Abstract
Melanoma Differentiation-Associated gene 5 (MDA5), encoded by the gene IFIH1, is a cytoplasmic sensor for viral double-stranded RNAs (dsRNAs). MDA5 activates the type I interferon signaling pathway upon detection of long viral dsRNA generated during replication of picornaviruses. Studies have shown that MDA5 forms a filament along the length of dsRNA and utilizes ATP-dependent filament dynamics to discriminate between self vs non-self on the basis of dsRNA length. This review summarizes our current understanding of how the MDA5 filament assembles and disassembles, how this filament dynamics are utilized in dsRNA length-dependent signaling, and how dysregulated filament dynamics lead to pathogenesis of immune disorders.
Introduction
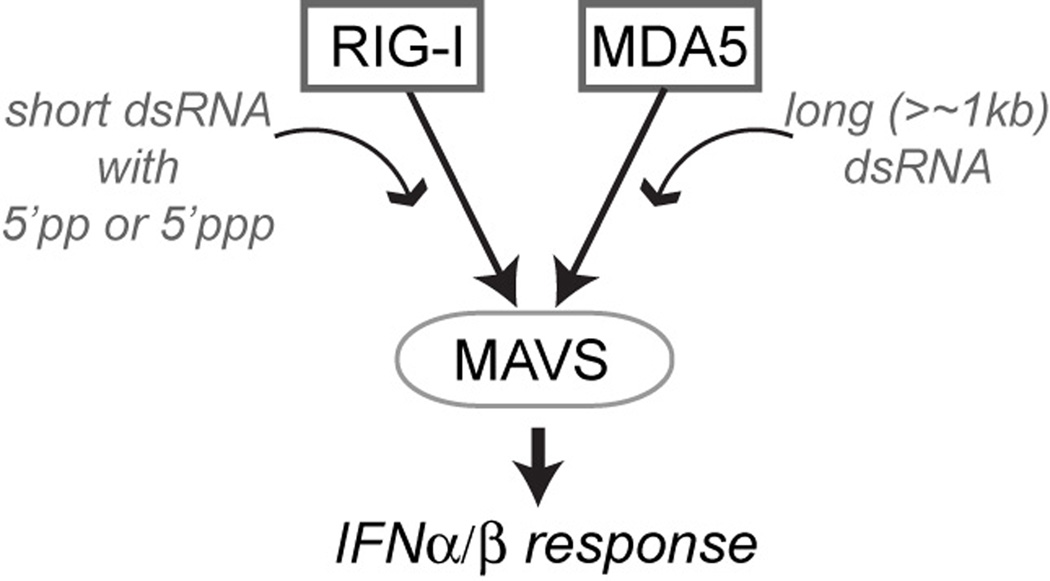
Melanoma Differentiation-Associated gene 5 (MDA5), encoded by the gene IFIH1, is a cytoplasmic sensor for viral double-stranded RNAs (dsRNAs). It is a paralog of Retinoic acid Inducible Gene-I (RIG-I), sharing the same domain architecture and activating the same interferon (IFN) signaling pathway through the common adaptor molecule, MAVS (Fig. 1). Earlier studies described MDA5 as a protein involved in tumor differentiation and apoptosis [1, 2]. In 2004, the Fujita and Randall groups independently showed that RIG-I and MDA5, respectively, are pattern recognition receptors that activate the type I IFN signaling pathway in response to viral dsRNA or its mimetic, polyinosinic-polycytidylic acid (polyIC) [3, 4]. Subsequent studies from multiple laboratories have demonstrated that RIG-I and MDA5 play non-redundant roles by recognizing largely distinct groups of viral RNAs [5–7]. While RIG-I recognizes relatively short dsRNA (< 1 kb) with 5’ppp or 5’pp often present in the genome of negative strand RNA viruses or their defective interfering particles [8–11], MDA5 was shown to detect long dsRNA replicative form of picornaviruses (Fig. 1) [12, 13]. The Akira group further showed that MDA5-mediated IFN signaling progressively increases with the length of dsRNA over ~1–7 kilobases (kb) [14]. This unusual length dependence has provided an explanation for how MDA5 selectively recognizes long viral dsRNA and discriminates against short cellular dsRNAs. However, it also raised the question of how MDA5 can measure the length of dsRNA that is orders of magnitude larger than the size of the protein itself. While some studies have suggested that MDA5 also detect other types of viral RNAs [15, 16], this review focuses on the molecular mechanism by which MDA5 selectively recognizes long vs. short viral dsRNA and the immune disorders caused by failure of self vs non-self discrimination.
Fig 1.
Schematic of viral RNA recognition and signal activation by RIG-I and MDA5. These receptors recognize distinct groups of viral RNAs but activate the IFNα/β signaling pathway through the common adaptor molecule, MAVS.
Cooperative Filament Assembly
Our initial investigation with human MDA5 showed that MDA5 binds to various types of nucleic acids, including ssRNA and dsDNA, with comparable affinities, but only with dsRNA it assembles into a filament along the dsRNA axis (Fig. 2) [17]. Filament formation was observed even in the presence of an excess amount of free dsRNA, clearly showing the cooperative nature of this assembly [17, 18]. No RNA sequence specificity has been observed so far for the filament assembly [17]. Similar filaments were observed with mouse MDA5 suggesting it is a conserved feature of MDA5 [19]. While the biological importance of the observed affinity for ssRNA/dsDNA is still unclear, the robust MDA5-stimulatory activity of dsRNA (not ssRNA or dsDNA) suggests that nucleic acid binding is insufficient, and filament formation plays an important role. In fact, filament formation is essential for dsRNA recognition by MDA5 as monomeric MDA5 binds to dsRNA with poor affinity (Fig. 2) [17]. Tight protein-protein interaction along the axis of dsRNA increases the affinity by >100 fold [17]. The instability of monomeric interaction with dsRNA explains the slow nucleation kinetics observed for the MDA5 filament [18].
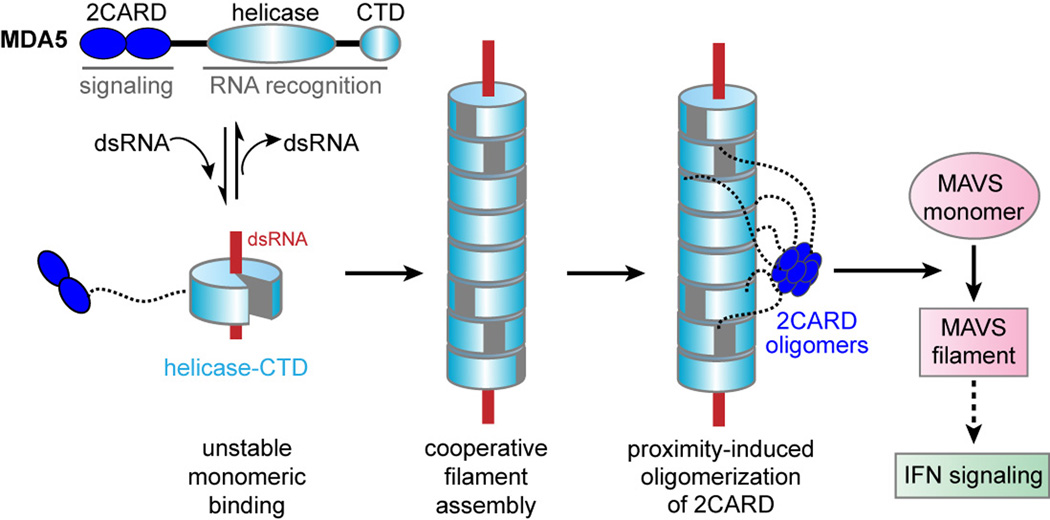
Fig 2.
Model of MDA5 filament formation and signal activation. MDA5 contains the 2CARD, helicase domain and CTD. Upon dsRNA binding, helicase-CTD forms a ring-like structure, which cooperatively stacks along dsRNA to assemble into a filament. Filament formation brings together neighboring 2CARDs into proximity and induces oligomerization of 2CARD alongside the filament. The 2CARD oligomer then triggers monomer-to-filament transition of MAVS and activates the downstream IFN signaling pathway.
MDA5 consists of the N-terminal signaling domain, tandem caspase activation recruitment domain (2CARD), the central DExD/H motif helicase domain and the C-terminal domain (CTD) (Fig. 2). Our crystal structure of 2CARD-truncation mutant in complex with 12 bp dsRNA showed that the helicase domain and CTD together function as a RNA recognition unit, forming a ring-like structure around dsRNA (Fig. 2) [20]. This ring architecture of MDA5 is similar to that of RIG-I [21–23], but differs in the orientation of CTD. In RIG-I, the CTD is tilted towards the dsRNA end, allowing CTD to sense the 5’ppp and blunt end of dsRNA. By contrast, CTD of MDA5 is parallel to the dsRNA axis, allowing dsRNA to pass through the ring structure without any steric clash. Thus, the differential orientation of CTD allows RIG-I and MDA5 to adopt the dsRNA end-capping and stem-binding modes, respectively, which in turn leads to divergent viral RNA recognition by these receptors.
Filament structure, as determined by protein-protein crosslinking [20] and electron microscopy (EM) helical reconstruction [24], showed that monomer is stacked head-to-tail with ~70° turn per monomer. There is an extensive monomer:monomer contact along the axis of the filament, explaining how filament formation compensates for the weak monomeric interaction with dsRNA. Mutations in the filament interface impair filament formation in vitro and IFN signaling in cells, further supporting the role of filament formation for dsRNA recognition and signal activation by MDA5.
How does the MDA5 filament activate MAVS and the downstream signaling pathway? Investigation of isolated 2CARD of MDA5 revealed that it oligomerizes in a concentration-dependent manner (> 10 µM), and that this oligomeric 2CARD induces the monomer-to-filament transition of MAVS CARD [20], a pre-requisite for the downstream IFN signal activation [25]. Since 2CARD within the full-length MDA5 filament would be at sufficiently high local concentrations, this observation provides the plausible model that filament assembly by the RNA-binding domain (helicase-CTD) induces proximity and subsequent oligomerization of 2CARD (Fig. 2). In fact, the linker (~100 residues) between 2CARD and the helicase domain is long enough for oligomerization of ~10–11 neighboring 2CARDs [20]. Unanchored K63-linked polyubiquitin chains (K63-Ubn) were also proposed to assist 2CARD oligomerization of MDA5 (as well as RIG-I) [26]. Future investigation is required to dissect the importance of K63-Ubn in the context of full-length MDA5 filament and the potential synergism between proximity- and K63-Ubn-mediated mechanisms.
Is the filament formation sufficient to explain the dsRNA length discrimination observed over the range of ~1–7 kb? This seems not to be the case. While dsRNA affinity of MDA5 generally increases with the length of dsRNA, this increase is most significant under ~100–500 bp and little increase was observed above this range [18]. In addition, 10–11 MDA5 molecules (which occupy ~150 bp) appear sufficient to form the signaling-competent 2CARD oligomer, raising the question of how MDA5 discriminates between, for example, 10 molecules of 150 bp dsRNA and 1 molecule of 1.5 kb dsRNA with equal number of MDA5 binding sites? Answers to these questions lie in the ATP-dependent dynamic instability of the MDA5 filament, as discussed below.
Dynamic Instability & dsRNA Length Discrimination
Analysis of the MDA5 filament stability with and without ATP revealed that ATP markedly increases the dsRNA length dependence above ~500 bp, to the extent that closely recapitulates the length dependence observed in its cellular signaling activity [18]. How does ATP increase the sensitivity of the MDA5 filament to dsRNA length? Based on a series of bulk and single-molecule kinetic analyses, we found that there are two properties of the MDA5 filament responsible for this enhanced length discrimination in the presence of ATP; ATP hydrolysis-driven end disassembly and rate-limiting nucleation kinetics [18].
While the MDA5 filament is stable in the absence of ATP, in the presence of ATP, it undergoes rapid cycles of assembly and disassembly (Fig. 3A) [17–19]. This is because ATP hydrolysis weakens the MDA5-dsRNA interaction, and thus accelerates filament disassembly. This dynamic instability of the filament is reminiscent of those of microtubule, actin filament [27] and RecA filament [28]. More detailed examination revealed that ATP hydrolysis occurs throughout the MDA5 filament, seemingly independent of the relative position within the filament, but dissociation from dsRNA occurs primarily at dsRNA ends as interior molecules are stabilized by the interaction with the adjacent molecules [18]. Since higher fraction of MDA5 molecules would be placed at filament termini on short dsRNA, this end-disassembly mechanism results in more rapid filament disassembly on short dsRNAs.
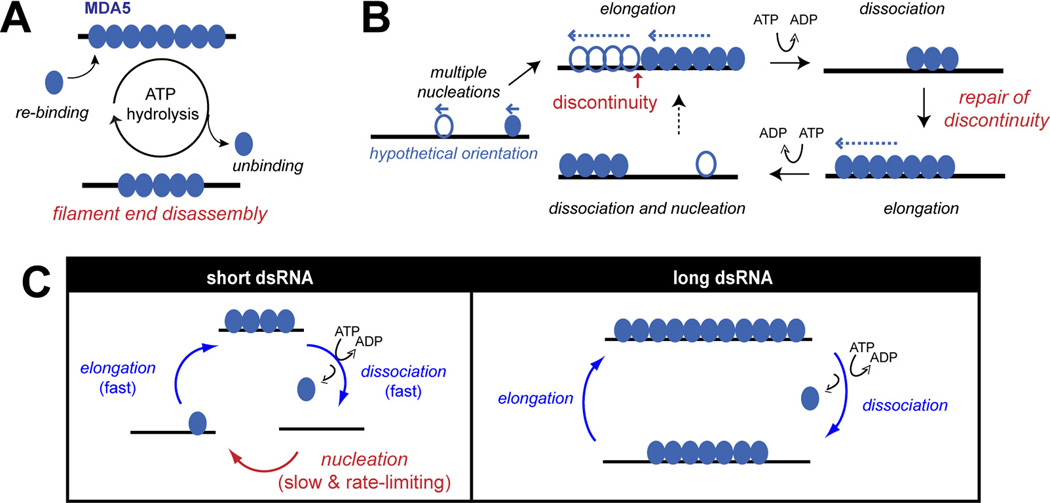
Fig 3.
Kinetic model for dsRNA length discrimination by the MDA5 filament. (A) ATP hydrolysis promotes end-disassembly of the MDA5 filament, which results in dsRNA length-dependent stability. (B) Multi-nucleated filament repairs gaps (or discontinuity) during ATP-mediated filament rearrangement. During dynamic equilibrium between assembly and ATP-driven end disassembly reactions, short filaments disassemble faster than longer ones, allowing continued elongation of longer filaments to more accurately reflect the length of underlying dsRNA scaffolds. (C) The role of slow nucleation kinetics in dsRNA length discrimination by MDA5. On short dsRNA, RNA binding occurs primarily by rate-limiting nucleation, whereas on long dsRNA, it is mediated by rapid filament elongation. Thus, slow nucleation selectively suppresses MDA5 binding to short dsRNA.
What happens if multiple filaments nucleate on the same dsRNA molecule? The single molecule analysis showed that multiple nucleation indeed occurs and is often required to coat the entire length of dsRNA (due to interior nucleation and unidirectional propagation of the filament) (Fig. 3B) [18]. Multiple filaments often co-exist on the same RNA molecule even when no gaps are visible by EM [18]. ATP-driven filament disassembly occurs at the ends of these individually nucleated filaments (rather than dsRNA ends), resulting in apparent interior breaking of the filament [18, 19]. Interestingly, when filament is formed in the presence of ATP, repeated cycles of assembly and disassembly repair gaps in the filament and promote formation of longer and more continuous filaments (Fig. 3B) [18]. Thus, MDA5 utilizes ATP hydrolysis to promote filament end-disassembly while repairing gaps on long dsRNA, which together contribute to the dsRNA length dependent stability of the filament.
We also found that nucleation of the MDA5 filament is slow and rate-limiting relative to filament elongation, and this further suppresses stable accumulation of MDA5 molecules on short dsRNA [18]. This is because MDA5 binds short dsRNA predominantly through de novo nucleation rather than filament elongation. Additionally, filament disassembly often occurs to completion on short dsRNA, which makes re-binding to again require de novo nucleation (Fig. 3C). On long dsRNA, on the other hand, MDA5 filaments cycle between partial disassembly and elongation, bypassing nucleation and allowing stable accumulation of MDA5 molecules on RNA (Fig. 3C).
Thus, the unique combination of the filament assembly and disassembly kinetics enables MDA5 to regulate the stability of the filament according to the length of dsRNA. Since 2CARD oligomerization requires MDA5 filament formation, this kinetic mechanism of the filament provides an explanation for the dsRNA length-dependent signaling activity of MDA5. Note that RIG-I also forms filaments [29, 30], but utilize distinct assembly and disassembly mechanisms, and thus adopt different dsRNA length specificity.
Dysregulation and Immune Disorders
Multiple Genome Wide Association (GWA) studies have shown that MDA5 is not only involved in antiviral immunity [31], but also in pathogenesis of several immune disorders, such as type 1 diabetes [32, 33], systemic lupus erythematosus [34, 35], multiple sclerosis [36] and rheumatoid arthritis [37]. Single nucleotide polymorphisms (SNPs) in the MDA5 gene (IFIH1), such as T946A, R843H, E627X and I923V, were shown to be complete or partial loss-of-function mutations and to be associated with protection from these diseases [17, 31, 38, 39]. Although linkage disequilibrium between MDA5 and other genes in the disease-associated region complicates pinpointing the specific causal variant [40], the importance of MDA5 in type I IFN signaling pathway and the known role of IFN in these diseases have raised the intriguing possibility that hyper-activation of MDA5 contributes to pathogenesis of these immune disorders.
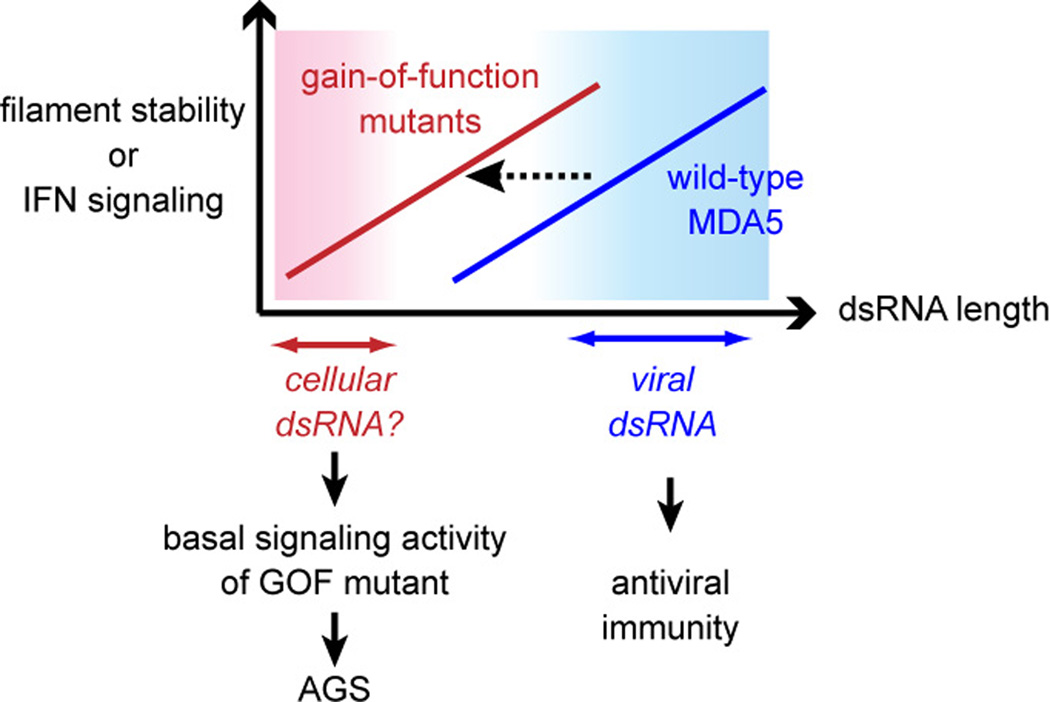
More clear demonstration of the causal role of MDA5 in inflammatory and autoimmune diseases came from studies involving mouse models and Mendelian genetic studies of monogenic inflammatory disease, Aicardi-Goutieres Syndrome (AGS). Transgenic mice bearing multiple copies of the MDA5 gene displayed systemic up-regulation of type I IFN [41]. While high basal expression of IFN was insufficient to cause overt inflammatory and autoimmune phenotypes in these mouse models, it exacerbated the autoimmune pathology in mice with the lupus-susceptible genetic background [41]. More recently, chemically induced mutation (G821S) in the MDA5 gene was shown to cause lupus-like nephritis with wide spread systemic inflammation [42]. Biochemical examination of G821S suggested that this mutation renders MDA5 unresponsive to dsRNA, but constitutively activates its signaling activity [42]. Finally, genetic analysis of AGS revealed that several gain-of-function mutations in MDA5 can cause a spectrum of neuro-inflammatory phenotypes with a type I IFN signature [43, 44]. These mutations over-stabilize the MDA5 filament in vitro (either by impairing ATP hydrolysis or by increasing dsRNA affinity), sensitize MDA5 to short dsRNAs (~150 bp), and increase the basal signaling activity in the absence of viral infection (Fig. 4) [43]. Additional analyses showed that this basal signaling activity is dependent on the mutants’ ability to bind dsRNA and assemble into filaments [43], suggesting the presence of endogenous dsRNAs stimulating these mutants (Fig. 4). Future research is required to determine the identity of the endogenous ligands for MDA5 and the mechanistic explanation for the brain specificity of the AGS phenotype.
Fig 4.
Model of how the gain-of-function (GOF) mutations in MDA5 cause Aicardi-Goutieres Syndrome (AGS). These mis-sense mutations over-stabilize the MDA5 filament, resulting in increased sensitivity to short dsRNAs and aberrant activation by as-yet-unidentified cellular RNAs.
Conclusion & Future Direction
We here summarize our current understanding of how the MDA5 filament assembles and disassembles, how its kinetic properties regulate self vs. non-self discrimination on the basis of dsRNA length, and how its dysregulation results in inflammatory and autoimmune pathology. Current data indicate at least three functional roles of MDA5 filament formation: (i) it enables tight binding of MDA5 to dsRNA to compensate for the low affinity interaction between monomeric protein and dsRNA, (ii) it allows MDA5 to regulate its interaction with dsRNA according to the length of dsRNA, and (iii) it brings together neighboring 2CARDs into proximity to induce 2CARD oligomerization and MAVS stimulation.
Several important issues remain to be investigated. First, ATP hydrolysis appears to play multiple roles in MDA5 function, in addition to promoting filament disassembly as described in this review. While some ATP hydrolysis-deficient mutants display gain-of-function properties (as expected from stabilization of the filament), others show loss-of-function phenotype [45], indicating that ATP hydrolysis activity per se does not determine the functional outcome. Specific conformation stabilized by each of these mutations may bear clues as to how these mutation have divergent consequence in IFN signaling. Second, the fate of MDA5 filament during the course of infection and IFN response is still unclear. Reported challenges in isolating the MDA5:dsRNA complex or the filament from cells suggest the dynamic, and potentially transient, nature of the filament. Some studies have suggested localization of MDA5 to stress granules during viral infection [46, 47], although the temporal order between this relocalization event and viral RNA recognition is still unclear. Direct visualization of MDA5 filaments in cells would provide additional insights. Finally, very few cellular proteins have so far been demonstrated to specifically interact with the MDA5 filament and regulate its function. One example is LGP2, a homologous helicase lacking 2CARD, which promotes MDA5 filament nucleation and stabilizes short filaments, thereby enhancing MDA5-mediated IFN induction [48]. Future studies aimed at identifying other such regulatory factors would reveal an additional layer of complexity in MDA5 function, and would be likely important for therapeutic targeting of MDA5 for both antiviral immunity and treatment of immune disorders.
Highlights.
-
▪
MDA5 cooperatively assembles into a filament along dsRNA.
-
▪
MDA5 filament undergoes end-disassembly during ATP hydrolysis.
-
▪
MDA5 filament dynamics regulate its stability according to dsRNA length.
-
▪
Mutations that over-stabilize the MDA5 filament cause inflammatory diseases.
Acknowledgements
YdTD was supported by fellowships from Novartis Foundation and Swiss National Science Foundation. BW was supported by Charles A. King Trust Fellowship. SH was a Pew Scholar and was funded by NIH R01 grant (AI106912 and AI111784).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kovacsovics M, et al. Overexpression of Helicard, a CARD Containing Helicase Cleaved during Apoptosis, Accelerates DNA Degradation. Curr. Biol. 2002;12:834–843. doi: 10.1016/s0960-9822(02)00842-4. [DOI] [PubMed] [Google Scholar]
- 2.Kang DC, et al. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Aad. Sci. USA. 2002;99(2):637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 4.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-promoter. Proc Natl Acad Sci U S A. 2004;101(49):17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 6.Loo MY, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82(1):335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103(22):8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hornung V, et al. 5'-Triphosphate RNA Is the Ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 9.Pichlmair A, et al. RIG-I–Mediated Antiviral Responses to Single-Stranded RNA Bearing 5'-Phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 10.Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5'-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;108:3092. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Q, et al. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Reports. 2012;29:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Triantafilou K, et al. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125(Pt 20):4761–4769. doi: 10.1242/jcs.103887.. * Refs. (12–13) suggest that MDA5 recognizes the replicative form of picornaviruses.
- 14. Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091.. ** This work provides the first evidence suggesting that MDA5 discriminates self vs. non-self RNAs on the basis of dsRNA length. They also report distinct dsRNA length-specificity of RIG-I.
- 15.Runge S, et al. In Vivo Ligands of MDA5 and RIG-I in Measles Virus-Infected Cells. PLoS Pathog. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deddouche S, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peisley A, et al. Cooperative Assembly and Dynamic Disassembly of MDA5 Filaments for Viral dsRNA Recognition. Proc Natl Acad Sci U S A. 2011;108(52):21010–21015. doi: 10.1073/pnas.1113651108.. ** This paper provides the first description of the MDA5 filament, its ATP-driven disassembly and the role of filament assembly in dsRNA length discrimination.
- 18. Peisley A, et al. Kinetic Mechanism for Viral dsRNA Length Discrimination by MDA5 Filament. Proc Natl Acad Sci U S A. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109.. ** This study reports the importance of filament end-disassembly and slow nucleation kinetics for dsRNA length-dependent stability of the MDA5 filament.
- 19. Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;7:1714–1726. doi: 10.1038/emboj.2012.19.. * This paper provides an independent finding of the MDA5 filament and its dynamic instability along with Ref. (17).
- 20. Wu B, et al. Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048.. ** This paper describes the crystal structure of MDA5 in complex with dsRNA and the molecular mechanism for divergent dsRNA recognition by MDA5 and RIG-I. They also provide the evidence supporting filament-induced (i.e. proximity-induced) oligomerization of MDA5 2CARD.
- 21.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 22.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479(7373):423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berke IC, et al. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci U S A. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:1–14. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X, et al. Ubiquitin-Induced Oligomerization of the RNA Sensors RIG-I and MDA5 Activates Antiviral Innate Immune Response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kueh HY, Michison TJ. Structural Plasticity in Actin and Tubulin Polymer Dynamics. Science. 2009;325:960–963. doi: 10.1126/science.1168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox MM. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 29.Peisley A, et al. RIG-I Forms Signaling-Competent Filaments in an ATP-Dependent, Ubiquitin-Independent Manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Patel JR, et al. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann F, et al. Polymorphisms in MDA-5 link protein function to clearance of hepatitis C virus. Hepatology. 2014 doi: 10.1002/hep.27344. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nejentsev S, et al. Rare Variants of IFIH1, a Gene Implicated in Antiviral Responses, Protect Against Type 1 Diabetes. Science. 2009;324(5925):387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smyth DJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800.. ** This is the first GWA study linking MDA5 and immune disorders. They report the risk-allele A946T, a common SNP in the MDA5 gene.
- 34.Molineros JE, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS Genetics. 2013;9:e1003222. doi: 10.1371/journal.pgen.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson T, et al. Autoimmune Disease Risk Variant of IFIH1 Is Associated with Increased Sensitivity to IFN-{alpha} and Serologic Autoimmunity in Lupus Patients. J. Immunol. 2011;187:1298–1303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez A, et al. IFIH1-GCA-KCNH7 locus: influence on multiple sclerosis risk. Eur J Hum Genet. 2008;16:861–864. doi: 10.1038/ejhg.2008.16. [DOI] [PubMed] [Google Scholar]
- 37.Cen H, et al. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity. 2013;46:455–462. doi: 10.3109/08916934.2013.796937. [DOI] [PubMed] [Google Scholar]
- 38.Chistiakov DA, et al. Loss-of-function mutations E627X and I923V of IFIH1 are associated with lower poly(I:C)–induced interferon-a production in peripheral blood mononuclear cells of type 1 diabetes patients. Hum. Immunol. 2010;71(11):1128–1134. doi: 10.1016/j.humimm.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Shigemoto T, et al. Identification of Loss of Function Mutations in Human Genes Encoding RIG-I and MDA5. J. Biol. Chem. 2009;284(20):13348–13354. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd JA. Statistical false positive or true disease pathway? Nat Genet. 2006;38:731–733. doi: 10.1038/ng0706-731. [DOI] [PubMed] [Google Scholar]
- 41.Crampton SP, et al. Ifih1 gene dose effect reveals MDA5-mediated chronic type I IFN gene signature, viral resistance, and accelerated autoimmunity. J. Immunol. 2012;188:1451–1459. doi: 10.4049/jimmunol.1102705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Funabiki M, et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014.. ** This paper reports an analysis of a mouse model with a gain-of-function mutation in MDA5 that causes systemic inflammation and autoimmune phenotypes.
- 43. Rice GI, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet. 2014;46(5):503–509. doi: 10.1038/ng.2933.. ** This study reports six distinct gain-of-function mutations in MDA5 that cause the neuro-inflamamtory disease (such as Aicardi Goutières Syndrome) in human.
- 44.Oda H, et al. Aicardi-Goutières syndrome is caused by IFIH1 mutations. Am J Hum Genet. 2014;95:121–125. doi: 10.1016/j.ajhg.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bamming D, Horvath CM. Regulation of Signal Transduction by Enzymatically Inactive Antiviral RNA Helicase Proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 2009;284(15):9700–9712. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langereis MA, Feng Q, van Kuppeveld FJ. MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon. J. Virol. 2013;87:6314–6325. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onomoto K, et al. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One. 2013;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bruns AM, et al. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell. 2014;55:771–781. doi: 10.1016/j.molcel.2014.07.003.. * This paper provides the mechanism of how LGP2 regulates the MDA5 filament assembly and disassembly processes and how this leads to up-regulation of MDA5 signaling.