of BMMCs. The plate reading result is consistent with our previous result obtained using flow cytometry.18
Drug Response Evaluation on OSBs and MMCs
After co-culture for 1 week, the response of OSBs and BMMCs to carfilzomib (CFZ) was evaluated at four CFZ concentrations (0 nM, 0.5 nM, 5 nM and 50 nM). CFZ is a selective proteasome inhibitor which has been recently approved for treating MM patients. Both clinical and in vitro studies showed that CFZ increases the expression of alkaline phosphatase (ALP) which is a bone-anabolic marker.19 After dosing CFZ for 6 h and further co-culture for 1 week, the fluorescent intensity of OSBs and BMMCs were measured by plate reading (see Supplementary Information 2 for details). The results in Fig. 2f suggested that CFZ might slightly induce OSB proliferation at the concentration less than 5 nM, but became toxic to OSB at 50 nM. The relative BMMC fluorescent intensity changes in Fig. 2g indicated the decreased viability of BMMCs at 50 nM. We are currently developing a fluorescent-based assay to differentiate MMC populations present in BMMCs.
The results obtained with the prototype device demonstrate the possibility of: (1) replicating the multi-cellular MM tumor niche with perfusion as an important microenvironmental factor and (2) characterizing cell responses to drugs using conventional flow cytometry and plate reading techniques. With this device, we are further developing fluorescence-based assays for studying the response of MMCs and OSBs to several proteasome inhibitors, which have emerged as an important therapeutic strategy for MM patients.22 We anticipate that this ex vivo tumor engineering approach may provide a new avenue for: (1) testing of personalized therapeutics for MM patients, (2) evaluation of new drugs without the need for costly animal models, and (3) studying the biology of MM and in particular the mechanisms responsible for drug resistance and relapse. Beyond multiple myeloma, our ability to conserve the endosteal niche is expected to be useful in studying solid tumors like breast and prostate cancers that metastasize to the bone through the same endosteal niche.
Reconstruction of 3D Cellular Network with Primary Murine Osteocytes and Microbeads
Osteocytes are the most abundant cells (>90%) that reside in mineralized extracellular matrix cavities (“lacunae”) in bones. Neighboring osteocytes are interconnected by extending dendritic processes through smaller channels (“canaliculi”) and forming gap junctions. Osteocytes in the resulting 3-dimensional (3D) cellular networks are known to function as mechanotransduction sensors and master regulators of homeostatic bone remodeling.23,24 It is known that the perfusion of lacunar and canalicular fluids and the mass transfer of molecules occur, as a result of compressive mechanical loading of bones which behave as “poroelastic sponges.” Osteocytes respond to perfusion-induced shear on cell membrane surfaces by activating neighboring OSBs to form new bone. Also, osteocytes have also been implicated with regulative contributions in metabolic demands for minerals25 and hematopoiesis.26 Despite these important roles of osteocytes, a significant challenge remains for in vivo studies of osteocytes due to difficulties in accessing and characterizing osteocyte networks deeply embedded in hard structures of mineralized bones. Furthermore, for in vitro studies, the in vivo phenotype of primary osteocytes could not have been maintained as they differentiate back to osteoblasts during conventional culture.27,28
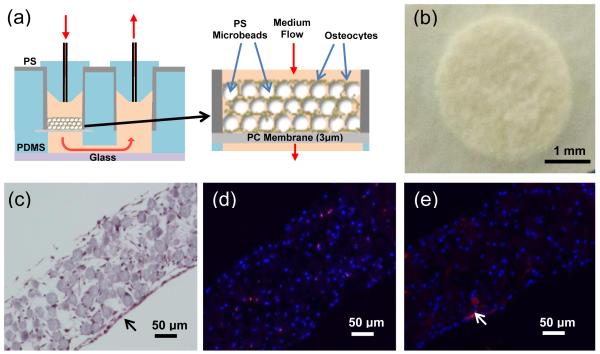
As illustrated in Fig. 3a, we recently found29 that a 3D osteocyte network can be reconstructed by the biomimetic assembly of early osteocytic cell line (MLO-A5) cells and microbeads within the physical confine of PDMS-based microfluidic culture wells fabricated on a glass slide. In this previous study, we used 25 μm microbeads of biphasic calcium phosphate (BCP) to: (1) allow a single cell to be placed within the interstitial space between the microbeads, (2) mitigate the proliferation of the entrapped cell due to its physical confinement in the interstitial site, and (3) control cell-to-cell distance to be 20-25 μm as observed in murine bones. The entrapped cells formed a 3D cellular network by extending their processes through openings between the microbeads and forming gap junctions within 3 days of culture. We also observed that the time-dependent osteocytic transitions of the MLO-A5 cells exhibited trends that were consistent with in vivo observations, particularly the expression of SOST gene which is the key osteocyte-specific marker and produces sclerostin as a major signaling molecule for the mechanotransduction function of osteocytes. In contrast, cells cultured in 2D well plates did not replicate in vivo trends.
Figure 3.
3D cellular network reconstruction with primary murine osteocytes: (a) schematic illustration of the device configuration and use, (b) tissue sample after 13 days of culture; (c) cross-section of the tissue with H&E stain showing the network formation of a 3D network of osteocytes after 13 days, and (d) cross-section of the tissue with immunofluorescence staining showing sclerostin production (red) by osteocytes (nucleus blue, DAPI) after 13 days, and (e) ALP immunofluorescence staining (red) after 13 days.
In the present study, the prototype device was used to further demonstrate the microbeads-guided reconstruction approach with primary murine osteocytes. Primary osteocytes were isolated from murine long bones (see Supplementary Information 3 for details). Isolated cells and BCP microbeads were mixed and placed into the culture wells to form ~200 μm-thick tissue sample. After 13 days of culture, the tissue samples were removed from the culture wells as mechanically integrated structures as shown in Fig. 3b. The cross-section histological image of the tissue with hematoxylin and eosin (H&E) staining in Fig. 3c suggested that cells were relatively evenly distributed throughout the tissue structure without proliferation, and thus emulating the lacunae-like structure of bones. Moreover, cells were attached and spread onto the surface of BCP microbeads. Neighboring cells became interconnected through developing their processes and thus forming a 3D cellular network.
The immunofluorescence staining of tissue samples cultured for 13 days showed that: (1) some cells inside of the 3D tissue structure produced sclerostin (Fig. 3d) and (2) some cells located on the outer tissue surface produced ALP as indicated by the arrow in Fig. 3e. It is well established that sclerostin can only be produced by mature osteocytes while ALP is an osteoblastic phenotypic marker. Therefore, our results suggested that the osteocyte phenotype of the harvested cells could be preferentially preserved inside of the tissue structure due the 3D confinement effect of microbeads as mentioned earlier. As it is beyond the scope of this device-centric paper, we report elsewhere30 in details that these reconstructed osteocytes express SOST gene which produces sclerostin, as determined by both PCR and histological immunostaining. In contrast, this key osteocytic gene expression is lost within a few weeks of conventional 2D culture, as the osteocytes differentiate to osteoblasts.
Significance of these results is that, for the first time to our best knowledge, we demonstrate the possibility of preserving the SOST gene expression of primary osteocytes ex vivo. For clinical relevance, the SOST expression is important since it produces sclerostin which is a major signaling molecule for mechanotransduction in bones.31,32 Also, it is an important new drug target for treating osteoporosis33 and osteolytic lesions caused by bone metastases.34 We are currently working on: (1) extending the biomimetic assembly approach for use with primary osteocytes harvested from discarded human bones from orthopaedic implant surgeries and (2) characterizing the mechanotransduction function of the reconstructed osteocyte network by applying perfusion-induced shear stress. From the device point of view, the above results demonstrate that the prototype device can be used to generate 3D tissue structures that can be characterized by conventional histology.
Circulation of Primary Murine T Cells through Primary Murine Intestine Epithelial Cells Maintained on Nanofibrous Mesh
Adoptive T cell therapy in the form of allogeneic blood and marrow transplantation (allo-BMT) has proven to be one of the few curative treatments for patients suffering from a number of drug-resistant hematological malignancies. However, the full exploitation of this clinical intervention is greatly limited by graft versus host disease (GVHD), as one of the major BMT complications. This disease is characterized by severe and potentially lethal tissue damage to skin, liver, and gut tissues of transplanted patients, mediated by donor T cells responding to host alloantigens.35–37 In particular, GVHD of the gastrointestinal tissues is closely associated with non-relapse mortality following allo-BMT.38 Currently, there is no way to predict which patient-donor pairs will develop GVHD after BMT. Our long-term interest is to explore the possibility of emulating the potential killing of patient-derived intestinal epithelial cells (IECs) by donor T cells, where IECs are the primary population targeted in GI GVHD.39,40 In native tissues, IECs reside on a thin fibrous basement membrane (BMa) consisting of the intermingled networks of laminins and collagens and provides cell anchoring and barrier functions. The membrane networks interact with cells through membranous integrin receptors and other plasma membrane molecules, influencing cell differentiation, migration, adhesion, phenotype, and survival.
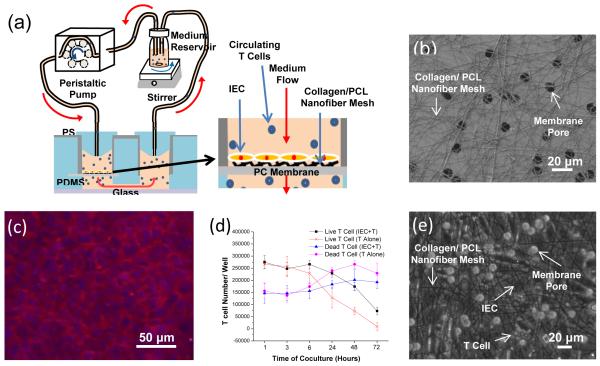
As an initial step towards this application, we used the prototype device to: (1) culture and maintain primary murine IECs isolated from the small intestine of a C57Bl/6-Tg(CAG-OVA)916 Jen/J mouse (B6-SIINFEKL) and (2) assess the device’s capability in supporting the circulation of primary murine T cells through the IECs (see Fig. 4a for conceptual illustration of the experimental approach and Supplementary Information 4 for experimental details). As shown by the scanning electron microscopic (SEM) image in Fig. 4b, nanofiber mesh was used to mimic the BMa of the epithelial tissue as well as to support the long-term viability of IECs. The latter role is particularly important, since primary IECs cannot be kept viable during conventional culture. The nanofiber mesh was produced by electronspining polycaprolactone (PCL)/type I collagen onto the PC membrane surface prior to the device assembly. The average open space in the nanofiber mesh was controlled to be about 6 μm (Fig. 4b) since the average diameter of T cells is about 5 μm. For the same reason, we also selected the average pore of the PC membrane to be 10 μm (Fig. 4b). With the use of nanofiber mesh, IECs was able to develop into a confluent layer and exhibit cobblestone morphology while remaining viable in the perfusion device for up to 7 days (Fig. 4c).
Figure 4.
Circulation of primary murine T cells through primary murine IECs: (a) schematic illustration of the device configuration and use; (b) SEM image showing collagen/ PCL nanofiber mesh on PC membrane with 10 μm pores; (c) merged fluorescent image showing IEC cytoskeleton (red, ActinRed) and nucleus (blue, DAPI) after Day 7 on collagen/PCL nanofiber mesh; (d) effect of IEC presence on T cell viability; and (e) bright field image showing IECs and T cells cultured for 6 h on nanofiber mesh.
After IECs became confluent (approximately 4 days post seeding), T cells harvested from an eGFP transgenic C57Bl6/J mouse were introduced and circulated through the culture chambers (2.5×105 cells per the chamber). As illustrated in Fig. 4a, a peristaltic pump was used to circulate T cells in RPMI complete medium. Visually, we did not see the entrapment of T cells in any part of the culture device and external circulatory pathways. T cell viability was quantified by sampling the culture medium at various time points and counting live and dead T cell numbers suspended in the medium. As shown in Fig. 4d, the overall viability of T cells decreased during the 72 h culture period. This was expected since it is well known that the viability of T cells cannot be maintained in vitro unless they are stimulated by antigen-presenting cells or maintained via the addition of cytokines like IL2.41 However, interestingly, there were more viable T cells when they were circulated through the IEC layer. The results suggest that T cells were activated by IECs, resulting in the increased viability of T cells. Although both B6-SIINFEKL IECs and T cells were of B6 background, it is likely that minor antigen differences between the B6-SIINFEKL and the eGFP-B6 strains could have elicited activation of the T cells. Fig. 4e shows that IECs were spreading on the nanofiber mesh surface, and were in physical contact with T cells. Since the membrane pores (10 μm) and opening spaces (>6 μm) between nanofibers were larger than T cells (5 μm), they were able to go through the IEC layer without getting trapped in the culture chamber.
These results suggest that the device can be used to: (1) support long-term culture of primary murine IECs through the enabling use of PCL/collagen nanofiber mesh and (2) circulate and maintain viable T cells without entrapment. With this initial basis, we plan to further optimize the use of the device in replicating the ex vivo alloreaction of T cells initially using established murine models and later using biospecimens obtained from patient-donor pairs. Furthermore, the ability to mimic the circulation non-adherent cells, as presented here, is a unique feature of the device that is paramount to studying the role of immune cells and other potentially circulating cells in health and diseased states. For example, beside the GVHD application, this approach may have an overarching application to other important immune related research areas such as autoinflammatory diseases that target gastrointestinal tissues, the interaction of circulating tumor cells with the bone marrow niche, and the interactions of T cells with tumors.
Considerations for Future Device and System Development
Through the above case studies, we demonstrate the prototype device’s design attributes for: (1) replicating 3D tissue and tumor microenvironments with appropriate biomaterials, stromal cells, and perfusion, (2) circulating non-adherent cells through the tissue culture wells, and (3) compatibility with common biological readouts. One of the novel features of the current design is the use of the membrane to hold the tissue structures while its transparency allows optical access for plate reading and the average hole size of the membrane could be varied for the specific applications (see Table 1). We used PC membranes mainly because of their commercial availability with various pore sizes that we have exploited in our case studies and while being biocompatible. Another practical feature is the use of removable plugs at the entrance of the culture wells to allow convenient and temporal placement of cells and biomaterials. In future, we can consider vascularizing 3D tissue structures. However, for applications not involving endothelium niches, perfusion through 200 μm thick tissues may be sufficient to replicate interstitial flow and mass transfer effects in tissue spaces between blood and lymphatic capillaries since average separation distance between these capillaries is 200 μm.
The current device, as a custom-made laboratory device, takes about two days to make. Also, at the current stage, device fabrication is quite laborious as described in detail in Supplementary Information 1 and Supplementary Figs. 1 through 3. The future device for wide use purposes should obviously be mass produced using standard industry practices while eliminating labor-intensive steps through automation and large-scale operation.
There are also concerns for the use of PDMS parts although they are not in direct contact with cells in our current design. First, PDMS has been reported to: (1) leach hydrophobic components from cell culture media including drugs, antibodies, growth factors, and lipids and (2) store them within the bulk structure of PDMS.42 This issue can potentially complicate the system reproducibility required to support the clinical trial and regulatory validation steps. Second, an industrial base does not exist yet to support manufacturing of PDMS-associated devices. For these reasons, it may be important to replace the PDMS parts in the current design with PS parts since PS has been the most accepted and widely used material for cell culture. The replacement is certainly feasible since the fabrication of entirely PS-based microfluidic devices has been previously demonstrated43,44 with the combination of photolithography and hot embossing capable of generating devices with feature sizes down to 10 nm.45,46 For our current device design, the minimum feature size is 2 mm in the thickness direction and 6 mm in the lateral directions (See Fig. 1b). These dimensional requirements can certainly be achieved even conventional PS molding, cutting, and bonding practices that are used today to manufacture well plates.
From the user perspective, a significant challenge remains with the need to use as many as 48 syringe pumps to apply and control flow to all 48 culture wells in the device. When precise flow control is not critically needed, we may need to consider gravity-based medium flow approaches that have been reported in the literature47 as part of our future device design. Also, we will need to consider automation of fluid and cell handling through integration with plate dispensers and washers. Also, 3D printing of multi-cellular tissue and biomaterial structures may be important in obtaining spatial control to replicate heterogeneous tissue and tumor structures.
Conclusions
The well plate-based perfusion culture device was designed, fabricated, and used to: (1) ascertain the role of perfusion in facilitating the expansion of human multiple myeloma cells, (2) preserve the physiological phenotype of murine osteocytes by reconstructing the 3D cellular network of murine osteocytes, and (3) circulate T cells through murine intestine epithelial cells. Through these case studies, we demonstrate the versatility of the device’s design features to support: (1) the convenient and temporal placement of cells and biomaterials into the culture wells of the device, (2) the replication of tissue and tumor microenvironments with perfusion, stromal cells, and biomaterials, (3) the circulation of non-adherent cells through the culture wells, and (4) conventional tissue cell characterization by plate reading, histology, and flow cytometry.
Supplementary Material
Acknowledgements
Research in this publication was supported in parts by the Provost Office of the Stevens Institute of Technology (Stevens), the John Theurer Cancer Center at Hackensack University Medical Center (HackensackUMC), the National Institutes of Health grants (1R21AR065032 and 1R21CA174543), and the National Science Foundation grant (DMR1409779). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and the National Science Foundation. CFZ was provided by ONYX Pharmaceuticals, an subsidiary, and drug evaluation studies were conducted in part with funding support from ONYX Pharmaceuticals. We thank Dr. David Siegel at HUMC for providing MM patient biospecimens and Dr. Peter Tolias at Stevens for useful discussions.
References
- 1.Tolias P. Personalized Medicine. 2012;9:463–6. doi: 10.2217/pme.12.51. [DOI] [PubMed] [Google Scholar]
- 2.Li A, Walling J, Kotliarov Y, Center A, Steed ME, Ahn SJ, Rosenblum M, Mikkelsen T, Zenklusen JC, Fine HA. Molecular cancer research: MCR. 2008;6:21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 3.Domcke S, Sinha R, Levin DA, Sander C, Schultz N. Nature communications. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandberg R, Ingemar E. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2052–7. doi: 10.1073/pnas.0408105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aggeler J, Park CS, Bissell MJ. Journal of dairy science. 1988;71:2830–42. doi: 10.3168/jds.S0022-0302(88)79879-3. [DOI] [PubMed] [Google Scholar]
- 6.Huh D, Hamilton GA, Ingber DE. Trends in cell biology. 2011;21:745–54. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasirayi G, Auger V, Scott SM, Rahman PKSM, Islam M, O’Hare L, Ali Z. Micro and Nanosystemse. 2011;3:137–160. [Google Scholar]
- 8.Lee JH, Wang H, Kaplan JB, Lee WY. Tissue engineering. Part C, Methods. 2011;17 doi: 10.1089/ten.TEC.2010.0285. [DOI] [PubMed] [Google Scholar]
- 9.Lee JH, Gu Y, Wang H, Lee WY. Biomaterials. 2012;33:999–1006. doi: 10.1016/j.biomaterials.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Domansky K, Inman W, Serdy J, Dash A, Lim MH, Griffith LG. lab on a chip. 2010;10:51–8. doi: 10.1039/b913221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang T, Mikheev AM, Monnat RJ, Rostomily RC, Folch A. 17th International Conference on Miniaturized Systems for Chemistry and Life Science; 2013.pp. 506–8. [Google Scholar]
- 12.Robbins NF, Rowley J, Quinto M, Hastings AZ, Towns BG, Snodgrass BR. 2006. US 7767446 B2.
- 13.Vukasinovic J. 2014. US 20140106452 A1.
- 14.Rajkumar SV. American journal of hematology. 2013;88:226–35. doi: 10.1002/ajh.23390. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Lee WY, Siegel DS, Tolias P, Zilberberg J. Tissue engineering. Part C, Methods. 2014;20:663–70. doi: 10.1089/ten.TEC.2013.0490. [DOI] [PubMed] [Google Scholar]
- 16.Roodman GD. Journal of bone and mineral metabolism. 2010;28:244–50. doi: 10.1007/s00774-009-0154-7. [DOI] [PubMed] [Google Scholar]
- 17.Roodman GD. Bone. 2011;48:135–40. doi: 10.1016/j.bone.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Gu Y, Sun Q, Siegel DS, Tolias P, Yang Z, Lee WY, Zilberberg J. PloS one. doi: 10.1371/journal.pone.0125995. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu B, Chen Y, Usmani SZ, Ye S, Qiang W, Papanikolaou X, Heuck CJ, Yaccoby S, Williams BO, Van Rhee F, Barlogie B, Epstein J, Qiang YW. PLoS One. 2013;8:1–15. doi: 10.1371/journal.pone.0074191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapur S, Baylink DJ, William Lau KH. Bone. 2003;32:241–51. doi: 10.1016/s8756-3282(02)00979-1. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein AS, Juarez TM, Helmke CD, Gustin MC, Mikos AG. Biomaterials. 2001;22:1279–88. doi: 10.1016/s0142-9612(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 22.Moreau P, Richardson PG, Cavo M, Orlowski RZ, San Miguel JF, Palumbo A, Harousseau J. Blood. 2012;120:947–59. doi: 10.1182/blood-2012-04-403733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dallas SL, Prideaux M, Bonewald LF. Endocrine reviews. 2013;34:658–90. doi: 10.1210/er.2012-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loiselle AE, Jiang JX, Donahue HJ. Bone. 2013;54:205–12. doi: 10.1016/j.bone.2012.08.132. [DOI] [PubMed] [Google Scholar]
- 25.Bonewald LF. Journal of Bone and Mineral Research. 2011;26:229–38. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asada N, Katayama Y, Sato M, Minagawa K, Wakahashi K, Kawano H, Kawano Y, Sada A, Ikeda K, Matsui T, Tanimoto M. Cell stem cell. 2013;12:737–47. doi: 10.1016/j.stem.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Schneider P, Meier M, Wepf R, Muller R. Bone. 2010;47:848–58. doi: 10.1016/j.bone.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 28.Boukhechba F, Balaguer T, Michiels JF, Ackermann K, Quincey D, Bouler JM, Pyerin W, Carle GF, Rochet N. Journal of Bone and Mineral Research. 2009;24:1927–35. doi: 10.1359/jbmr.090517. [DOI] [PubMed] [Google Scholar]
- 29.Gu Y, Zhang W, Sun Q, Hao Y, Zilberberg J, Lee WY. Journal of Materials Chemistry B. 2015 doi: 10.1039/C5TB00421G. DOI: 10.1039/C5TB00421G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Q, Gu Y, Zhang W, Zilberberg J, Lee WY. Submitted.
- 31.Klein-Nulend J, Nijweide PJ, Burger EH. Current osteoporosis reports. 2003;1:5–10. doi: 10.1007/s11914-003-0002-y. [DOI] [PubMed] [Google Scholar]
- 32.Heino TJ, Hentunen TA, Väänänen HK. Journal of cellular biochemistry. 2002;85:185–97. doi: 10.1002/jcb.10109. [DOI] [PubMed] [Google Scholar]
- 33.Burge R, Dawson-Hughes B, Solomon DH, et al. Journal of Bone and Mineral Research. 2007;22:465–75. doi: 10.1359/jbmr.061113. [DOI] [PubMed] [Google Scholar]
- 34.Roodman GD. New England Journal of Medicine. 2004;350:1655–64. doi: 10.1056/NEJMra030831. [DOI] [PubMed] [Google Scholar]
- 35.Ferrara JL, Levine JE, Reddy P, Holler E. Lancet. 2009;373:1550–61. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung AD, Chao NJ. Clinical haematology. 2013;26:285–92. doi: 10.1016/j.beha.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sung AD, Chao NJ. Stem cells translational medicine. 2013;2:25–32. doi: 10.5966/sctm.2012-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris AC, Levine JE, Ferrara JL. Clinical haematology. 2012;25:473–478. doi: 10.1016/j.beha.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanash AM, Dudakov JA, Hua G, O’Connor MH, Young LF, Singer NV, West ML, Jenq RR, Holland AM, Kappel LW, Ghosh A, Tsai JJ, Rao UK, Yim NL, Smith OM, Velardi E, Hawryluk EB, Murphy GF, Liu C, Fouser LA, Kolesnick R, Blazar BR, van den Brink MRM. Immunity. 2013;37:339–50. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, Drachenberg CB, Hadley GA. The Journal of experimental medicine. 2005;201:1647–57. doi: 10.1084/jem.20041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marrack P, Kappler J. Annual review of immunology. 2004;22:765–87. doi: 10.1146/annurev.immunol.22.012703.104554. [DOI] [PubMed] [Google Scholar]
- 42.Wang JD, Douville NJ, Takayama S, ElSayed M. Annals of biomedical engineering. 2012;40:1862–73. doi: 10.1007/s10439-012-0562-z. [DOI] [PubMed] [Google Scholar]
- 43.Young EWK, Berthier E, Guckenberger DJ, Sackmann E, Lamers C, Meyvantsson I, Huttenocher A, Beebe DJ. Analytical Chemistry. 2011;83:1408–1417. doi: 10.1021/ac102897h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CS, Breslauer DN, Luna JI, Grimes A, Chin W, Leeb LP, Khine M. Lab on a chip. 2008;8:622–4. doi: 10.1039/b719029h. [DOI] [PubMed] [Google Scholar]
- 45.Chou SY, Krauss PR. Microelectronic Engineering. 1997;35:237–40. [Google Scholar]
- 46.Chou SY, Krauss PR, Renstrom PJ. Science. 1996;272:85–87. [Google Scholar]
- 47.Sung JH, Kam C, Shuler ML. Lab on a chip. 2010;10:446–55. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 48.Wells AD, Gudmundsdottir H, Turka LA. Journal of Clinical Investigation. 1997;100:3173–83. doi: 10.1172/JCI119873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, Ogbolu KR, Wang H. Journal of Experimental Nanoscience. 2008;3:329–45. [Google Scholar]
- 50.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, Haddad BR, Rhim JS, Dritschilo A, Riegel A, McBride A, Schlegel R. The American journal of pathology. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.