Abstract
Hydrogen exchange (HX) mass spectrometry (MS) of complex mixtures requires a fast, reproducible, and high peak capacity separation prior to MS detection. The current paradigm relies on liquid chromatography (LC) with fast gradients performed at low temperatures to minimize back exchange. Unfortunately, under these conditions, the efficiency of LC is limited due to resistance to mass transfer, reducing the capability to analyze complex samples. Capillary electrophoresis (CE), on the other hand, is not limited by resistance to mass transfer, enabling very rapid separations that are not adversely affected by low temperature. Previously, we have demonstrated an integrated microfluidic device coupling CE with electrospray ionization (ESI) capable of very rapid and high efficiency separations. In this work, we demonstrate the utility of this microchip CE-ESI device for HX MS. High speed CE-ESI of a bovine hemoglobin pepsin digestion was performed in 1 minute with a peak capacity of 62 versus a similar LC separation performed in 7 minutes with peak capacity of 31. A room temperature CE method performed in 1.25 minutes provided similar deuterium retention as an 8.5 minute LC method conducted at 0 °C. Separation of a complex mixture with CE was done with considerably better speed and nearly triple the peak capacity than the equivalent separation by LC. Overall the results indicate the potential utility of microchip CE-ESI for HX MS.
INTRODUCTION
Proteins play a critical role in a number of biological processes including cellular signaling, gene expression, biochemical reaction catalysis, and apoptosis. Understanding the molecular basis of these and many other biological functions requires a thorough comprehension of the structure, dynamics and conformational changes of the proteins and protein complexes involved. This challenging task necessitates experimental techniques that can probe the fundamental characteristics of proteins and help elucidate the link between protein structure and function. In the past decades, this topic has been a great focus in the literature, and a variety of different approaches have been described including nuclear magnetic resonance, X Ray crystallography, small angle X-ray scattering, and cryo-electron microscopy. These techniques have been extremely valuable for many areas, but they are not able to characterize all proteins including those that will not crystallize, those that are highly dynamic in solution, and especially those at and in membranes. Other techniques capable of probing protein conformation and dynamics are therefore extremely valuable in protein analysis. Hydrogen exchange (HX) mass spectrometry (MS), offers great potential for analyses of protein complexes and large protein systems as it provides access to proteins other techniques struggle to analyze.1-2 HX MS does not necessitate protein crystallization, requires very little sample, is amenable to studying proteins that are difficult to purify, and can reveal conformational changes on a wide time scale.3 As a technique, however, it is not without its own set of challenges.
The basic strategy for HX MS derives from the original description of Rosa and Richards4 and consists of labeling proteins in their native state with deuterium, digesting the protein into peptides and quantifying the amount of deuterium uptake at different points along the amino acid backbone of the protein by observing a shift in mass. In proteins, the rate of hydrogen deuterium exchange is governed by essentially four factors: pH, temperature, solvent accessibility, and hydrogen bonding.3 As pH and temperature are experimentally controlled, the measured deuterium uptake indicates the degree to which that part of the molecule was exposed to the solvent and the amount of hydrogen bonding; thereby providing information about the folded structure of the molecule and interactions between regions of the protein and other molecules in solution.5 To conserve the information generated during the labeling step, it is necessary to perform the digestion and separation steps under quench conditions.6,7 This is typically achieved by lowering the pH to 2.5 and the temperature to 0 °C. While these conditions significantly slow the rate of H/D exchange, they do not completely stop it, requiring the digestion and separation to be performed as quickly as possible, generally in less than 10 minutes. Further delay causes significant information loss through H/D back exchange. Liquid chromatography (LC) is the most common separation method for HX MS, and is often performed at low temperatures with fast gradients to minimize the amount of observed back exchange.
HX MS using LC as the separation method is robust for small proteins (<40-50 kDa) and for simple complexes of 1-2 components. However, it’s estimated that nearly every major biological process performed in cells is carried out by protein machines containing at least 10 protein molecules.8 The vast complexity of such targets requires new methodology and technology advances to truly uncover the relationship between structure, function, and dynamics of protein complexes. To date, larger proteins and protein complexes (>150 kDa and 3+ members) remain difficult to study via HX MS because of the huge number of peptides that are generated upon digestion. The LC step does not generally provide adequate separation power (commonly quantified as peak capacity) to allow the large number of peptides generated to be clearly identified and characterized during a deuterium exchange experiment. Because the limiting factor preventing the analysis of increasingly complex samples lies with the limited resolving power of LC at low temperatures and fast run times, improving the separation power is pivotal for expanding the scope of analyzing biological systems using HX MS.
While it has been used for HX MS out of convenience and availability, liquid chromatography is not ideally suited to fast separations at low temperature because it is limited by resistance to mass transfer (the van Deemter C-term). Even the most state of the art, modern Ultra Performance Liquid Chromatography (UPLC), performed under quench conditions, cannot achieve a peak capacity much greater than about 60.9 Higher peak capacity could be obtained with a longer LC separation, but only at the expense of greater deuterium loss, which is not an acceptable alternative. One way to improve these LC separations without increasing the run time would be to use smaller stationary phase particles, but the back pressure required to operate such LC columns would be excessively high and difficult to achieve with commercial systems.10 Another possibility is to keep separation time constant while increasing the flow rate and making the gradient more shallow. Again, this increase in flow rate leads to higher backpressure that may not be easy to achieve. If a different separation modality were available, with superior performance at low temperate, HX MS separations could be improved.
Free zone capillary electrophoresis (CE) separations have been used for HX MS characterization of standard peptides,11 however its application to protein digests remains to be investigated. CE is very fast, efficient, and is not limited by resistance to mass transfer. In the absence of extra-column band-broadening and joule heating (both of which are minimized in microfluidic devices), the efficiency of a CE separation is determined only by the voltage applied and the temperature.12,13 Equation 1 illustrates the relationship between the number of theoretical plates, N, and the applied voltage, V, where D is the diffusion coefficient of the molecule and μ is the electrophoretic mobility.12 It is theoretically possible to achieve the same separation efficiency and peak capacity in any amount of time by varying the length of the separation column while holding constant the voltage applied.
| (1) |
Equation 2, the Stokes-Einstein equation, defines a spherical molecule’s diffusion coefficient, where k is Boltzmann’s constant, η is the solvent viscosity, r is the radius, and T is the temperature.14 Equation 3 defines the electrophoretic mobility of a molecule, where q is the charge of the molecule.15
| (2) |
| (3) |
Substituting Equations 2 and 3 into Equation 1 results in Equation 4, revealing the inverse temperature dependence of the separation efficiency for a CE separation.
| (4) |
As Equation 4 reveals, cooling a CE separation will improve the separation efficiency. However, at constant applied voltage, the overall run time will increase at lower temperatures due to an increase in solvent viscosity.16,17 Increasing the voltage to offset the increased viscosity will reduce the migration time, keeping run time constant and further improving the efficiency of the separation.
Recently, we have demonstrated a microfluidic platform for performing very fast and highly efficient CE separations coupled with electrospray ionization (ESI).18,19 The improved speed and peak capacity of this platform offers many potential benefits for HX MS such as expanding the types of proteins and protein complexes able to be analyzed and reducing back exchange. Furthermore, microfluidic technology is extremely well suited towards integration of multiple functional elements,20,21 such as online digestion and peptide concentration, resulting in the prospect of a fully integrated microchip CE-ESI system for HX MS analysis.22
In this work, we demonstrate the fundamental utility of applying microchip CE-ESI devices towards HX MS. Microchip CE-ESI was used to analyze a pepsin digestion of the model protein bovine hemoglobin. The speed, peak capacity, and reproducibility of the separation were optimized with all the limitations of HX MS in mind. Each parameter was characterized and these metrics were compared to the separation of an identical mixture using a UPLC system. Data independent MS/MS was also performed to determine how peptide identification would perform with such short duration peaks from the microchip CE-ESI method. The deuterium recovery of the CE and LC methods were compared. Finally, the utility of the method for large complex systems was examined by using microchip CE-ESI to separate a 3 protein mixture consisting of 270 kDa of unique sequence.
EXPERIMENTAL SECTION
Reagents and Materials
LC-MS grade acetonitrile and formic acid (99.99%) were acquired from Fisher Chemical (Fairlawn, NJ). Water was purified with a Nanopure Diamond water purifier (Barnstead International, Dubuqe, IA). (3-Amino)di-isopropylethoxysilane (APDIPES) was acquired from Gelest (Morrisville, PA). Bovine hemoglobin, pepsin enzyme, cyanoborohydride coupling buffer, sodium phosphate dibasic, trichloro(1H,1H,2H,2H-perfluorooctyl)silane, and deuterium oxide (99.9%) were acquired from Sigma-Aldrich (St. Louis, MO). AL-20 POROS beads were purchased from Applied Biosystems (Carlsbad, CA). N-hydroxylsuccinimide functionalized polyethylene glycol (NHS-PEG) with 450 polymer units (MW = 20 kDa) was purchased from Nanocs, Inc. (Boston, MA). Human [Glu1] fibrinopeptide was purchased from American Peptide Company (Sunnyvale, CA).
Microchip Design, Fabrication, and Surface Coating
A schematic of the CE-ESI microchip used in this work is shown in Figure S1 of the Supporting Information, and is similar to devices published previously for CE-ESI.23,18 Following fabrication, the microchips were coated with APDIPES in the gas phase and modified with an NHS-PEG reagent as described previously.19,25
Hydrogen Deuterium Exchange Sample Preparation and Workflow
For HX MS, a stock of intact bovine hemoglobin was prepared in 20 mM TRIS buffer, H2O, pH 7.5. For CE-ESI, the stock concentration was 120 mg/mL while for LC-MS, the stock concentration was 24 mg/mL. A 5 μL aliquot of the stock solution was diluted 20-fold in either 20 mM TRIS buffer, H2O, pH 7.5 (undeuterated control) or 20 mM TRIS buffer, D2O, pD 7.5. The intact bovine hemoglobin was exposed to the deuterated buffer for four different time points (10 seconds, 1 minute, 10 minutes, and 60 minutes) and the deuteration reaction quenched by diluting an aliquot of the labeled protein 5-fold in ice cold 0.5% formic acid which lowered the solution pH to 2.5. The protein was immediately digested by placing the quenched protein solution onto a Spin-X 0.22 μm centrifuge tube filter (Corning Incorporated, Corning, NY) loaded with 40 μL of pepsin bead slurry. Pepsin was immobilized via aldehyde coupling.24 The centrifuge tube was vortexed for 30 seconds at room temperature, and then the peptides were separated from the immobilized pepsin beads by 20 seconds of centrifugation at room temperature (CE) or 4 °C (LC).
Microchip CE-ESI Operation
Following sample preparation, the bovine hemoglobin peptide solution was mixed 1:1 with acetonitrile and loaded into the sample reservoir of the CE-ESI microchip. Immediately after loading the sample on the microchip, voltage was applied to the microchip. After waiting 15 seconds to ensure that the sample had reached the injection cross, the sample was injected onto the separation channel where the bovine hemoglobin digest underwent CE separation and detection via ESI-MS. The total time between quenching the deuteration reaction and injecting the sample onto the separation channel was between 100 and 110 seconds.
CE-ESI microchips were operated by the application of voltages to the solvent reservoirs, as previously described.18,19 Home built reservoir caps containing platinum wire electrodes (Alpha Aesar, Ward Hill, MA) were used to form voltage connections to each reservoir. A home-built power supply was used containing five individual power supply modules from Ultravolt Inc. (Ronkonkoma, NY). Three of the modules (20A24-P15) supplied 0 to +20 KV while the other two modules (10A12-P4) could supply 0 to +10 KV. The power supply was controlled via an SCB-68 breakout box connected to a PC via a PCI 6713 DAQ card. A LabVIEW program was used to control the voltage outputs. During microchip operation, voltages of +20, +20, +18, and +1 kV were applied to the S, B, W, and EO reservoirs, respectively. The applied voltages resulted in a field strength of 1500 V/cm. These conditions yielded a flow rate of approximately 630 nL/min exiting the ESI emitter of the microchip. To inject sample into the separation channel, the applied voltages were changed to +20, +19, +19, +1 kV at the S, B, W, and EO reservoirs, respectively. The length of the injection was 0.3 seconds. The background electrolyte for all CE separations was comprised of 50% acetonitrile, 0.5% formic acid, 49.5% water, pH 2.5. 10 nM glu-fibrinopeptide in BGE was placed in the EO pump reservoir to serve as a lock-mass reference compound (785.852+ ion). Lock-mass spectra were acquired before and after each 1 minute CE separation. The lock-mass scan time was 1 second with an interval of 70 seconds. Lock-mass data were acquired but not applied in real time; 3 scans were averaged with a mass window of ± 0.5 Da.
The CE-ESI device was mounted on a custom built stage with the ESI corner 5 mm from the sample cone inlet of a Synapt G2 mass spectrometer (Waters Corporation, Milford, MA). A copper clad circuit board with 0.5 kV applied to it was used to shield the ESI emitter from the voltages applied to the microfluidic device as described previously.19 The instrument was run in resolution mode, with a summed scan time of 50 ms and an interscan delay of 24 ms. For the undeuterated controls, data-independent MS/MS was used to fragment and identify the peptides utilizing the Waters MSE function. In MSE, the collision cell rapidly switches between low and high collision cell energy, producing alternate precursor and fragment ion information.25 The low collision energy precursor scans had a trap energy equal to 6 V while the high energy fragmentation scans had a trap energy of 32 V. MSE data were acquired from 50 – 1200 m/z. For the labeled time points, MS only data were acquired from 300 – 1200 m/z.
LC-MS Operation
LC-MS Hb deuterium exchange samples were prepared in a similar manner as those for CE-ESI, however the stock Hb concentration was reduced to 24 mg/mL. To minimize carryover, the pepsin digest was diluted 1:1 with 1:4 20 mM TRIS:0.5% formic acid solution prior to injection. The sample (50 μL) was injected into a Waters M-class nanoACQUITY UPLC with HX MS technology. The total time from quenching the labeling reaction and injection onto the instrument was 75 seconds. Peptides were trapped and desalted for 3 min at 100 μL/min using a Waters VanGuard BEH C18 1.7 μm trap column. Chromatographic separation was performed using a 1 × 50 mm Waters ACQUITY UPLC C18 (HSS T3) reversed-phase column at 0 °C. Peptides were eluted over 3 min with a linear 5-35% acetonitrile gradient at 100 μL/min. Mass spectra were obtained using a Waters Synapt G2Si operating in positive ion mode at a capillary voltage of 3 kV and a desolvation temperature of 175 °C. For MSE, the low collision energy precursor scans had a trap energy of 4 V and high energy fragmentation scans had a trap energy of 32 V. The instrument was run with a summed scan time of 200 ms and an interscan delay of 24 ms. Human Glu-fibrinopeptide was infused at 5 μL/min through the reference probe and scanned every 10 s, with the 785.852+ ion used as a lockmass.
Complex Mixture Sample Preparation and Separation
A mixture containing 48 pmol phosphorylase b, 95 pmol Rpn1, and 82 pmol Ubp6 in 20 mM Tris was mixed 1:1 with 0.8 M GdnHCl, 0.8% formic acid. Digestion was initiated via addition of 7.5 μL immobilized pepsin, and proceeded for 5 min on ice. Resultant peptides were isolated by filtering through a Spin-X 0.22 μm centrifuge tube filter (Corning Incorporated, Corning, NY), centrifuged at 7800 g for 30 s. For LC-MS, the sample was analyzed in a similar manner to the Hb samples, however peptides were eluted from a 1 × 100 mm Waters ACQUITY UPLC C18 (BEH) column using a linear 5-40% acetonitrile gradient over 9 min.
For CE-ESI, following digestion, the mixture sample was concentrated and desalted using an Oasis HLB solid phase extraction cartridge (Waters Corporation, Milford, MA). 300 μL of 3 μM digest material was loaded on the cartridge, washed with 500 μL of 0.5% formic acid, and eluted with 100 μL of 90% acetonitrile, 0.05% formic acid. The sample was separated using a 23 cm CE-ESI microchip. The chip operation was identical to that already described with the following exceptions: The BGE was 50% acetonitrile, 1% formic acid. Voltages of +22, +22, +20, and +1.5 kV were applied to the S, B, W, and EO reservoirs, respectively, resulting in a field strength of 767 V/cm. To inject sample into the separation channel, the applied voltages were changed to +22, +21, +21, +1.5. The length of the injection was 0.8 seconds. The Synapt G2 was run in resolution mode with a summed scan time of 100 ms with an m/z range of 300 - 1200.
Data Processing
The software program Peakfinder (previously available from PNNL) was used to calculate the separation window, median 4σ peak width, and peak capacity for all separations. Peptides from both CE and LC experiments were identified using ProteinLynx Global Server 3.0 and deuterium incorporation was calculated with DynamX 2.0 software. The following parameters were utilized by DynamX to filter the PLGS output data prior to calculating deuterium uptake, protein sequence coverage, and peptide redundancy: 0.3 products per amino acid, 3 consecutive products, 10 ppm mass error on parent ion, identified in all undeuterated control runs.
RESULTS AND DISCUSSION
CE-ESI Separation Performance
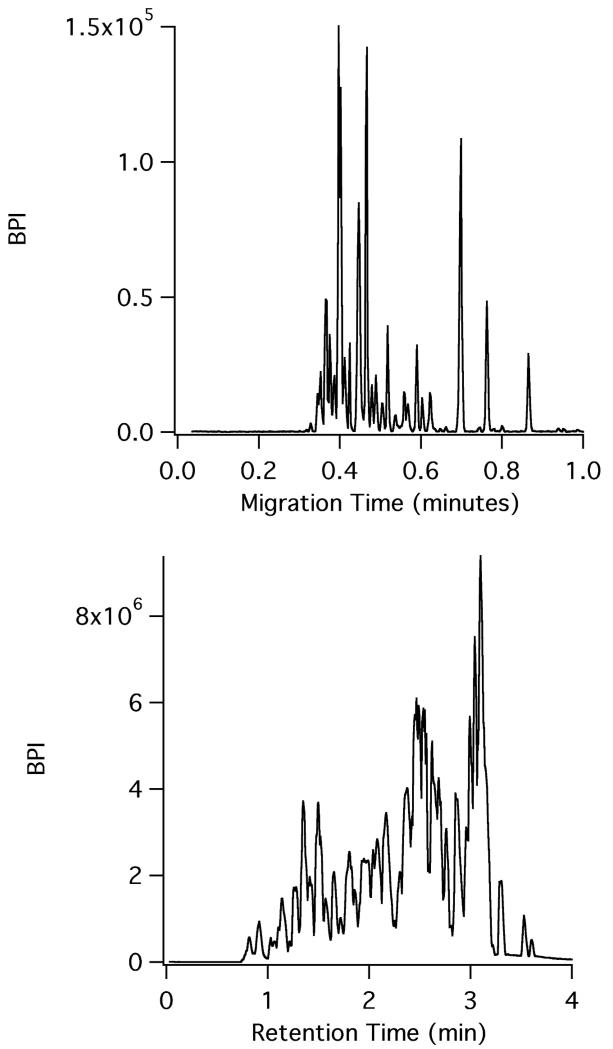
To demonstrate the validity of CE-ESI for separations under conditions that would be found in a hydrogen exchange mass spectrometry experiment, CE-ESI microchips were used to analyze a pepsin digest of bovine hemoglobin. The microchips utilized a surface coating and chip operation scheme recently described for intact protein separations;26 this is the first report of using these chips for analyzing peptide digests. The surface coating reduces the electroosmotic flow in the separation channel to near zero; the movement of analytes is primarily due to electrophoretic migration. Figure 1 compares a microchip CE-ESI-MS electropherogram of a bovine hemoglobin pepsin digest to the LC-MS separation of the same sample using a state-of-the-art UPLC system for HX MS experiments, the Waters M-Class. As demonstrated by the figure, the CE separation of the hemoglobin digest, which took place at room temperature, is very fast and completed in less than 1 minute. Run-to-run reproducibility was high. Three replicate injections of the hemoglobin digest were utilized to assess the run-to-run migration time reproducibility (data not shown). For three representative peptides spanning the window of the electropherogram, the migration time % relative standard deviation (RSD) was calculated to be 0.75%. The observed median width at base for the peaks in CE (n = 29) was 0.61 seconds, corresponding to a peak capacity of 62. For such a rapid separation, this represents an extremely high peak capacity. The metric of peak capacity/minute represents how quickly a separation technique can generate resolving power. After accounting for the time between sample loading and injection on the microchip, the hemoglobin digest CE-ESI separation has a calculated peak capacity/minute of 53.4. In comparison, Busnel et. al. utilized a sheathless porous tip to perform CE-ESI of protein digests and in a 60 minute separation, a peak capacity of 193 was reported (using 4σ peak width), corresponding to a peak capacity/minute of 3.2.27 By contrast, a typical LC separation peak capacity generally ranges from 100 – 400, but can take several hours to achieve these results, yielding peak capacity/minute values in the range of 1.5 – 3.9.28,10
Figure 1.
CE-MS (top) versus LC-MS (bottom) of the same bovine hemoglobin pepsin digest. CE-ESI was performed at room temperature while LC-MS was performed at 0 °C.
To assess the separation performance of the CE-ESI microchip relative to the current paradigm for HX MS experiments, we compared the microchip CE separation at room temperature to a UPLC separation performed at 0 °C of the same bovine hemoglobin digest. As illustrated by Figure 1, the LC separation run time is just under 4 minutes. After adding the 3 minute trapping time prior to the LC separation, the time between sample loading and detection is 7 minutes for the last eluting species in the LC run. The observed peak capacity of the chromatogram was 31 with a median peak width (n = 16) of 5.35 seconds. For the hemoglobin digest sample, the LC chromatogram has a peak capacity/minute of 4.4. This comparison highlights the speed and resolving power of microchip CE compared to LC; the peak capacity/minute of the CE electropherogram is more than an order of magnitude greater than the LC separation. Table 1 compares figures of merit for the CE-ESI and LC-MS methods, highlighting differences in speed, peak width, and separation performance. The table further illustrates the improved performance coupled with faster run times, demonstrating the potential for utilizing CE-ESI for HX MS experiments. We note that in one of the early reports6 of the fragment separation method for HX analysis of hemoglobin, then detected by scintillation counting, the HPLC run time was in excess of 90 minutes. The microchip CE-ESI separation time described in this work is 100-times faster.
Table 1.
Comparison of CE and LC figures of merit for bovine hemoglobin pepsin digest separation
| Characteristic | LC | CE |
|---|---|---|
| Separation speed (min) | 3-12 | 1-4 |
| Peak width (s) | 5-10 | <1 |
| Injection amount (fmol) | ~50000 | ~5 |
| Flow rate (μL/min) | 50-100 | <1 |
| Hb peak capacity/min | 4.4 | 62 |
| Retention/Migration time %RSD |
0.4 | 1.1 |
| Commercial availability | Waters M-Class UPLC with HDX Technology |
None |
| MS modifications required | None | Moderate source changes |
MS Performance
Data-independent MS/MS (MSE) of a hemoglobin pepsin digest was performed to identify the peptic peptides separated by fast CE-ESI. To the best of our knowledge, this work represents the fastest ever utilization of MSE. In 2010, Bonn et al. reported LC-MSE of metabolites with a summed scan time of 100 ms.29 In this manuscript, the summed scan speed was 50 ms, sampling twice as fast. Figure S2 in Supporting Information shows representative peaks from both the microchip CE-ESI electropherogram and the LC-MS chromatogram, illustrating the number of MS data points across each peak. For the CE separation, a summed scan time of 50 ms and an interscan delay of 24 ms correspond to a data acquisition rate of approximately 13.5 Hz. While performing the MSE function, data points are alternated between low and high collision energy; therefore, for each scan, the MS sampling rate decreases to roughly 6 Hz per trace. Given the average peak width at base in the electropherogram of 0.61 seconds, the average CE peak will have 3.8 data points per peak at each collision energy. As a comparison, the average LC peak from the hemoglobin chromatogram will have 12 data points per peak at each collision energy. This indicates one of the challenges associated with coupling high speed separations to MS. Although mass spectrometers have improved greatly over the past decades, coupling to extremely fast separations remains challenging, and often mass spectrometers struggle to maintain adequate sampling of narrow microchip CE peaks.30 The 3.8 data points per peak results in peak undersampling, artificially broadening the reported peak widths. Therefore, a faster acquisition rate would perhaps reveal even greater separation performance than that reported here. While there is concern that the observed peak undersampling would diminish the ability to identify peptides with MSE, the preliminary results from this work seem to indicate that this is not the case. The data could still be processed by commercially available software to identify peptides in the electropherogram.
Accurate mass measurements are crucial for reliable peptide identification.31 For ESI, this is most often accomplished utilizing internal calibration from a reference compound, or ‘lock-mass’.32 There are multiple ways of introducing the lock-mass compound into the mass spectrometer. Typically, the lock-mass compound is either introduced post-column at a ‘T’ junction,33 which can introduce band broadening to the separation, or multiple emitters are utilized,34 introducing instrumental complexity and data sampling concerns.35 Another strategy is to mix the reference compound directly with the mobile phase of an LC separation, however, interaction with the stationary phase can limit the choice of an acceptable lock-mass compound and may have implications on the separation performance. Microchip CE-ESI offers a unique solution to this problem. We simply added a very low concentration (10 nM) of a lock-mass compound (glu-fibrinopeptide B) to the BGE in the EO pump reservoir of the microchip. Based on the applied voltages, fluid from the EO pump reservoir flows towards the ESI emitter, comprising the bulk fluid flow necessary for ESI generation. Lock-mass spectra were acquired at the beginning of each CE-ESI electropherogram. The short CE analysis time eliminates the need to make further lock-mass acquisitions. This strategy avoids both band-broadening and data sampling concerns and provides a much simpler solution than using dual emitters. This approach is only possible using the described microchip CE-ESI platform, and would not be amenable for use with an LC-MS system. Using this strategy, we observed a mass error of less than 5 ppm for the identified peptide parent ions. In the absence of the lock-mass compound, a mass error of near 50 ppm was typically observed (data not shown).
Sequence Coverage and Limitations
For HX MS experiments aimed at analyzing intact proteins or protein complexes, maximizing the structural information contained in the peptide digest data is paramount. Protein sequence coverage and peptide redundancy are important metrics when evaluating MS data for HX MS. Following peptide identification with PLGS, the data were filtered with DynamX prior to calculating sequence coverage and peptide redundancy. For the microchip CE-ESI separation of bovine hemoglobin pepsin digest, the sequence coverage of the α and β chain was 48.2% and 84.8% with a redundancy score of 3.29 and 1.29, respectively. For the LC-MS method, the sequence coverage was 97.9% and 92.4% with redundancy scores of 6.62 and 5.20. While initially concerned that the mass accuracy may be contributing to the poor sequence coverage, analysis of the data revealed that the mass accuracy was quite comparable between the two methods. The average mass error for the CE and LC methods was ± 1.5 ppm and ± 3.0 ppm respectively for identified peptides. While the data shown here indicate that microchip CE-ESI has less than ideal sequence coverage, low coverage at this early stage of development should not detract from the potential of utilizing high speed CE separations for HX MS. The decreased sequence coverage and redundancy scores are likely the result of the several factors discussed below, including changes to separation speed, sensitivity, injection volume and preconcentration. Many of these aspects can be addressed in future method development.
Speed
Increasing the speed of separation while maintaining, or enhancing, separation performance necessitates short duration peaks. As discussed earlier, the average peak width of the CE separation is roughly an order of magnitude narrower than the LC peaks (0.6 seconds v 5.35 seconds). If all else were equal, this would adversely affect the sensitivity of the CE method as the mass spectrometer has approximately one-tenth the time to acquire spectra. Furthermore, in certain cases the narrow peak width may have diminished the ability of the PLGS software to identify the peaks regardless of their concentration. While many more peptides were identified with the LC-MS method, certain peptides of corresponding m/z and charge state were also observed in the CE method, but were not positively identified by the software. In most cases, these peptides were above the intensity thresholds set for PLGS, therefore, we attribute the missing data to the narrow peak widths.
Sensitivity
The LC data used for comparison were acquired using a more sensitive mass spectrometer (Synapt G2Si) than the CE data (Synapt G2), a difference easily remedied. The difference in mass spectrometer scan speed (200 ms scan time for the LC v. 50 ms for the CE) between the two methods exacerbates the sensitivity variation between the two mass spectrometers.
Injection volume
In order to perform an extremely rapid, high peak capacity separation, the injection volume must be as low as possible in order to minimize the injection broadening contribution to the peak width. For the microchip CE separation, the injection volume was 600 pL, compared to the 50 μL injected onto the LC column. The difference in initial protein concentration was done intentionally to partially offset the difference in injection volume; however, when comparing amount injected, the LC method used 17,000 times more sample than the CE method. For the initial protein stock concentration of 120 mg/mL, the amount of sample injected onto the CE separation capillary was approximately 350 pg. For the LC method, an initial protein stock concentration of 24 mg/mL corresponded to 6 μg injected on column, more than 4 orders of magnitude greater than the amount utilized for the CE method.
Preconcentration
LC-MS as implemented for this comparison has a significant advantage in that there is an integrated sample pre-concentration step resulting from the use of a trap column. By loading a large volume of sample and concentrating at the head of the LC column, a significant increase in peak concentration can be observed. Unfortunately, this type of sample processing is more difficult to integrate with CE-ESI and was not done in the current comparison. Although a significant focus in the literature, a robust method for coupling sample processing with CE-ESI remains challenging.36,37 For the LC method, concentrating a large sample at the head of the column and eluting a narrow injection band resulted in a peak concentration increase, boosting sensitivity. The injection volume of the LC sample was 50 μL; the average peak volume at detection was 8.9 μL, resulting in a 5.6-fold peak concentration. Conversely, the injection volume of the CE separation was 600 pL, while the average peak volume at detection was 1.15 nL, resulting in a nearly 2-fold dilution. It is possible to overcome the sample preconcentration limitations of the microchip CE-ESI system described here, thereby eliminating the sensitivity issues and peptide coverage difficulties apparent in this initial comparison. Furthermore, integrating sample processing onto the CE-ESI microchip will likely improve the compatibility of the method with the high concentration of salt, reducing agents, and denaturants commonly employed in HX MS workflows. Such developments will be the focus of a future report.
Deuterium Recovery
Deuterium recovery must be addressed in any new HX MS method. After demonstrating the separation power and MS performance of the CE-ESI microchip with undeuterated protein, a deuterium labeling experiment of the bovine hemoglobin digest was performed to compare deuterium uptake between the CE and LC methods. Peaks between the control and deuterated time points were matched based on either retention or migration time, underscoring the need for an extremely reproducible separation. Over the course of the labeling experiment, the CE method had a migration time reproducibility of 1.14% RSD. For the LC separations, the % RSD for retention time was 0.38%. Unlike the LC method, the CE separation was not temperature controlled, which may have contributed to the observed migration time variance. Despite the extremely narrow CE peaks, the commercial Waters HX software (DynamX) was capable of matching peaks between the undeuterated control run and the labeled time points in order to quantify the changes in deuterium uptake.
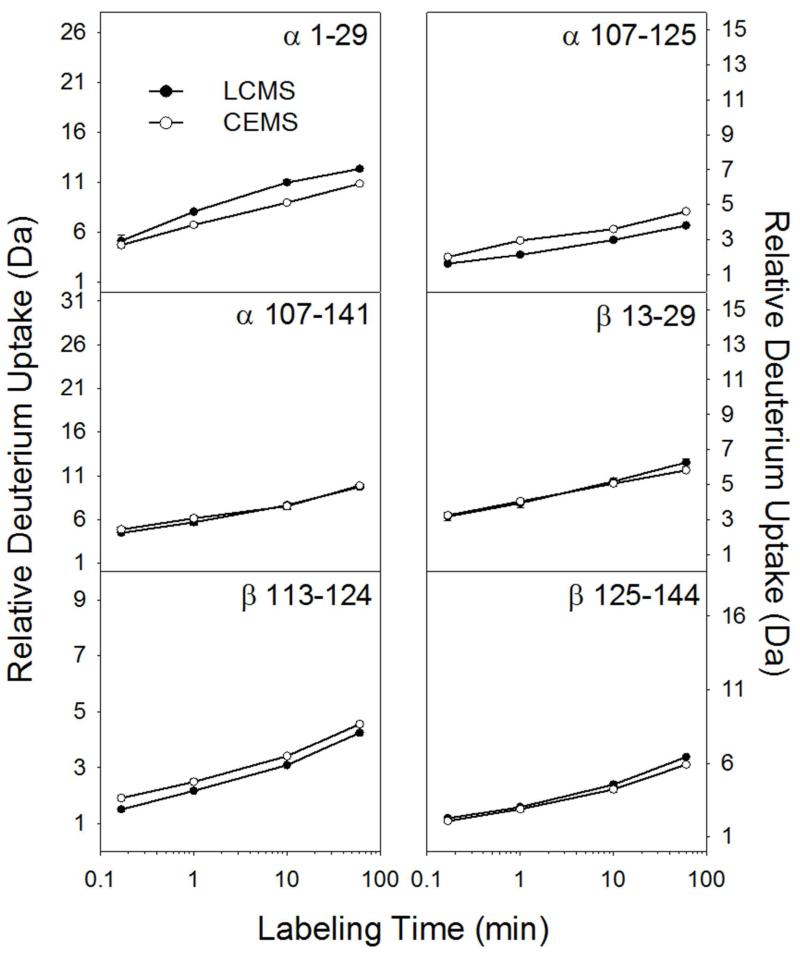
Figure 2 illustrates the comparison of deuterium uptake between CE and LC for 6 hemoglobin peptides, 3 from the α chain and 3 from the β chain. Note that the separations occurred at different temperatures: the CE separation was performed at room temperature while the LC separation was performed at 0 °C (cooling the CE-ESI chip to 0 °C could be done with a Peltier device and this addition will be the subject of a future report). Decreasing the temperature to 0 °C will reduce the rate of H/D back exchange approximately 10-fold versus room temperature.38 Given this difference in temperature, we would expect the CE method to result in roughly 10 times more deuterium loss (because it was done at higher temperature) than the LC method. However, the CE method was faster overall, reducing the time available for back exchange. We define the time window for back exchange as the time between quenching the deuterium labeling reaction and completing the separation. For the CE method, the time window for back exchange was 2.5 minutes (1 minute separation plus 1.5 minutes of digestion and sample preparation). For the LC separation, the time window was 8.5 minutes (4 minute separation, 3 minutes of trapping, 1.5 minutes of digestion and sample prep). Weighing the differences in time and temperature between CE and LC, the amount of deuterium should be similar,38 as Figure 2 confirms. Three peptides (α107-141, β13-29, and β125-144) displayed nearly identical deuterium uptake, α1-29 retained more deuterium using the LC method, and α107-125 and β113-124 slightly more deuterium using the CE method. These small differences were reconciled by accounting for experimental temperature and peptide elution time.38
Figure 2.
Measured deuterium level comparison for six representative peptic peptides from bovine hemoglobin. Error bars, representing standard deviation of duplicate measurements, are present but in most cases too small to see.
Complex Mixture Analysis
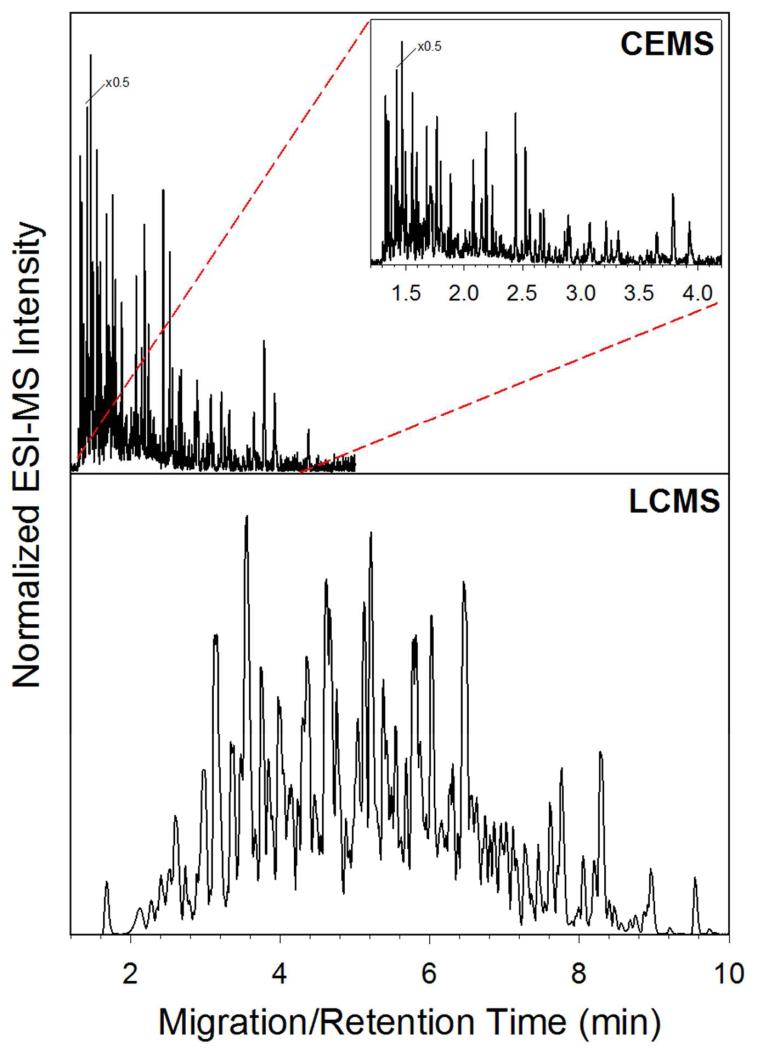
The analysis of protein complexes and complicated systems serves as one of the primary reasons for pushing microchip CE-ESI forward as a method for HX MS. In order to demonstrate the improved speed and separation power of CE-ESI, a complex mixture of 270 kDa was separated using both CE-ESI and LC-MS. Figure 3 illustrates the electropherogram (top) and chromatogram (bottom) of the sample. As illustrated by the figure, the CE separation is much faster and efficient compared to the LC separation. The CE run time is around 4 minutes, while the LC run time is near 14 minutes. For the CE electropherogram, the observed median peak width was 0.79 seconds (n = 36), resulting in a peak capacity of 195. For the LC chromatogram, the median peak width was 7.6 seconds (n = 47), resulting in an observed peak capacity of 77. This comparison illustrates the potential for CE-ESI as a platform for HX MS experiments; the CE separation is over 3 times faster with more than double the peak capacity for a protein complex mixture.
Figure 3.
Comparison of CE-ESI electropherogram (top) and LC-MS chromatogram (bottom) of 270 kDa complex mixture separation.
CONCLUSIONS
In this report, we have demonstrated that microchip CE-ESI is fast, reproducible, and offers increased resolving power for HX MS experiments. Microchip CE-ESI of a bovine hemoglobin pepsin digest resulted in vastly superior peak capacity (62 in less than 60 seconds) compared to a state of the art UPLC separation of the same sample (peak capacity of 31 in 7 minutes). Faster separation means higher deuterium recovery, all with improved separative performance. These innovations are possible due to implementing the CE-ESI monolithically on a microchip. In the simple tests shown here, the CE method observed similar amounts of deuterium uptake when compared against the LC method even though the temperature and time parameters were different. Future reports will describe both the operation of microchip CE-ESI at low temperatures as well as integrating digestion and sample processing directly on a microchip to further minimize experiment processing time and back exchange and increase automation. An additional benefit of integrating these functionalities onto a single microchip could be to reduce the overall sample consumption, which could provide a significant advantage in a sample limited setting. While the sequence coverage for the CE method left room for improvement, the results were attributed to the speed of the CE separation and the decreased sensitivity when compared to the LC method. Integrating pre-concentration and sample processing with microchip CE-ESI will be critical in improving the sensitivity of the method in order to increase the observed sequence coverage. The improved separation performance for complex mixtures was clear in the CE-ESI method; not only was the separation faster, but the peak capacity was higher. Even more complex systems could therefore be within reach, including large protein machines and systems. Overall, we believe that microchip CE-ESI presents a promising platform for HX MS. A fully integrated and automated CE-ESI microchip could provide a very powerful tool for the HX MS analysis of complex mixtures, greatly expanding the scope of biological systems that can currently be analyzed.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the University of North Carolina at Chapel Hill Center for Biomedical Microdevices and the National Institutes of Health (Grant R01-GM101135) and a research collaboration between NEU and Waters Corporation for support of this work. The authors would like to acknowledge Dan Finley for providing Rpn1 and Ubp6 proteins and Keith Fadgen at Waters Corporation for assistance with DynamX software.
Footnotes
SUPPORTING INFORMATION
Supporting information includes further details regarding microchip design, fabrication, and surface coating as well as figures illustrating the microchip CE-ESI schematic and representative data sampling peak profiles for bovine hemoglobin digest. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Wales TE, Engen JR. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- (2).Konermann L, Pan J, Liu YH. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- (3).Marcsisin SR, Engen JR. Anal Bioanal Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Rosa JJ, Richards FM. J. Mol. Biol. 1979;133:399–416. doi: 10.1016/0022-2836(79)90400-5. [DOI] [PubMed] [Google Scholar]
- (5).Engen JR, Smith DL. Anal Chem. 2001;73:256A–265A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- (6).Englander JJ, Rogero JR, Englander SW. Anal. Biochem. 1985;147:234–244. doi: 10.1016/0003-2697(85)90033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhang ZQ, Smith DL. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Alberts B. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- (9).Wales TE, Fadgen KE, Gerhardt GC, Engen JR. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jorgenson JW. Annu Rev Anal Chem (Palo Alto Calif) 2010;3:129–150. doi: 10.1146/annurev.anchem.1.031207.113014. [DOI] [PubMed] [Google Scholar]
- (11).Lau SSM, Stainforth NM, Skellern GG, Wren SAC, Tettey JNA. Electrophoresis. 2008;29:393–400. doi: 10.1002/elps.200700368. [DOI] [PubMed] [Google Scholar]
- (12).Jorgenson JW, Lukacs KD. Clin Chem. 1981;27:1551–1553. [PubMed] [Google Scholar]
- (13).Jacobson SC, Hergenroder R, Koutny LB, Ramsey JM. Analytical Chemistry. 1994;66:1114–1118. [Google Scholar]
- (14).Young ME, Carroad PA, Bell RL. Biotechnol Bioeng. 1980;22:947–955. [Google Scholar]
- (15).Landers JP. Handbook of capillary electrophoresis. 2nd ed. CRC Press; Boca Raton: 1997. p. 894. [Google Scholar]
- (16).Nelson RJ, Paulus A, Cohen AS, Guttman A, Karger BL. J Chromatogr. 1989;480:111–127. [Google Scholar]
- (17).Knox JH, Mccormack KA. Chromatographia. 1994;38:207–214. [Google Scholar]
- (18).Mellors JS, Gorbounov V, Ramsey RS, Ramsey JM. Anal Chem. 2008;80:6881–6887. doi: 10.1021/ac800428w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Batz NG, Mellors JS, Alarie JP, Ramsey JM. Anal Chem. 2014;86:3493–3500. doi: 10.1021/ac404106u. [DOI] [PubMed] [Google Scholar]
- (20).Chambers AG, Mellors JS, Henley WH, Ramsey JM. Anal Chem. 2011;83:842–849. doi: 10.1021/ac102437z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ramsey JD, Jacobson SC, Culbertson CT, Ramsey JM. Anal Chem. 2003;75:3758–3764. doi: 10.1021/ac0264574. [DOI] [PubMed] [Google Scholar]
- (22).Rob T, Gill PK, Golemi-Kotra D, Wilson DJ. Lab Chip. 2013;13:2528–2532. doi: 10.1039/c3lc00007a. [DOI] [PubMed] [Google Scholar]
- (23).Mellors JS, Jorabchi K, Smith LM, Ramsey JM. Anal Chem. 2010;82:967–973. doi: 10.1021/ac902218y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ahn J, Jung MC, Wyndham K, Yu YQ, Engen JR. Anal Chem. 2012;84:7256–7262. doi: 10.1021/ac301749h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK. Rapid Commun Mass Sp. 2006;20:1989–1994. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- (26).Redman EA, Batz NG, Mellors JS, Ramsey JM. Anal Chem. 2015;87:2264–2272. doi: 10.1021/ac503964j. [DOI] [PubMed] [Google Scholar]
- (27).Busnel JM, Schoenmaker B, Ramautar R, Carrasco-Pancorbo A, Ratnayake C, Feitelson JS, Chapman JD, Deelder AM, Mayboroda OA. Anal Chem. 2010;82:9476–9483. doi: 10.1021/ac102159d. [DOI] [PubMed] [Google Scholar]
- (28).Gilar M, Daly AE, Kele M, Neue UD, Gebler JC. J Chromatogr A. 2004;1061:183–192. doi: 10.1016/j.chroma.2004.10.092. [DOI] [PubMed] [Google Scholar]
- (29).Bonn B, Leandersson C, Fontaine F, Zamora I. Rapid Commun Mass Spectrom. 2010;24:3127–3138. doi: 10.1002/rcm.4753. [DOI] [PubMed] [Google Scholar]
- (30).Evans CR, Jorgenson JW. Anal Bioanal Chem. 2004;378:1952–1961. doi: 10.1007/s00216-004-2516-2. [DOI] [PubMed] [Google Scholar]
- (31).Bristow AW. Mass Spectrom Rev. 2006;25:99–111. doi: 10.1002/mas.20058. [DOI] [PubMed] [Google Scholar]
- (32).Liu T, Belov ME, Jaitly N, Qian WJ, Smith RD. Chem Rev. 2007;107:3621–3653. doi: 10.1021/cr068288j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Charles L. Rapid Commun Mass Spectrom. 2003;17:1383–1388. doi: 10.1002/rcm.1060. [DOI] [PubMed] [Google Scholar]
- (34).Jiang L, Moini M. Anal Chem. 2000;72:20–24. doi: 10.1021/ac990777e. [DOI] [PubMed] [Google Scholar]
- (35).Chambers AG, Ramsey JM. Anal Chem. 2012;84:1446–1451. doi: 10.1021/ac202603s. [DOI] [PubMed] [Google Scholar]
- (36).Puig P, Borrull F, Calull M, Aguilar C. Anal Chim Acta. 2008;616:1–18. doi: 10.1016/j.aca.2008.03.062. [DOI] [PubMed] [Google Scholar]
- (37).Ramautar R, Jong GJ, Somsen GW. Electrophoresis. 2012;33:243–250. doi: 10.1002/elps.201100453. [DOI] [PubMed] [Google Scholar]
- (38).Bai Y, Milne JS, Mayne L, Englander SW. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.