Abstract
Woodchuck hepatitis virus (WHV) is often used as surrogate to study mechanism of HBV infection. Currently, most infections are conducted using strains WHV7 or WHV8 that have very high sequence identity. This study focused on natural strain WHVNY that is more genetically distant from WHV7. Three naive adult woodchucks inoculated with WHVNY developed productive acute infection with long lasting viremia. However, only one of two woodchucks infected with WHV7 at the same multiplicity demonstrated productive liver infection. Quantification of intracellular WHV RNA and DNA replication intermediates; percentages of core antigen-positive hepatocytes; and serum relaxed circular DNA, showed that strains WHVNY and WHV7 displayed comparable replication levels and capacities to induce acute infection in naive adult woodchucks. Strain WHVNY was therefore validated as valuable reagent to analyze the mechanism of hepadnavirus infection, especially in co- and super-infection settings, which required discrimination between two related virus genomes replicating in the same liver.
Keywords: Hepadnavirus, woodchuck hepatitis virus strain, WHVNY, WHV7, acute WHV infection, replication markers
1. INTRODUCTION
Human hepatitis B virus (HBV) is a prototype member of Hepadnaviridae family, and it belongs to a subfamily of orthohepadnaviruses. It is an enveloped double-stranded DNA virus that infects hepatocytes (Schaefer, 2007; Seeger and Mason, 2000). HBV remains a significant health risk. Approximately 400 million individuals around the world are chronically infected with HBV. Chronic HBV infection is a number one risk factor for development of hepatocellular carcinoma (HCC). More than fifty percent of all HCC cases are associated with chronic HBV infection (Akbar et al., 2006; Di Bisceglie, 2009; Dienstag, 2008; Lupberger and Hildt, 2007; McMahon, 2004; Nguyen et al., 2009; Seeger and Mason, 2000). There is no cure for HBV, and current anti-HBV drugs provide only a temporary relief. The number of HBV-related targets used for antiviral interventions is very limited. Currently available anti-HBV drugs are (i) the nucleoside/nucleotide analogs (entecavir, lamivudine, adefovir dipivoxil, tenofovir and telbivudine) that target only the reverse transcription; and (ii) versions of interferon alpha (including alpha-2b interferon and pegylated alpha-2a interferon) that are beneficial only to a subset of infected individuals (Asselah et al., 2007; Lam et al., 2011; Lok et al., 2007; Papatheodoridis and Hadziyannis, 2004; Papatheodoridis et al., 2012; Qiu et al., 2013). For comparison, unlike HBV, anti-HIV therapies work against four different kinds of virus-specific targets. These include viral entry, reverse transcription, integrase, and protease (Laskey and Siliciano, 2014). Clearly, search for new HBV-specific therapeutic targets for novel antiviral interventions remains a priority. The understanding of mechanism and determinants of the maintenance of chronic HBV infection is very important for identification of novel anti-HBV targets and strategies. The tools and reagents that facilitate further understanding of the mechanism of chronic hepadnavirus infection are therefore welcomed. Among such helpful tools are diverse natural variants of hepadnaviruses. Woodchuck hepatitis virus (WHV) is another member of Hepadnaviridae family. Like HBV, it belongs to a subfamily of orthohepadnaviruses. In nature, WHV is found in woodchucks (Marmota monax) (Schaefer, 2007; Seeger and Mason, 2000). In vivo infection of woodchuck livers caused by HBV-related WHV is a an invaluable surrogate model to study mechanism of HBV infection (Cote et al., 2000; Glebe et al., 2009; Kew et al., 1993; Menne and Cote, 2007). In the current study, we examined a natural WHV strain, WHVNY (Kew et al., 1993), in terms of its ability to induce productive acute infection in naive adult woodchucks. Currently, in US, majority of laboratory WHV infections are conducted using well-known strain WHV7, or less frequently - another strain, WHV8, which has a very high degree of sequence identity to WHV7 (Cote et al., 2000; Glebe et al., 2009; Kew et al., 1993; Menne and Cote, 2007). In fact, the genomes of WHV7 and WHV8 differ only in 14 nucleotides, which equals to 0.42% of sequence diversity. As we reported recently, the degree of sequence diversity (119 nucleotides including 15 nts deletion, which is 119/3323=3.58% of sequence difference) and unique nucleotide polymorphisms of WHVNY (as compared to WHV7) were sufficient for development of the sensitive WHV-strain-specific assays that were able to discriminate between WHV7 and WHVNY in the complex mixtures containing the sequences of both strains. The development and optimization of the WHV strain-specific assays made feasible the use of WHVNY along with WHV7 in super-infection experiments and investigate, whether the cell-to-cell spread of hepadnavirus and super-infection can continue to occur during chronic state of hepadnavirus infection, and therefore, virus spread/super-infection may potentially represent determinants of the maintenance of chronic infection, which can be possibly targeted by antivirals (Rodrigues et al., 2015). The above experiments, outcomes of which suggested that a limited cell-to-cell spread of hepadnavirus continues during chronic infection (Rodrigues et al., 2015), are instrumental in efforts to resolve a long standing argument in HBV research field, which suggests that during chronic hepadnavirus infection (this includes WHV), virus spread and super-infection are unlikely events, and the chronic state of infection can be maintained solely by division of hepadnavirus-infected hepatocytes in the absence of the spread/super-infection (Litwin et al., 2005; Mason et al., 2007; Mason et al., 2008; Mason et al., 2009b; Mason et al., 2010; Walters et al., 2004; Xu et al., 2007; Zhou et al., 1999; Zoulim and Locarnini, 2009). Although, WHVNY was described in 1993, its replication and infection properties were not characterized in detail (Kew et al., 1993). Current study represents the first report, in which replication parameters and potentials to induce acute infection of strains WHVNY and WHV7 were compared using in vivo woodchuck model.
2. MATERIALS AND METHODS
2.1. Woodchucks and infections
Three WHV-negative (naive) adult woodchucks, F6678, M6543 and F6541 were infected with strain WHVNY (Kew et al., 1993), while two other naive adult woodchucks, F6693 and F6671 were inoculated with strain WHV7 (Cote et al., 2000). The multiplicity of infection (MOI) was 1.66×108 WHV genome equivalents (GE)/animal. For WHVNY inoculations, the serum of woodchuck 2761 (Kew et al., 1993) was used, while the serum from woodchuck 3922 was used for WHV7 infections. Infected animals were monitored for 14 weeks post-infection. Serum samples were collected on the day of infection (week 0), and each week thereafter. Liver biopsies were obtained immediately prior to infection (week 0), and at weeks +6 and +13 post-infection. Liver tissues were also collected during necropsy at week +14.
2.2. Quantification of WHV rcDNA in serum samples
In this study, the assays that measured rcDNA and other WHV replication intermediates were not strain-specific, and were based on sequences of WHV7 and WHVNY that were either identical or had sufficiently high degree of identity (Rodrigues et al., 2015). For WHV rcDNA isolation, each aliquot of 50 μl of serum was mixed with 450 μl of lysis buffer (0.01 M Tris-HCl, pH 8.0, 0.01 M EDTA, 0.1 M NaCl, 0.2% SDS) containing 1 mg/ml of Pronase (Roche). After two hours of incubation at 37°C, DNA was first extracted with an equal volume of phenol and then with a mixture of 24:1 chloroform/isoamyl alcohol. Finally, DNA was precipitated by adding 2 μl of dextran (MP Biomedicals) solution (10μg/μl solution in DNase-/RNase-free water), 0.1 volume of 3.0 M sodium acetate, pH 5.5 and 2 volumes of 100% ethanol. For quantification of rcDNA copy numbers by qPCR, primers #23 (2504-AGAAGACGCACTCCCTCTCCT-2524 (all positions of the primers/probes are based on WHV7 genome sequence (Rodrigues et al., 2015)), #24 (2579-TGGCAGATGGAGATTGAGAGC-2559) and TaqMan probe #28 (2531-/6-FAM/AGAAGATCTCAATCACCGCGTCGCAG/3BHQ_1/−2556) were used. The 6-FAM indicates the 6-carboxyfluorescein located at 5′-end of the probe, while BHQ_1 reflects a Black Hole Quencher 1 placed at 3′-end of the probe. The quantification was done using a 10-fold dilution series of plasmid pUC-CMVWHV linearized with NheI (the standard curve was in a range from 20 to 2.0×107 GE of WHV). The qPCR assay was performed using the TaqMan Gene Expression Mastermix, and 7500 RT-PCR System (Applied Biosystems) as described earlier (Freitas et al., 2012).
2.3. Immunohistochemistry
Liver sections were stained for WHcAg as previously described (Freitas et al., 2012). For quantifications of the average percentages of WHV core-positive cells, five to seven images per tissue section were analyzed using 40x magnification. The core-positive cells were identified manually and quantified using TMARKER software (Schüffler et al., 2013).
2.4. Measurements of the surface antigens of WHV (WHsAg) and anti-WHsAg antibodies
WHsAg and anti-WHsAg antibodies in serum samples were assayed at a 1:100 dilution and compared to woodchuck sera with known concentration of WHsAg or anti-WHsAg antibodies, respectively, by using quantitative ELISA (Cote et al., 1993; Liu et al., 2011). The cut-off values for detection of WHsAg and anti-WHsAg antibodies were 50 ng/ml, and 100 standard units/ml (U/ml), respectively.
2.5. Determination of the fraction of serum WHV bound to natural anti-WHsAg antibodies
Aliquots of serum were incubated with 100 μl of Pansorbin suspension (Calbiochem) in 1ml of Williams’ Medium E for 3 hours at 4°C on a rocking platform. Pansorbin-associated immune complexes of WHV bound to anti-WHsAg antibodies were then precipitated by centrifugation at 13,000 rpm for 1 min, washed four times with ice-cold phosphate buffer saline (PBS) containing 0.5% Nonidet P-40 (Fisher) and used for DNA isolation and quantification of WHV rcDNA by qPCR (Freitas et al., 2012). For woodchucks F6693, F6678, F6541 and M6543, the percentages of serum WHV bound to anti-WHsAg antibodies were measured for weeks +6, +8, +10, +12 and +14. For animal F6671, the measurements were done for weeks +3, +4, +5 and +6. As a negative control, we used a preparation of HBV-enveloped human hepatitis delta virus (HDV) that was produced in Huh7 cells and therefore, was free of virus particles bound to antibodies against HBV envelope proteins.
2.6. Quantification of RI-DNA and cccDNA of WHV
Liver tissues obtained either during biopsies or at necropsy were homogenized in the volume of 0.6 to 1.5 ml of TE buffer (10:10, i.e., 10 mM Tris-HCl pH 7.6, 10 mM EDTA) (Rodrigues et al., 2015) using glass grinders. To assay for RI-DNA (Mason et al, 2009a), total DNA was isolated from the obtained homogenate. Briefly, 210 μl of the obtained homogenate was mixed with 390 μl of TE buffer (10:10) and 600 μl of buffer containing 0.01 M Tris-HCl, pH 7.6, 0.01 M EDTA, 0.2% SDS and 4 mg/ml Pronase. After 2 hours of incubation at 37°C, total DNA was extracted twice with an equal volume of phenol/chloroform and precipitated by adding 0.1 volumes of 3.0 M sodium acetate, pH 5.5 and 2 volumes of 100% ethanol. After centrifugation (13,000 rpm for 30 min at 4°C), the pellets were washed twice with 100% ethanol and re-suspended in 25–50 μl DNase-/RNase-free water. Quantification of RI-DNA copy numbers was performed using qPCR as described above for measurements of serum rcDNA. For cccDNA isolation, 210 μl of the above mentioned homogenate was mixed with 930 μl of TE buffer (10:10) and 60 μl of 10% SDS. Samples were vortexed and 300 μl of 2.5 M KCl was then added. After that, samples were mixed by vortexing again, incubated for at least 30 min at room temperature, and then clarified by centrifugation at 13,000 rpm for 30 min at 4°C. The supernatants were collected and subjected to phenol extraction followed by phenol/chloroform extraction. The recovered aqueous phase was precipitated by adding two volumes of 100% ethanol and 2 μl of 10 μg/μl solution of dextran (MP Biomedicals) in DNase-/RNase-free water. After an overnight incubation at room temperature samples were centrifuged and DNA pellets were washed twice with 70% ethanol and once with 100% ethanol, and then re-suspended in 50 μl of DNase-/RNase-free water. 40 μl of each sample was then incubated overnight at 37°C with 150 U of Plasmid-Safe ATP-Dependent DNase (Epicentre) in a final volume of 500 μl containing 1x NEB 4 buffer (New England Biolabs (NEB)), 1 mM ATP, 40 μg/ml of RNase A and 50 U/ml of HpaI (NEB). HpaI does not cut WHV DNA, and was used to cut host DNA and thus facilitate its digestion with Plasmid-Safe ATP-Dependent DNase. After the incubation, cccDNA was further purified by extraction with phenol, phenol/chloroform and ethanol precipitation (Rodrigues et al., 2015). The cccDNA levels were quantified by qPCR using primers #290 (1701-GGTCCGTGTTGCTTGGTCT-1719), #291 (1977-GGACATGGAACACAGGCAAAAACA-1954) and TaqMan probe #292 (1846-/6-FAM/AATGGGAGGAGGGCAGCATTGATCCT/3BHQ_1/−1871) (Freitas et al., 2012). The copy numbers were measured using a 10-fold dilution series of plasmid pUC-CMVWHV linearized with NheI (range from 20 to 2.0×107 GE of WHV). Total DNA concentration in total DNA preparations was measured using Hoechst 33258 dye and used to obtain normalized RI-DNA and cccDNA values in copy numbers/per μg of total DNA (Rodrigues et al., 2015).
2.7. Quantification of pgRNA of WHV
Total RNA from liver tissue samples was isolated using TRI reagent (Molecular Research Center). For animals F6678, F6541, M6543 and F6693, 200 ng of isolated total RNA were treated with Turbo DNase (Life Technologies). The synthesis of WHV cDNA was conducted using 16.6 ng of DNase-treated RNA (dtRNA). For woodchuck F6671, 8 μg of total RNA were treated with Turbo DNase and then re-extracted with TRI Reagent. The synthesis of WHV cDNA was performed using 2 μg of dtRNA. The reverse transcription reactions were done using High Capacity cDNA Reverse Transcription Kit (Life Technologies) using primer #850 (2602-TGTACCCATTGAAGATCAGCAGTT-2579). For the subsequent qPCR, forward primer #23, reverse primer #24 and TaqMan probe #28 were used as it was done for rcDNA measurements. The quantification was done using a 10-fold dilution series of in vitro transcribed and gel-purified RNA standard (in a range from 20 to 2.0×106 GE of WHV RNA). The WHV RNA that served as a standard was transcribed using as a template the plasmid pJSWHV7-2C6 (that encodes the RNA of WHV7) linearized with XhoI (Rodrigues et al., 2015). The pgRNA copy numbers were expressed per μg of total RNA.
3. RESULTS
3.1. Kinetics of WHV relaxed circular DNA (rcDNA) accumulation in sera of infected woodchucks
The patterns of acute WHV infection induced by two different strains of WHV were compared using in vivo infection of WHV-negative (naive) adult woodchucks. Strain WHV7 is currently used for the majority of experimental infections in laboratories within US. Natural strain WHVNY was described in 1993, but it was not yet extensively characterized and compared to WHV7 (Cote et al., 2000; Kew et al., 1993). Three naive adult woodchucks, F6678, M6543 and F6541, were inoculated with the strain WHVNY. Two other naive adult woodchucks, F6671 and F6693, were infected with WHV7 (Cote et al., 2000). Each animal received the dose of 1.66×108 GE of WHV. Both kinds of inoculum were analyzed for the percentage of WHV virions bound to the antibodies that were able to recognize the surface antigens of WHV (anti-WHsAg antibodies). For WHV7 inoculum, the fraction of the virions bound to anti-WHsAg antibodies was 62.6%. The percentage of antibody-bound virions in WHVNY inoculum was 37.9%.
Serum samples collected from five above mentioned woodchucks immediately before infection (week 0), and every week during 14 weeks post-infection (weeks +1 through week +14), were first analyzed for virion-associated rcDNA of WHV. The results are summarized in Fig. 1. As expected, WHV rcDNA was not detected in the samples collected prior to infection (not shown). For all five animals, it was possible to quantify serum titers of WHV starting the week +1. Out of two woodchucks infected with WHV7, only animal F6693 developed a productive acute infection, in which the titers exceeded the value of 1.0×109 GE/ml by week +5, reached the maximum levels at week +7, and then displayed a plateau-like type of WHV7 accumulation, with the virions’ levels between 1.37×109 GE/ml and 7.29×109 GE/ml. The pattern of viremia observed in woodchuck F6693 is a typical example of a productive infection caused by WHV7 in naive adult woodchucks similar to previously observed infection profiles (Cote et al., 2000; Glebe et al., 2009; Menne and Cote, 2007), and thus can be used as an appropriate reference for comparison to infection profiles induced by WHVNY. For woodchuck F6671, the titers were quantifiable during weeks +1 through +6. The highest levels of viremia were observed at weeks +3 and +4, however overall WHV7 titers remained low within the range of 4.76×102 to 7.09×104 GE/ml. Between weeks +7 and +14, the titers were below detection limits of our real-time PCR (qPCR) assay. The animal F6671 had the lowest titers observed among the five woodchucks. It was not unusual to observe short time viremia with relatively low titers for naive adult woodchucks inoculated with WHV7 inoculum. For example, in a recently published paper, only 2/6 woodchucks inoculated with WHV7 developed productive acute infections with relatively high viremia, while the third woodchuck had a long lasting viremia with moderate WHV titers, and the rest of animals apparently cleared the initial infection relatively fast (as judged by serum WHV levels) and had only brief viremia with relatively low titers (Glebe et al., 2009).
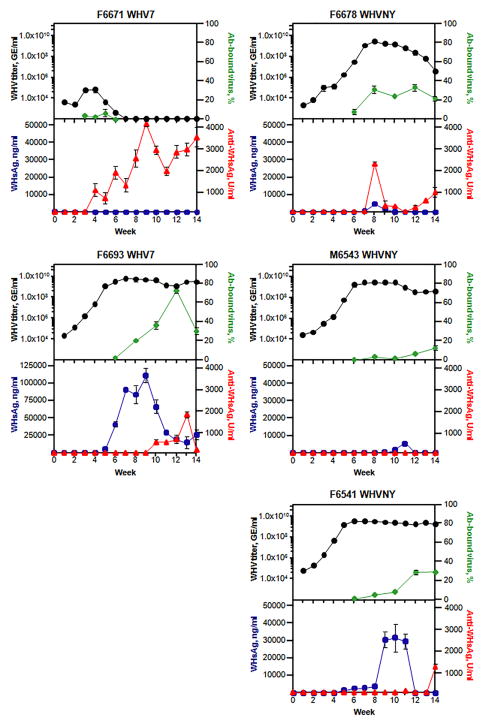
Fig 1. Time courses of acute infections induced in naive adult woodchucks by WHV strains WHV7 and WHVNY.
Woodchucks F6671 and F6693 were infected with strain WHV7. Woodchucks F6678, M6543 and F6541 were infected with strain WHVNY. Infected animals were monitored for 14 weeks post-inoculation. The serum titers of WHV were assayed weekly using qPCR as previously described (Freitas et al., 2012). The data for WHV titers are shown as black circles. The results of WHV titers’ measurements are expressed in WHV GE/ml of serum (logarithmic scale). The WHsAg in sera was examined weekly as well using ELISA as described before (Cote et al., 1993; Liu et al., 2011). The values for WHsAg are displayed in ng of WHsAg/ml of serum (blue rectangles). The anti-WHsAg antibodies in sera were also assayed using ELISA (Cote et al., 1993; Liu et al., 2011) on weekly basis. The results are shown as red triangles. The numbers obtained are displayed in standard units/ml of serum (U/ml). Also shown are the percentages of WHV virions bound to anti-WHsAg antibodies relative to total numbers of serum virions (green diamonds). The identification number and infecting WHV strain for each woodchuck is shown above each graph. The X axis represents time post-infection, weeks. The time of inoculation is shown as week 0. Each Y axis is labeled accordingly to the parameter it represents. All the assays are described in Materials and Methods. The standard errors of the means are indicated.
All three WHVNY-infected woodchucks developed productive acute infection profiles with long lasting viremia and high titers. Animal F6678 reached WHVNY titer above 1.0×109 GE/ml at week +7, maintained titers between 1.26×109 and 3.15×109 GE/ml at weeks +7 through +10, and then showed gradual decline in serum rcDNA to the level of 4.27×106 GE/ml by week +14. Unlike woodchuck F6678 that displayed a peak of viremia, animals M6543 and F6541 had the plateau of WHVNY titers, starting at about week +6. For F6541, the titers remained above 1.0×109 GE/ml between weeks +5 and +14. For M6543, the titers were above 1.0×109 GE/ml between weeks +6 and +10 post-infection, then there was a somewhat decline to the values within a range of 2.95×108 GE/ml to 9.35×108 GE/ml between weeks +11 through +14. Thus, none of WHVNY-infected woodchucks showed any signs of quick resolution of infection (Fig. 1). Overall, strain WHVNY did not demonstrate any noticeable deficiency in ability to induce productive WHV infection in naive adult woodchucks as judged by comparison to (i) the course of WHV7 viremia in woodchuck F6693 (Fig. 1) in the current study, and to (ii) previously published data regarding WHV7 infections (Cote et al., 2000; Glebe et al., 2009; Menne and Cote, 2007).
Interestingly, while we monitored the animals for 14 weeks (less than six months) after infection, three woodchucks, F6693, F6541 and M6543, demonstrated the plateau-like accumulation patterns of serum WHV (with high concentration of virions) later in infection, which was similar to infection patterns that were observed for animals that were infected as neonates and then eventually progressed to the chronicity state (Menne and Cote, 2007). However, currently the progression to chronic infection in woodchucks infected with WHV as adults is considered rare (less than 5%) (Menne and Cote, 2007).
3.2. WHV surface antigens, anti-WHsAg antibodies and virions bound to anti-WHsAg antibodies in serum samples
Next, the accumulation of (i) WHV envelope proteins (surface antigens or WHsAg), (ii) antibodies against WHsAg, and (iii) circulating in blood immunocomplexes of WHV virions bound to anti-WHsAg antibodies were considered (Fig. 1). For woodchuck F6671 that did not develop productive WHV7 infection, no WHsAg in serum was detected during the entire time of the experiment (the cut-off of the ELISA was 50 ng/ml of WHsAg). However, anti-WHsAg antibodies were detectable between weeks +4 and +14, within the range of 734 to 4,156 U/ml with levels almost always exceeding 2,000 U/ml after week +8. The cut-off of the assay for anti-WHsAg antibodies was 100 U/ml. In case of F6671, the earliest production of anti-WHsAg antibodies among all five animals studied was observed. Between weeks +3 and +5, the fraction of antibody-bound WHV7 was between 2.26 and 6.21 %, and became undetectable by week +6. Interestingly, while early appearance of anti-WHsAg antibodies and their highest accumulation levels among the five woodchucks coincided with fast suppression of initial WHV7 infection (as judged by serum WHV7 titers (Fig. 1)), we did not observe a significant portion of circulating serum antibody-bound virions within the time frame, when WHV7 titers were detectable (Fig. 1). For another WHV7-infected animal, F6693, serum WHsAg was detected starting week +5. The levels of WHsAg were relatively high for the rest of the monitoring period, with the peak of 115,553 ng/ml observed at week +9. After week +6, WHsAg levels remained above 16,000 ng/ml. In fact, at all times when WHsAg was detectable (between weeks +5 and +14), its levels were the highest compared to that of other four woodchucks. However, the observed B cell-mediated production of anti-WHsAg antibodies appeared rather weak. The accumulation of anti-WHsAg antibodies was observed during weeks +10 through +14, and was at relatively low levels of 175 to 1,903 U/ml. That highest amount of 1,903 U/ml was observed at week +13. Nevertheless, it was very surprising to find that regardless of high titers of WHV7 and low levels of anti-WHsAg antibodies, the fraction of antibody-bound WHV7 virions was above 20% between weeks +8 and +14, and reached about 73% at week +12. Thus, there was no apparent correlation between the levels of anti-WHsAg antibodies and the fraction of antibody-bound WHV virions. The low levels of anti-WHsAg antibodies did not affect the concentration of virions in the blood, regardless of the binding to the majority of virus particles at week +12 (Fig. 1). Given low levels of accumulation of anti-WHsAg antibodies, we can speculate that binding of the majority of WHV virions at week +12 might be explained by the high affinity of the antibodies towards WHsAg.
For WHVNY-infected woodchuck, F6541, the production of WHsAg has been observed between weeks +5 and +12. The levels of the envelope proteins’ accumulation were in the range of 425 (week +12) to 32,786 ng/ml (peak of WHsAg accumulation at week +10). Relatively high WHsAg levels over 30,000 ng/ml were observed between weeks +9 and +11. The accumulation of anti-WHsAg antibodies was very low. No anti-WHsAg antibodies were detected before week +11, and also at weeks +12 and +13. A level of 107 U/ml was measured for week +11, while by week +14, the amounts of the accumulated antibodies were 1,360 U/ml. The percentage of antibody-bound WHV virions went from 0.65% (week +6) to about 30% by week +12 and remained at this level at week +14. Overall, the anti-envelope antibodies did not exert a considerable regulatory effect on WHVNY infection during first 14 weeks after inoculation (Fig. 1). For animal M6543, WHsAg was only measurable during weeks +9 to +11, with the highest level of 6,236 ng/ml at week +11, which actually was a very low value as compared to the numbers measured for woodchucks F6693 and F6541. No anti-WHsAg antibodies were detected during the entire time course period (Fig. 1). However, small fractions of antibody-bound virions (between 0.2% and 13%) were detected at weeks +6 through +14. The portion of these serum immunocomplexes reached about 13.0% by week +14. It needs to be taken into consideration, that ELISA directed against anti-WHsAg antibodies had the cut-off of 100 U/ml, and for detection of anti-WHsAg antibody-bound virions we used immunoprecipitation coupled with sensitive qPCR assay, which may explain higher sensitivity of the latter approach. In case of WHVNY-infected animal F6678, WHsAg was observed at weeks +7 through +10. The peak of 5,017 ng/ml was measured at week +8. Relatively low levels of anti-WHsAg antibodies were detectable between weeks +7 and +14. The peak of the antibodies’ concentration at 2,351 U/ml was observed at week +8, then levels of anti-WHsAg antibodies declined to 90 U/ml by week +11, and after that their levels increased and reached 1,038 U/ml by week +14. The antibody-bound virus was measurable between weeks +6 and +14, with highest levels of 31.45% at week +8, and 33.71% at week +12. At weeks +10 and +14, smaller but similar levels of the antibody-virus complexes of 24.4 and 21.94%, respectively, were quantified (Fig. 1). Overall, most of the time (with possible exception of F6671) no obvious correlation was observed between the viremia patterns and the levels of either (i) WHsAg, (ii) anti-WHsAg antibodies, or (iii) percentage of antibody-bound WHV. It also became apparent that production/accumulation of anti-WHsAg antibodies did not correlate with the fraction of virions bound to these antibodies (Fig. 1).
3.3. Spread of the virus through the liver and fractions of WHV core-positive cells
Hepatocytes positive for WHV core antigen (WHcAg) were quantified using immunohistochemistry approach. The samples of liver tissues were harvested at weeks 0, +6, +13 and +14. The tissue sections were stained using the antibodies against WHcAg (Freitas et al., 2012). The results are summarized in Table 1. As expected, all the samples collected at week 0 were negative for WHcAg. For WHV7-infected animal F6693, the percentage of core-positive cells were 75% by week +6, and declined to 31.8% by week +13, and remained at a similar level of 38.1% at week +14 (Table 1). The data was consistent with observed high levels of WHV7 in serum at the corresponding time points (Fig. 1). In case of F6671, which did not develop high and long lasting viremia (Fig. 1), our immunostaining was unable to detect WHcAg-positive hepatocytes at weeks +13 and +14, while only a small fraction of core-positive cells (0.3%) was found at week +6 (Table 1).
Table 1.
The fraction of WHV core antigen-positive cells at different time points post-infectiona.
| Woodchuckb | Strainb | Time post-infection, weeks | |||
|---|---|---|---|---|---|
| week 0 | week +6 | week +13 | week +14 | ||
| F6693 | WHV7 | 0.0% (0/1500) | 75.0% (1192/1590) | 31.8% (652/2049) | 38.1% (720/1889) |
| F6671 | WHV7 | 0.0% (0/1500) | 0.3% (5/1801) | 0.0% (0/1500) | 0.0% (0/1500) |
| F6678 | WHVNY | 0.0% (0/1500) | 1.0% (17/1675) | 48.9% (1414/2894) | 22.4% (539/2411) |
| M6543 | WHVNY | 0.0% (0/1500) | 70.7% (1840/2602) | 10.8% (367/3402) | 49.2%(1315/2672) |
| F6541 | WHVNY | 0.0% (0/1500) | 66.0% (1221/1849) | 30.2% (768/2539) | 41.7% (800/1920) |
Staining of liver tissue samples for WHV core antigen was done as described previously (Freitas et al., 2012).
Woodchucks used in the current study, the strains of WHV used for infection, and harvesting of liver tissue samples for analysis are described in Materials and Methods.
WHVNY-infected woodchuck F6678 displayed somewhat delayed spread of the virus throughout the liver. Regardless of the relatively high titer of WHVNY in serum by week +6 (Fig. 1), the portion of core-positive cells was approximately just 1.0%. However, the fraction of WHcAg-positive cells was about 49% at week +13, and declined to 22.4% by week +14. The other two WHVNY-infected animals, M6543 and F6541, have the numbers of WHcAg-positive cells mostly similar to those of F6693. Both woodchucks had about 70% of core-positive cells by week +6. For animal F6541, the amounts of WHcAg -positive cells became approximately 30% and 42% at weeks +13 and +14, respectively. For woodchuck M6543, we quantified only about 11% of cells positive by immunostaining at week +13, while at week +14, the percentage of positive cells was measured at the level of 49.2% (Table 1). We speculate that during biopsy taken at week +13, the part of the liver chosen for harvest had particularly low number of hepatocytes replicating WHV. WHV infection is known to be not uniform, which means that different parts of the same infected liver can contain considerably different number of hepatocytes that can replicate WHV at sufficiently different rates, which would be reflected by the staining for WHcAg (Mason et al., 2009a; Summers et al., 2003). Woodchucks, F6693, F6541 and M6543 as noted above also displayed comparable patterns of WHV viremia (Fig. 1).
Previously reported patterns of the spread of WHV through the livers of naive adult woodchucks usually include the infection of practically of the entire liver with WHV (>95% of core-positive hepatocytes) within a few weeks after the inoculation with subsequent substantial decrease of the numbers of WHV-core positive cells within several weeks after the infection of a whole liver was observed (Mason et al., 2007, Mason et al., 2009a). Based on the data in Table 1 and previous reports (Mason et al., 2007, Mason et al., 2009a), we may reasonably speculate that (i) there was no efficient infection of the entire liver for animal F6671, while (ii) for F6693, M6543 and F6541, the infection of virtually the entire livers likely occurred around week +6 post-infection, and for woodchuck F6678, the spread was delayed and the infection of practically the entire liver likely took place between weeks +6 and +13.
3. 4. Comparison of DNA and RNA replication markers of WHV7 and WHVNY
The levels of the viral intracellular replicative DNA intermediates (RI-DNA) (Mason et al., 2009a), covalently closed circular DNA (cccDNA) and pre-genomic/pre-core RNA (pgRNA) of WHV7 and WHVNY in liver tissues were quantified using previously described qPCR assays (Freitas et al., 2012). The results are summarized in Tables 2 and 3.
Table 2.
Quantity of WHV RI-DNA and cccDNA at different time points after infectiona.
| Woodchuckb | Strainb | Time post-infection, weeks | |||
|---|---|---|---|---|---|
| week 0 | week +6 | week +13 | week +14 | ||
| RI-DNA measurementsc | |||||
|
| |||||
| F6693 | WHV7 | und | (1.09+/−0.15)×108 | (2.19+/−0.43)×107 | (4.27+/−0.27)×107 |
| F6671 | WHV7 | und | (7.49+/−1.24)×102 | (5.72+/−1.67)×102 | (5.20+/−2.88)×102 |
| F6678 | WHVNY | und | (1.87+/−0.33)×106 | (1.75+/−0.34)×106 | (3.88+/−1.21)×106 |
| M6543 | WHVNY | und | (2.95+/−0.53)×107 | (7.69+/−4.86)×106 | (1.00+/−0.12)×107 |
| F6541 | WHVNY | und | (3.37+/−0.81)×107 | (1.29+/−0.38)×107 | (2.57+/−0.43)×107 |
|
| |||||
| ave NYd | 2.17×107 | 0.74×107 | 1.32×107 | ||
|
| |||||
| (ave NY/7 (F6693)) e | 0.20 | 0.34 | 0.31 | ||
|
| |||||
| cccDNA measurementsf | |||||
|
| |||||
| F6693 | WHV7 | und | (2.64+/−0.04)×105 | (5.89+/−1.09)×104 | (1.09+/−0.08)×105 |
| F6671 | WHV7 | und | (4.72+/−2.44)×101 | (4.34+/−0.19)×101 | (1.25+/−0.25)×102 |
| F6678 | WHVNY | und | (1.08+/−0.07)×105 | (2.48+/−0.25)×104 | (4.39+/−2.44)×104 |
| M6543 | WHVNY | und | (4.48+/−1.00)×105 | (7.61+/−0.17)×104 | (1.23+/−0.45)×105 |
| F6541 | WHVNY | und | (3.33+/−0.48)×105 | (9.26+/−1.25)×104 | (1.05+/−0.13)×105 |
|
| |||||
| ave NYd | 2.96×105 | 6.45×104 | 9.06×104 | ||
|
| |||||
| (ave NY/7 (F6693)) e | 1.12 | 1.10 | 0.83 | ||
The DNA isolation procedures and qPCR assays used for measurements of RI-DNA and cccDNA in harvested liver tissue samples are described in Materials and Methods.
Woodchucks used, the strains of WHV used for infection, as well as collection of liver tissue samples for analysis are detailed in Materials and Methods.
The levels of RI-DNA of WHV are expressed in WHV GE/μg of total DNA (+/− standard deviation). The DNA concentration values in total DNA preps were used for calculations.
The ave NY represent the average number for a given parameter (i.e., RI-DNA or cccDNA) obtained for a particular time point post-infection using the corresponding data for three WHVNY-infected woodchucks (F6541, M6543 and F6678).
The (ave NY/7 (F6693)) ratio values were calculated using the determined ave NY values and numbers obtained for WHV7-infected woodchuck F6693. Only animals that developed productive acute WHV infection (Fig. 1) were considered for the calculating the ratio values.
The data for cccDNA was shown as WHV GE/μg of total DNA (+/− standard deviation). The numbers were calculated per μg of total DNA in the corresponding total DNA preps.
For both, RI-DNA and cccDNA, “und” stands for undetected.
Table 3.
Quantity of WHV pgRNA and pgRNA/cccDNA ratio at different times post-infectiona.
| Woodchuckb | Strainb | Time post-infection, weeks | |||
|---|---|---|---|---|---|
| week 0 | week +6 | week +13 | week +14 | ||
| pgRNA measurementsc | |||||
|
| |||||
| F6693 | WHV7 | und | (1.85+/−0.26)×108 | (1.04+/−0.62)×108 | (2.16+/−1.38)×108 |
| F6671 | WHV7 | und | (2.32+/−2.27)×103 | (1.21+/−0.04)×103 | (1.13+/−1.01)×103 |
| F6678 | WHVNY | und | (4.32+/−1.07)×107 | (7.04+/−2.95)×107 | (1.04+/−0.18)×108 |
| M6543 | WHVNY | und | (3.86+/−0.19)×108 | (1.57+/−1.43)×108 | (7.93+/−2.60)×107 |
| F6541 | WHVNY | und | (3.82+/−0.39)×108 | (2.99+/−0.22)×108 | (3.31+/−0.13)×108 |
|
| |||||
| ave NYd | 2.70×108 | 1.75×108 | 1.71×108 | ||
|
| |||||
| (ave NY/7 (F6693)) e | 1.46 | 1.68 | 0.79 | ||
|
| |||||
| ratio (pgRNA/cccDNA)f | |||||
|
| |||||
| F6693 | WHV7 | 7.01×102 | 1.77×103 | 1.98×103 | |
| F6671 | WHV7 | 4.92×101 | 2.79×101 | 9.04×100 | |
| F6678 | WHVNY | 4.00×102 | 2.84×103 | 2.37×103 | |
| M6543 | WHVNY | 8.62×102 | 2.06×103 | 6.45×102 | |
| F6541 | WHVNY | 1.15×103 | 3.23×103 | 3.15×103 | |
|
| |||||
| ave NYd | 8.04×102 | 2.71×103 | 2.06×103 | ||
|
| |||||
| (ave NY/7 (F6693))e | 1.15 | 1.53 | 1.04 | ||
The RNA isolation and qPCR assay used for measurements of WHV pre-core/pre-genomic RNA (pgRNA) in liver tissue samples are described in Materials and Methods. The cccDNA values are shown in Table 3.
Animals used in the current study, the strains of WHV employed for infection, as well as collection of liver tissue samples for analysis are detailed in Materials and Methods.
The quantities of WHV pgRNA are expressed in GE/μg of total RNA (+/− standard deviation). The “und” stands for undetected.
The ave NY is the average number for a particular parameter (i.e., for pgRNA or (pgRNA/cccDNA) ratio) obtained for a given time point using the data for three WHVNY-infected woodchucks, F6541, M6543 and F6678.
The (ave NY/7 (F6693)) ratios were calculated using the ave NY values and numbers obtained for WHV7-infected woodchuck F6693. Only woodchucks, which developed productive acute WHV infection (Fig. 1), were considered for the calculating the ratio values.
The ratio (pgRNA/cccDNA) that is indicative of transcriptional activity of cccDNA and stability of produced pgRNA was calculated (for weeks +6, +13 and +14) using pgRNA numbers from this Table, and cccDNA numbers from Table 2.
First, the RI-DNA measurements were considered. As anticipated, in the tissues harvested prior to infection, no RI-DNA was detected. Consistent with very low WHV7 serum titers (Fig. 1), woodchuck F6671 displayed measurable, but considerably low RI-DNA levels between 5.20×102 and 7.49×102 GE/μg of total DNA, and did not show major variations between different time points. These levels of accumulated RI-DNA were the lowest among the five animals studied. For the second WHV7-infected woodchuck F6693, the highest RI-DNA numbers were measured for week +6, and were 1.09×108 GE/μg of total DNA. Later in infection, by weeks +13 +14, the RI-DNA levels went somewhat down to approximately (2.20÷4.30)×107 GE/μg of total DNA, which were about 2.5 to 5.0 fold lower than that of week +6 (Table 2).
The group of WHVNY-infected woodchucks demonstrated consistent relatively high RI-DNA levels. For F6678, RI-DNA levels were between 1.75×106 to 3.88×106 GE/μg of total DNA. Animal M6543 had RI-DNA numbers also within not a wide range from 7.69×106 to 2.95×107 GE/μg of total DNA. Similarly, for F6541, the levels of RI-DNA were between 1.29×107 and 3.37×107 GE/μg of total DNA. Overall, the average RI-DNA values obtained for three WHVNY-infected animals were in a range between about 0.20 to 0.34 (ave NY/7 ratio for RI-DNA) of the values measured for WHV7-infected animal F6693, which developed a productive acute infection with high viremia (Fig. 1 and Table 2). Although the average RI-DNA numbers obtained for WHVNY-infected animals were somewhat lower than that of woodchuck F6693, the observed differences were mostly within about 3-fold range, and thus, were not considered as profound. The only exception was week +6, when because of lower RI-DNA value for F6678, the average NY/7 ratio was 0.20 (Table 2).
Second replication intermediate analyzed was cccDNA. No cccDNA of WHV were detected in the samples harvested prior to infection. As expected, animal F6671 had very low accumulation of cccDNA that was between 4.0×101 and 5.0×101 GE/μg of total DNA at weeks +6 and +13, and was somewhat higher at the level of 1.25×102 GE/μg of total DNA by week +14. The cccDNA accumulation for another WHV7-infected animal, F6693, was in a range of 5.89×104 to 2.64×105 GE/μg of total DNA without dramatic fluctuations (Table 2).
Very similar cccDNA numbers were measured for three WHVNY-infected animals. For F6678, cccDNA levels were slightly above 1.0×105 GE/μg of total DNA at week+6, then later went somewhat down to about (2.5÷4.4)×104 GE/μg of total DNA at weeks +13 and +14. Woodchuck M6543 displayed cccDNA numbers within a range of 7.61×104 to 4.48×105 GE/μg of total DNA, while the range for F6541, was between 9.26×104 and 3.33×105 GE/μg of total DNA with the lowest number observed at week +13 (Table 2). Overall, if like in case of RI-DNA values’ comparison, we again consider only the data obtained for animal F6693 versus the average values obtained for WHVNY-infected woodchucks F6678, M6543 and F6541, all of which displayed the profiles of productive acute WHV infections, then the cccDNA ratio NY/7 would be 1.12 (week+6), 1.10 (week +13), and 0.83 (week +14). These numbers suggest that the rates of WHVNY and WHV7 replication during the course of productive acute WHV infection in naive adult woodchucks were quite comparable.
The above conclusion was supported by the measurements of the levels of pgRNA in liver tissues samples (Table 3). As expected, no pgRNA of WHV was found in tissues harvested before the infection. Animal F6671 displayed very low levels of pgRNA within a narrow range between 1.13×103 and 2.32×103 GE/μg of total RNA. Animal F6693, as anticipated, had considerable accumulation of pgRNA between 1.04×108 to 2.16×108 GE/μg of total RNA. For this animal, no significant variations were observed between different time points.
The WHVNY-infected woodchucks demonstrated levels of pgRNA comparable to that of F6693. They were within a range of 4.32×107 to 3.86×108 GE/μg of total RNA. For each WHVNY-infected animal, the variations observed between different time points were not dramatic (Table 3). The pgRNA ratio ave NY/7 (F6693) (when WHV7-infected woodchuck F6693 was compared to the average values calculated for three WHVNY-infected animals) was 1.46 for week +6, 1.68 for week +13, and 0.79 for week +14. These values again confirmed similar replication levels of WHVNY and WHV7 in productively infected woodchuck livers.
Finally, we analyzed the ratio pgRNA/cccDNA, which can be considered as being reflective of the transcriptional activity of cccDNA and stability of pgRNA during hepadnavirus replication (Freitas et al., 2012; Lesmana et al., 2014; Pollicino et al., 2011; Wieland et al., 2003). The numbers are summarized in Table 3 as well. As anticipated, WHV7-infected woodchuck F6671 had the lowest ratio values between 9.0 and 49.2, with numbers declining from week +6 to week +14. As expected, woodchuck F6693 showed the numbers that were mostly similar to those of WHVNY-infected animals. The ratios ave NY/7 for the pgRNA/cccDNA values (when F6693 was compared to averages calculated for F6678, M6543 and F6541) were 1.15 (week +6), 1.53 (week +13), and 1.04 (week +14).
Overall, the data support the conclusion that WHVNY and WHV7 exhibit similar replication parameters without dramatic differences in the setting of acute infection in naive adult woodchucks, which was observed over a course of 14 weeks monitoring after the infection. Many of the parameters were close, and most of the time the observed differences did not exceed 3-fold range. Only for week +6, the NY/7 ratio for the RI-DNA was 0.2 (5-fold difference).
3.5. Biostatistical analysis of the infection profiles of WHV7 versus WHVNY
First, the serum WHV titers were compared. Taking in consideration that woodchuck F6671 inoculated with strain WHV7 did not develop efficient WHV infection, the comparison was done using only the data obtained for woodchucks F6693 (infected with WHV7) and F6678, M6543 and F6541 (all infected with WHVNY), which displayed the patterns of productive acute WHV infection, and demonstrated long lasting high viremia. Given a good amount of available data in literature on infection patterns induced in naive adult woodchucks by inoculation with strain WHV7, it became apparent (i) that animal F6693 is a typical example of efficient acute WHV7 infection as it was observed on many previous occasions (Cote et al., 2000; Glebe et al., 2009; Menne and Cote, 2007), and (ii) that the data generated for this animal can be used as the representative set to conduct the comparison of acute infection induced by strain WHV7 to the infection patterns generated by inoculation of adult naive woodchucks with another strain, WHVNY. Therefore, animal F6693 was compared to three WHVNY-infected woodchucks using a two-sided t-test on log-transformed data (Ostle, 1964). The null hypothesis was that titer values of woodchuck F6693 have the same distribution as those from the three WHVNY-infected woodchucks. The alternative was that they did not. The results of the analysis are shown in Table 4. The smallest and only p-value less than 0.05 (i.e., 0.030) was obtained at week +8. Since there was a 50% chance that at least one of our 14 tests will be significant though rcDNA levels were statistically equivalent at every week tested, we used the Bonferroni adjustment (Miller, 1966) in order to protect against claiming significance spuriously due to multiple testing. Therefore, the value 0.030 when adjusted for the multiplicity of comparisons (n=14) is not significant (a p-value less than 0.05/14 = 0.0036 would be significant at the 5% level using the Bonferroni adjustment (Table 4)). In addition, Fig. 1 shows that at weeks +3, +5, +7 through +10, and also at weeks +13 and +14, the animal F6693’s titer values were somewhat (within (2.04÷4.31)-fold range) higher than the average titers for WHVNY-infected animals. However, these titer values of F6693 differed from the corresponding numbers of the animal F6541 in less than two-fold range, which is not a profound difference. Overall, it became apparent that based on the WHV titers’ values, WHV7-infected woodchuck F6693 is not greatly different from the group of animals infected with WHVNY.
Table 4.
Statistical analysis of WHVNY and WHV7 serum titers (serum rcDNA) over the course of 14 weeks post-infectiona.
| Time post-infection (weeks) | serum rcDNA p-value |
|---|---|
| +1 | 0.806 |
| +2 | 0.550 |
| +3 | 0.490 |
| +4 | 0.548 |
| +5 | 0.506 |
| +6 | 0.600 |
| +7 | 0.232 |
| +8 | 0.030b |
| +9 | 0.108 |
| +10 | 0.138 |
| +11 | 0.660 |
| +12 | 0.502 |
| +13 | 0.440 |
| +14 | 0.488 |
The data for WHV7-infected animal F6693 was compared to the data for WHVNY-infected woodchucks, F6678, M6543 and F6541. The animal F6671 was not included in comparison, because, unlike other animals, it did not develop productive acute infection after inoculation with WHV7. Two-sided t-tests and Bonferroni adjustment were used (Ostle, 1964; Miller, 1966). The null hypothesis was that WHV7-infected woodchuck F6693 statistically belongs to the same group as three woodchucks infected with WHVNY in terms of the observed serum WHV titer values. Alternative hypothesis is that animal F6693 is significantly different from three WHVNY-infected woodchucks.
The p-value of 0.030 cannot be considered significant considering multiple (n=14) comparisons (see text).
Second, the values of RI-DNA, cccDNA and pgRNA determined at weeks +6, +13 and +14 were compared for the same above mentioned four woodchucks that developed productive acute WHV infections (Fig. 1), using two-sided t-tests as well. The outcome of the statistical analysis is summarized in Table 5. As in the case of viral titers, none of the above measurements differed significantly between animals infected either with WHVNY or with WHV7 (Table 5). In conclusion, the statistical comparisons confirmed our previous interpretations that no profound differences were observed between the patterns of acute WHV infections induced by either WHVNY or WHV7 for the woodchucks that displayed long lasting viremia with considerably high titers within 14 weeks post-inoculation (Fig. 1).
Table 5.
Statistical analysis of accumulation of RI-DNA, cccDNA and pgRNA for three WHVNY-infected animals (F6678, M6543 and F6541) and WHV7-infected woodchuck F6693, all of which developed productive WHV infections.
| Time post-infection (weeks) | RI-DNA p-value | cccDNA p-value | pgRNA p-value |
|---|---|---|---|
| +6 | 0.668 | 0.710 | 0.999 |
| +13 | 0.714 | 0.954 | 0.964 |
| +14 | 0.315 | 0.372 | 0.366 |
The data reflects the probability that the WHV7-infected animal F6693 values came from the same population as the values of 3 WHVNY-infected animals. Two-sided t-test was based on null hypothesis that the two populations of values (three categories of numbers, RI-DNA, cccDNA and pgRNA, were considered for each population), i.e., values for woodchuck F6693 and the corresponding sets of data for three WHVNY-infected woodchucks (F6678, M6543 and F6541) belong to the same distributions of the values.
4. DISCUSSION
The current study is the first and only complete report that presents the extensive characterization of natural strain WHVNY and its comparison to well-known strain WHV7, which is currently used for the majority of experimental infections of woodchucks in US. Although, strain WHVNY has been described in 1993 (Kew et al., 1993), it was not evaluated in detail in terms of the capacity to induce productive WHV infection in naive adult woodchucks. We infected WHV-negative adult woodchucks with either WHV7 or WHVNY using the same MOI, and then compared time courses of acute WHV mono-infections. The comparison included the quantification of the (i) levels of relaxed circular DNA (rcDNA) in serum; (ii) levels of pre-genomic/pre-core RNA (pgRNA), replicative intermediate DNA (RI-DNA) and covalently closed circular DNA (cccDNA) in infected livers; (iii) fractions of WHcAg-positive cells in livers; (iv) levels of serum WHsAg and anti-WHsAg antibodies; and (v) fractions of circulating WHV virions that were bound to anti-WHsAg antibodies. To our knowledge, none of the published studies that used woodchuck experimental model contained all the experimental approaches characterizing WHV infection that were used by us. One of WHV7-infected animals displayed relatively low virus titers detectable only during first six weeks post-infection, while other three WHVNY-infected animals and second WHV7-infected woodchuck demonstrated well-developed infections with high and long lasting viremia (Fig. 1). The WHV7-infected animal F6693 demonstrated a typical pattern of productive acute infection with long lasting viremia, which was similar to the patterns of acute WHV infection that were previously observed in adult woodchucks inoculated with strain WHV7 (Cote et al., 2000; Glebe et al., 2009; Menne and Cote, 2007). Therefore, woodchuck F6693 served further as a reference for comparison with WHVNY-infected animals. Among the four animals (F6693, F6678, M6543 and F6541) that demonstrated profiles of productive WHV infection, the levels of serum accumulation of WHV virions (rcDNA) were found to be comparable. This observation was further confirmed by the biostatistical analysis (Fig. 1 and Table 4). Importantly, all three WHVNY-infected woodchucks showed the evidence of good level acute WHV infection. Two animals, F6541 and M6543 displayed plateaus in accumulation of serum WHVNY, which was similar to that of WHV7-infected animal F6693 (Fig. 1). Although, it was not unusual that one of two WHV7-infected woodchucks, F6671, did not develop long lasting high level WHV viremia (similar outcomes were observed previously for WHV7-infected woodchucks (Glebe et al., 2009)), it was interesting to observe that none of three WHVNY-infected woodchucks showed any signs of quick resolution/suppression of infection during the 14 weeks of monitoring. For WHVNY- and WHV7-infected woodchucks that display profiles of well-developed acute infection (F6693, F6678, M6543 and F6541), there was no apparent correlation between the detected levels of WHsAg and levels of serum virus (Fig. 1). In addition, there was also no correlation between the levels of serum anti-WHsAg antibodies and either (i) levels of serum WHV, or (ii) levels of circulating complexes of WHV bound to anti-WHsAg antibodies (Fig. 1). Apparently, the B cell-mediated immune response had a very little influence if any on the observed WHV infection/replication patterns during 14 weeks of monitoring period (this, however, does not exclude the possibility that the immune response may have been mounted at later times post-infection). In addition, we found that for the accumulation levels of the three categories of WHV replication intermediates (RI-DNA, cccDNA and pgRNA), the differences between values determined for WHV7-infected animal (F6693) and the average values determined for three WHVNY-infected woodchucks (F6678, M6543 and F6541) were almost always within three fold range, and thus were not considered significant. Although, for week +6, the difference between WHVNY and WHV7 in terms of RI-DNA accumulation was about 5-fold in favor of WHV7 (Table 2). The absence of significant differences in replication parameters between livers productively infected by WHVNY or by WHV7, was also confirmed by the statistical analysis of the experimental data (Table 5). The similarities between the numbers of infected hepatocytes were also found for animals infected with different WHV strains. The data on WHV core antigen staining in the liver of WHV7-infected animal F6693 was very similar to that of WHVNY-infected woodchuck F6541, and similar to a somewhat lesser degree to the data for another WHVNY-infected animal, M6543 (because of the week +13 data) (Table 1). The third WHVNY-inoculated woodchuck F6678 had somewhat delayed WHVNY spread through the liver, which however did not correlate with the levels of accumulation of DNA and RNA replication intermediates of WHV (Table 1, 2 and 3). Overall, several above discussed lines of evidence allowed us to conclude that strains WHV7 and WHVNY are comparable in their replication parameters and in terms of the capacity to induce productive acute WHV infection in naive adult woodchucks.
We recently published a paper, in which the consensus sequences of WHV7 and WHVNY were compared, and unique sequence features that can be instrumental in developing WHV strain-specific assays were indicated. Using the super-infection approach, when woodchucks that were chronic carriers of WHV7 were then super-infected with WHVNY, and employing a number of (developed by us) novel WHV strain-specific assays, which were able to exclusively detect only one strain in the complex mixtures of WHV7 and WHVNY sequences, we obtained the evidence that livers chronically infected with a hepadnavirus (WHV) were able to support a super-infection with a different WHV strain. The obtained results were consistent with the occurrence of continuous, but limited hepadnavirus cell-to-cell spread/super-infection during the chronic state of hepadnavirus infection. The generated data, therefore, advanced our understanding of the complex mechanism of the chronic hepadnavirus infection (Rodrigues et al., 2015). The findings of the current study provide important piece of novel and critical information that complements the above mentioned publication by presenting the detailed evidence for considerable similarity of WHV7 and WHVNY in terms of replication rates and capacity to cause productive acute infection in naive adult woodchucks. These data served as the basis of one of the important conclusions of the recent report (Rodrigues et al., 2015), which suggested that (i) the observed limitations of the WHV super-infection/spread could be likely explained by the properties of hepatocytes and actions of the immune system, and (ii) were not related to the properties of the super-infecting virus (WHVNY).
In addition, the results of the current study further justify the use of different WHV strain, WHVNY, as a useful reagent for co- and super-infection in vivo studies in woodchuck model, which would examine the replication of WHV7 and WHVNY in the same liver/same nuclei, and also would facilitate better understanding of the mechanisms of transient and chronic hepadnavirus infections.
Two WHV strains, WHVNY and WHV7, were compared in the settings of monoinfection of naive adult woodchucks
We compared the percentages of WHV core-positive hepatocytes, and quantified serum rcDNA; and RI-DNA, cccDNA and pgRNA in liver tissues
We measured WHsAg, anti-WHsAg antibodies and WHV bound to anti-WHsAg antibodies in serum samples
Strains WHV7 and WHVNY appeared quite comparable in terms of replication parameters and capacity to induce acute infection in naive adult woodchucks
Acknowledgments
We thank Bud Tennant of Cornell University for providing the serum of woodchuck 2761 that contained the strain WHVNY. We also thank the Histology laboratory of the Lombardi Cancer Center of the Georgetown University for the assistance with the staining of woodchuck liver tissues. SOG and SM were supported by NIH grant NCI R01CA166213. SOG was also supported by NIH grant NCRR P20RR016443, and by the University of Kansas Endowment Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Natalia Freitas, Email: nfreitas@kumc.edu.
Tetyana Lukash, Email: tlukash@kumc.edu.
Megan Dudek, Email: mdudek@kumc.edu.
Sam Litwin, Email: Samuel.Litwin@fccc.edu.
Stephan Menne, Email: sm923@georgetown.edu.
Severin O. Gudima, Email: sgudima@kumc.edu.
References
- Akbar SM, Horiike N, Onji M. Immune therapy including dendritic cell based therapy in chronic hepatitis B virus infection. World J Gastroenterol. 2006;12:2876–2883. doi: 10.3748/wjg.v12.i18.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselah T, Lada O, Moucari R, Martinot M, Boyer N, Marcellin P. Interferon therapy for chronic hepatitis B. Clin Liver Dis. 2007;11:839–849. doi: 10.1016/j.cld.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Cote PJ, Roneker C, Cass K, Schödel F, Peterson D, Tennant B, De Noronha F, Gerin J. New enzyme immunoassays for the serologic detection of woodchuck hepatitis virus infection. Viral Immunol. 1993;6:161–169. doi: 10.1089/vim.1993.6.161. [DOI] [PubMed] [Google Scholar]
- Cote PJ, Korba BE, Miller RH, Jacob JR, Baldwin BH, Hornbuckle WE, Purcell RH, Tennant BC, Gerin JL. Effects of age and viral determinants on chronicity as an outcome of experimental woodchuck hepatitis virus infection. Hepatology. 2000;31:190–200. doi: 10.1002/hep.510310128. [DOI] [PubMed] [Google Scholar]
- Di Bisceglie AM. Hepatitis B and hepatocellular carcinoma. Hepatology. 2009;49 (5 Suppl):S55–S60. doi: 10.1002/hep.22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstag JL. Hepatitis B virus infection. N Eng J Med. 2008;359:1486–1500. doi: 10.1056/NEJMra0801644. [DOI] [PubMed] [Google Scholar]
- Freitas N, Salisse J, Cunha C, Toshkov I, Menne S, Gudima SO. Hepatitis delta virus infects the cells of hepadnavirus-induced hepatocellular carcinoma in woodchucks. Hepatology. 2012;56:76–85. doi: 10.1002/hep.25663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebe D, Lorenz H, Gerlich WH, Butler SD, Tochkov IA, Tennant BC, Cote P, Menne S. Correlation of virus and host response markers with circulating immune complexes during acute and chronic woodchuck hepatitis virus infection. J Virol. 2009;83:1579–91. doi: 10.1128/JVI.01934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew MC, Chestnut T, Baldwin BH, Hornbuckle WE, Tennant BC, Purcell RH, Miller RH. Heterogeneity of the woodchuck hepatitis virus genome in a chronically infected woodchuck. Virus Res. 1993;27:229–237. doi: 10.1016/0168-1702(93)90035-l. [DOI] [PubMed] [Google Scholar]
- Lam YF, Yuen MF, Seto WK, Lai CL. Current antiviral therapy of chronic hepatitis B: efficacy and safety. Curr Hepatitis Rep. 2011;10:235–243. doi: 10.1007/s11901-011-0109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey SB, Siliciano RF. A mechanistic theory to explain the efficacy of antiretroviral therapy. Nat Rev Microbiol. 2014 doi: 10.1038/nrmicro3351. [DOI] [PubMed] [Google Scholar]
- Lesmana CR, Jackson K, Lim SG, Sulaiman A, Pakasi LS, Gani RA, Hasan I, Sulaiman AS, Lesmana LA, Hammond R, Revill P, Locarnini S, Bowden SD. Clinical significance of hepatitis B virion and SVP productivity: relationships between intrahepatic and serum markers in chronic hepatitis B patients. United European Gastroenterol J. 2014;2:99–107. doi: 10.1177/2050640614525151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin S, Toll E, Jilbert AR, Mason WS. The competing roles of virus replication and hepatocyte death rates in the emergence of drug-resistant mutants: Theoretical consideration. J Clin Virol. 2005;34(Suppl 1):S96–S107. doi: 10.1016/s1386-6532(05)80018-6. [DOI] [PubMed] [Google Scholar]
- Liu KH, Ascenzi MA, Bellezza CA, Bezuidenhout AJ, Cote PJ, Gonzalez-Aseguinolaza G, Hannaman D, Luxembourg A, Evans CF, Tennant BC, Menne S. Electroporation enhances immunogenicity of a DNA vaccine expressing woodchuck hepatitis surface antigen in woodchucks. J Virol. 2011;85:4853–4862. doi: 10.1128/JVI.02437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok AS, Zoulim F, Locarnini S, Bartholomeusz A, Ghany MG, Pawlotsky JM, Liaw YF, Mizokami M, Kuiken C. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–265. doi: 10.1002/hep.21698. [DOI] [PubMed] [Google Scholar]
- Lupberger J, Hildt E. Hepatitis B virus-induced oncogenesis. World J Gastroenterol. 2007;13:74–81. doi: 10.3748/wjg.v13.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason WS, Litwin S, Xu C, Jilbert AR. Hepatocytes turnover in transient and chronic hepadnavirus infection. J Viral Hepat. 2007;14(Suppl 1):22–8. doi: 10.1111/j.1365-2893.2007.00911.x. [DOI] [PubMed] [Google Scholar]
- Mason WS, Litwin S, Jilbert AR. Immune selection during chronic hepadnavirus infection. Hepatol Intl. 2008;2:3–16. doi: 10.1007/s12072-007-9024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason W, Xu C, Low HC, Saputelli J, Aldrich CE, Scougall C, Grosse A, Colonno R, Litwin S, Jilbert AR. The amount of hepatocytes turnover that occurred during resolution of transient hepadnavirus infections was lower when virus replication was inhibited with entecavir. J Virol. 2009a;83:1778–1789. doi: 10.1128/JVI.01587-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason WS, Low HC, Xu C, Aldrich CE, Scougall CA, Grosse A, Clouston A, Chavez D, Litwin S, Peri S, Jilbert AR, Lanford RE. Detection of clonally expanded hepatocytes in chimpanzees with chronic hepatitis B virus infection. J Virol. 2009b;83:8396–8408. doi: 10.1128/JVI.00700-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason WS, Liu C, Aldrich CE, Litwin S, Yeh MM. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J Virol. 2010;84:8308–8315. doi: 10.1128/JVI.00833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon BJ. The natural history of chronic hepatitis B virus infection. Semin Liver Dis. 2004;24(Suppl 1):17–21. doi: 10.1055/s-2004-828674. [DOI] [PubMed] [Google Scholar]
- Menne S, Cote PJ. The woodchuck as an animal model for pathogenesis and therapy of chronic hepatitis B virus infection. World J Gastroenterol. 2007;13:104–124. doi: 10.3748/wjg.v13.i1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RG. Simultaneous Statistical Inference. 1. McGraw-Hill Book Company; New York: 1966. [Google Scholar]
- Nguyen VT, Law MG, Dore GJ. Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat. 2009;16:453–63. doi: 10.1111/j.1365-2893.2009.01117.x. [DOI] [PubMed] [Google Scholar]
- Ostle B. Statistics in Research: basic concepts and techniques for research workers. 2. Iowa State University Press; Ames, Iowa: 1964. 2nd print. [Google Scholar]
- Papatheodoridis GV, Hadziyannis SJ. Review article: current management of chronic hepatitis B. Aliment Pharmacol Ther. 2004;19:25–37. doi: 10.1046/j.1365-2036.2003.01810.x. [DOI] [PubMed] [Google Scholar]
- Papatheodoridis G, Buti M, Cornberg M, Janssen HL, Mutimer D, Pol S, Raimondo G, Dusheiko G, Lok A, Marcellin P. EASL clinical practice guidelines: management of chronic hepatitis B virus. J Hepatol. 2012;57:167–185. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Pollicino T, Raffa G, Santantonio T, Battista G, Iannello G, Alibrandi A, Squadrito G, Cacciola I, Calvi C, Colucci G, Levrero M, Raimondo G. Replicative and transcriptional activities of hepatitis B virus in patients coinfected with hepatitis B and hepatitis delta viruses. J Virol. 2011;85:432–439. doi: 10.1128/JVI.01609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu LP, Chen L, Chen KP. Antiviral B therapy: a review of current medications and novel small molecule inhibitors. Fundam Clin Pharmacol. 2013;28:364–381. doi: 10.1111/fcp.12053. [DOI] [PubMed] [Google Scholar]
- Rodrigues L, Freitas N, Kallakury BV, Menne S, Gudima SO. Superinfection with woodchuck hepatitis virus strain WHVNY of livers chronically infected with strain WHV7. J Virol. 2015;89:384–405. doi: 10.1128/JVI.02361-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13:14–21. doi: 10.3748/wjg.v13.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüffler PJ, Fuchs TJ, Ong CS, Wild PJ, Rupp NJ, Buhmann JM. TMARKER: A free software toolkit for histopathological cell counting and staining estimation. J Pathol Inform. 2013;4(Suppl):S2. doi: 10.4103/2153-3539.109804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Jilbert AR, Yang W, Aldrich CE, Saputelli J, Litwin S, Toll E, Mason WS. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc Natl Acad Sci USA. 2003;100:11652–11659. doi: 10.1073/pnas.1635109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KA, Joyce MA, Addison WR, Fischer KP, Tyrrell DL. Superinfection exclusion in duck hepatitis virus infection is mediated by large surface antigen. J Virol. 2004;78:7925–7937. doi: 10.1128/JVI.78.15.7925-7937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland S, Spangenberg HC, Thimme R, Purcell RH, Chisari F. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proc Natl Acad Sci USA. 2003;101:2129–2134. doi: 10.1073/pnas.0308478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yamamoto T, Zhou T, Aldrich CE, Frank K, Cullen JM, Jilbert AR, Mason WS. The liver of woodchucks chronically infected with the woodchuck hepatitis virus contains foci of virus core antigen-negative hepatocytes with both altered and normal morphology. Virology. 2007;359:283–294. doi: 10.1016/j.virol.2006.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Saputelli J, Aldrich CE, Deslauriers M, Mason WS. Emergence of drug-resistant populations of woodchuck hepatitis virus in woodchucks treated with the antiviral nucleoside lamivudine. Antimicrob Agents Chemother. 1999;43:1947–1954. doi: 10.1128/aac.43.8.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]