Abstract
Canonical histones are synthesized with a peak in S-phase, whereas histone variants are formed throughout the cell cycle. Unlike messenger RNA (mRNA) for all other genes with a poly(A) tail, canonical histone mRNAs contain a stem-loop structure at their 3’-ends. This stem-loop structure is the binding site for the stem-loop binding protein (SLBP), a protein involved in canonical histone mRNA processing. Recently, we found that arsenic depletes SLBP by enhancing its proteasomal degradation and epigenetically silencing the promoter of the SLBP gene. The loss of SLBP disrupts histone mRNA processing and induces aberrant polyadenylation of canonical histone H3.1 mRNA. Here, we present new data supporting the idea that the lack of SLBP allows the H3.1 mRNA to be polyadenylated using the downstream poly(A) signal. SLBP was also depleted in arsenic-transformed bronchial epithelial cells (BEAS-2B), which led us to hypothesize the involvement of SLBP and polyadenylated H3.1 mRNA in carcinogenesis. Here, for the first time, we report that overexpression of H3.1 polyadenylated mRNA and knockdown of SLBP enhances anchorage-independent cell growth. A pcDNA-H3.1 vector with a poly(A) signal sequence was stably transfected into BEAS-2B cells. Polyadenylated H3.1 mRNA and exogenous H3.1 protein levels were significantly increased in cells containing the pcDNA-H3.1 vector. A soft agar assay revealed that cells containing the vector formed significantly higher numbers of colonies compared to wild-type cells. Moreover, small hairpin RNA for SLBP (shSLBP) was used to knockdown the expression of SLBP. Cells stably transfected with the shSLBP vector grew significantly more colonies in soft agar than cells transfected with a control vector. This data suggests that upregulation of polyadenylated H3.1 mRNA holds potential as a mechanism to facilitate carcinogenesis by toxicants such as arsenic that deplete SLBP.
Keywords: Stem-loop binding protein, histone H3.1, Arsenic, Histone mRNA, polyadenylation, cell transformation
1. Introduction
Canonical histones
Nuclear DNA is tightly packed in chromatin about 10,000-fold [1, 2]. Nucleosomes are key players in the compaction of chromatin where they assemble 147 base pairs of DNA around their core octamer, although this number changes in nucleosomes containing histone variants [3, 4]. Each nucleosome is composed of an octamer containing two of each of the canonical histones- histone 2A (H2A), histone 2B (H2B), histone 3 (H3), and histone 4 (H4). Histone 1 (H1) is also a canonical histone, often called a linker histone because it binds and protects linker DNA that connects two nucleosomes [4, 5]. Here, canonical means “standard” or “typical”, referring to the most common histones found in the nucleus. Canonical histones are encoded by replication-dependent histone genes and expressed with a peak in S phase. Canonical histones within the nucleosome can be replaced by variant histones, such as H2A.X, H2A.Z and H3.3, to alter chromatin structure and influence gene expression [6]. Variant histones are encoded by replication-independent histone genes and expressed throughout the cell cycle. In mammalian cells, there are two canonical histones for H3, i.e. H3.1 and H3.2. H3.2 differs from H3.1 by only one amino acid and this single amino acid change is not known to impart different functional properties to the two isoforms. On the other hand, histone variant H3.3, which differs from H3.1 by 5 amino acids, gives rise to distinctive nucleosome properties and regulates gene expression differently than H3.1 [6].
Canonical histone mRNA processing
Histone genes have remained highly conserved during evolution and vertebrates have retained multiple copies of these genes. Humans, mice and chickens, for example, have about 10–15 copies of each of the canonical histone genes [7]. Interestingly, the replication-dependent canonical histone genes are the only metazoan genes whose messenger RNA (mRNA) does not terminate at the 3’-end with a poly(A) tail. Instead, the histone mRNAs end in a conserved 26-nucleotide sequence that can form a stem-loop structure, consisting of a four-nucleotide loop and a six base-pair stem. These 26 highly conserved nucleotides are the binding site for the stem loop binding protein (SLBP) [7]. Like the canonical histone genes, SLBP expression is limited to S-phase. SLBP is critical for canonical histone pre-mRNA processing and histone mRNA translation [8].
SLBP binds to the stem-loop structure at the 5’-end, while a 3’–5’exonuclease known as 3’hExo binds to and trims the 3’ end.. SLPB and 3’hExo bind cooperatively to the stem-loop structure, binding of one protein induces a structural change that increases the affinity for the other to bind [9]. SLBP binding occurs in the nucleus and it accompanies the mRNA transcript into the cytoplasm. SLBP is a component of the histone messenger ribonucleoprotein particle (mRNP), where it participates in efficient translation of histone mRNA [10]. The N-terminal domain of SLBP interacts with the Slo-interacting protein 1 (SLIP1) to initiate translation [11]. Expression of H3.1 mRNA can occur outside of S-phase. While H3.1 is nomrally expressed only during S phase, it may also be expressed in other phases of the cell cycle if the cell is responding to DNA damage. However, this low level, outside of S-phase expression of H3.1, is still coupled with DNA synthesis and the histone mRNA still contains a stem-loop structure [12].
SLBP degradation
SLBP accumulation is a dynamic process and is regulated by the cell cycle. As cells enter S-phase, SLBP levels increase more than 20-fold; Levels rapidly fall at the S/G2 boundary due to proteasomal degradation [13, 14]. SLBP proteasomal degradation is initiated by phosphorylation of two threonine residues, Thr61 and Thr60. Cyclin A/Cdk1 phosphorylates Thr61 and their concentrations increase at the end of S-phase. This modification primes Thr60 to be phosphorylated by Casein Kinase 2 (CK2) [15]. Peptidyl-prolyl cis/trans isomerase (Pin1) and protein phosphatase 2 (PP2A) work together to dephosphorylate a phosphothreonine in a conserved region of the RNA binding domain of SLBP. This dephosphorylation releases SLBP from the mRNA in the cytoplasm. Pin1 is also involved in SLBP polyubiquitination by interacting with Ser20 and Ser23 in the N-terminus of SLBP [16].
Because SLBP regulates a unique mechanism for processing and translation of mRNA, SLBP loss is likely to have important consequences. Mutations or depletion of SLBP can result in misprocessing of the canonical histone mRNAs, leading to the aberrant expression of polyadenylated mRNA from each of the canonical histone genes [17–19]. This review will discuss recent findings on the function of SLBP and how the cell compensates when SLBP is not present. Focus will be placed on data presented at the 8th International Conference on Metal Toxicity & Carcinogenesis in Albuquerque, NM that illustrates the ability of arsenic, a carcinogenic metal, to deplete SLBP levels which lead to aberrant polyadenylation of canonical histone mRNA. Recent investigations following up on the data presented at the meeting have revealed exciting insights into the significance of increased poly(A) H3.1 mRNA. Here, for the first time, we report that either overexpression of H3.1 polyadenylated mRNA or knockdown of SLBP induces cell transformation.
2. Arsenic exposure causes aberrant polyadenylation of H3.1 mRNA by decreasing SLBP levels
Arsenic toxicity
Arsenic is a metalloid listed on the Agency for Toxic Substances and Disease Registry (ATSDR) Priority List of Hazardous Substances and it is a human carcinogen. Naturally occurring inorganic arsenic, in particular, has been identified as the causal agent in human skin, lung, bladder, liver and prostate cancers [20–24]. Furthermore, arsenic exposure has also been associated with non-carcinogenic health outcomes, including cardiovascular disease, neurologic deficits, neuro-developmental deficits in childhood, and hypertension [25–29]. While arsenic displays both acute and chronic toxicity, it has no known biological function. The most common mode of human exposure to inorganic arsenic is through contaminated drinking-water. It affects more than 140 million people in more than 70 countries [30].
The metabolism of inorganic arsenic is known to generate oxidative stress, which is currently one of the most widely studied mechanisms of arsenic-induced toxicity and carcinogenicity [31]. It is a widely occurring phenomenon in biological systems that can be principally described as an imbalance between the production of reactive oxygen species (ROS) and the system’s ability to promptly detoxify the reactive intermediates and/or repair the subsequent cellular damage [32]. The disruption of the normal redox potential in a cellular environment via the production of ROS, such as superoxide anion (O2−), hydroxyl radical (•OH), hydrogen peroxide (H2O2), singlet oxygen (1O2), can have detrimental effects on all cellular components, including proteins, lipids and DNA [33–35]. Arsenic is an excellent generator of oxidative stress. It generates ROS by interfering with antioxidants, such as glutathione, and increasing inflammation, resulting in the accumulation of free radicals in cells.
Arsenic induces polyadenylation of H3.1 mRNA
Peripheral blood mononuclear cells (PBMCs) were exposed to 0–1 µM of sodium meta-arsenite (NaAsO2) for 48 hours. One uM arsenic is equivalent to 75 ppb arsenic. People in Bangledesh get exposed to 500 ppb or more [36]. Gene expression profiles were analyzed using 1.0 ST Affymetrix gene chips and results validated by Real-time reverse polymerase chain reaction (qRT-PCR). Surprisingly, we found that 90% of all histone genes were induced in the arsenic-treated PBMCs, and 8% of all the induced genes were histone genes [37].
These findings led us to inquire how the histone genes were being increased by arsenic. The increase in histone mRNA could be due to increased gene transcription or increased mRNA stability. As mentioned earlier, canonical histone mRNAs do not contain a poly(A) tail, which provides a feature that increases the half-life and stability of the mRNA. To investigate if histone mRNAs were acquiring a poly(A) tail due to arsenic exposure, a Northern blot was performed to examine H3.1 mRNA structure [37]. Bronchial epithelial lung cells (BEAS-2B) were utilized because of the known association of arsenic exposure and lung cancer [38] as well as the ability of this cell line to undergo malignant transformation after metal exposure [39–42]. The Northern blot revealed that the amount of polyadenylated H3.1 mRNA doubled after a 48-h exposure to 1 uM arsenic. qRT-PCR confirmed these results showing an arsenic-induced increase in polyadenylated H3.1 mRNA in BEAS-2B cells, as well as two other cell lines- BL41 (human lung carcinoma cells) and PBMCs [37]. To investigate if the increased presence of H3.1 mRNA would result in higher histone H3 levels, we measured H3 levels by Western blot in BL41 cells treated with 1 uM arsenic for 48-h. H3 protein levels were significantly higher in the arsenic-treated cells [37].
Arsenic decreases SLBP by epigenetically regulating the SLBP promoter and enhancing its proteasomal degradation
The mechanisms behind the aberrant polyadenylation of H3.1 mRNA induced by arsenic were unclear. However, several studies have reported that polyadenylated canonical histone mRNA occurs in cells that have a mutation in the SLBP gene or have lost SLBP protein expression [17]. This led us to investigate if SLBP depletion was behind these observations.
A Western blot analysis displayed decreased SLBP levels in arsenic-treated BEAS-2B cells. Arsenic-transformed BEAS-2B clones also displayed decreased SLBP levels compared to control clones. To investigate the mechanism causing the arsenic-induced depletion of SLBP, we first examined arsenic’s effect on SLBP protein degradation, which is mediated by the proteasome. We found that inhibiting the proteasome by treatment with MG-132, a known proteasomal inhibitor, recovered SLBP levels in arsenic-treated BEAS-2B cells [37]. The mechanisms behind arsenic-induced SLBP degradation have not been identified. It is possible that arsenic may directly or indirectly affect PIN1, PP2A or other proteins involved in dissociating SLBP from the mRNA creating free SLBP that is susceptible to ubiquitination and subsequent proteasomal degradation.
Given arsenic’s ability to alter gene expression, we investigated if the arsenic-induced depletion of SLBP was also due to arsenic affecting SLBP at the transcriptional level. qRT-PCR on arsenic-treated BEAS-2B displayed a decrease in SLBP mRNA [37]. Arsenic is known to exert its carcinogenicity and toxicity via epigenetic mechanisms. To investigate if the arsenic-induced decrease in SLBP mRNA was mediated by epigenetic mechanisms, we co-exposed BEAS-2B cells with arsenic and inhibitors of the epigenetic machinery. Cells exposed to either Na-butyrate, a histone deacetylase inhibitor, or 5-Azacytidine, a DNA methyltransferase inhibitor, displayed increased levels of SLBP mRNA restoring SLBP to levels found in untreated cells [37]. These results suggested that arsenic was inducing histone deacetylation and increasing DNA methylation at the SLBP promoter, two epigenetic events that promote gene silencing. Certain histone modifications affect gene expression in a positive or negative manner and arsenic has been shown to alter histone modifications to influence gene expression. To study if arsenic was altering histone modifications at the SLBP promoter, a ChIP assay was performed on arsenic-treated BEAS-2B cells. The results displayed a 50% decrease in H3K4me3 levels, an activating histone mark, at the SLBP promoter [37].
These results demonstrate that arsenic-induced depletion of SLBP occurs at both the protein and transcriptional levels. Mechanisms underlying arsenic’s effects at the SLBP gene promoter as well as induction of proteasomal degradation have yet to be identified. Other studies have demonstrated arsenic’s ability to alter global levels of histone modifications [43] as well as affect epigenetic enzymes [23]. Future studies should investigate the levels and activity of the epigenetic machinery that may be responsible for the arsenic-induced SLBP silencing, such as mixed-lineage leukemia protein 1 (MLL), DNA methyltransferase (DNMT1), and others.
H3.1 poly(A) mRNA exists outside of S-phase
As mentioned earlier, canonical histone mRNA expression is limited to S-phase of the cell cycle; however, poly(A) histone mRNA is not subjected to the normal histone mRNA degradation processes that occur at the end of S-phase. The poly(A) tail stabilizes the histone mRNA and aids in its evasion of degradation [44]. This led us to inquire if arsenic-induced polyadenylation of H3.1 mRNA would allow for its presence outside of S-phase. To test this hypothesis, we examined H3.1 poly(A) mRNA levels in cell cycle synchronized BEAS-2B cells after arsenic treatment. There was a significant increase in polyadenylated H3.1 mRNA in the later stage of S-phase where canonical histone mRNA levels normally fall dramatically. Conversely, SLBP mRNA was shown to be significantly decreased at the G1 to mid S-phase where it normally rises rapidly. The most interesting finding was that H3.1 polyadenylated mRNA was present in mitosis at almost 3 times the level found in control cells [37]. Presence of canonical histones outside of S-phase affects chromosomal segregation in mitosis [45] and holds potential to disrupt histone variant deposition, which will influence gene expression. These data are also in line with results from Sullivan et al., which demonstrate that in Drosophila dSLBP mutant embryos stable polyadenylated canonical histone mRNA accumulates in non-replicating cells [18].
Mechanism of arsenic-induced polyadenylation of H3.1 mRNA
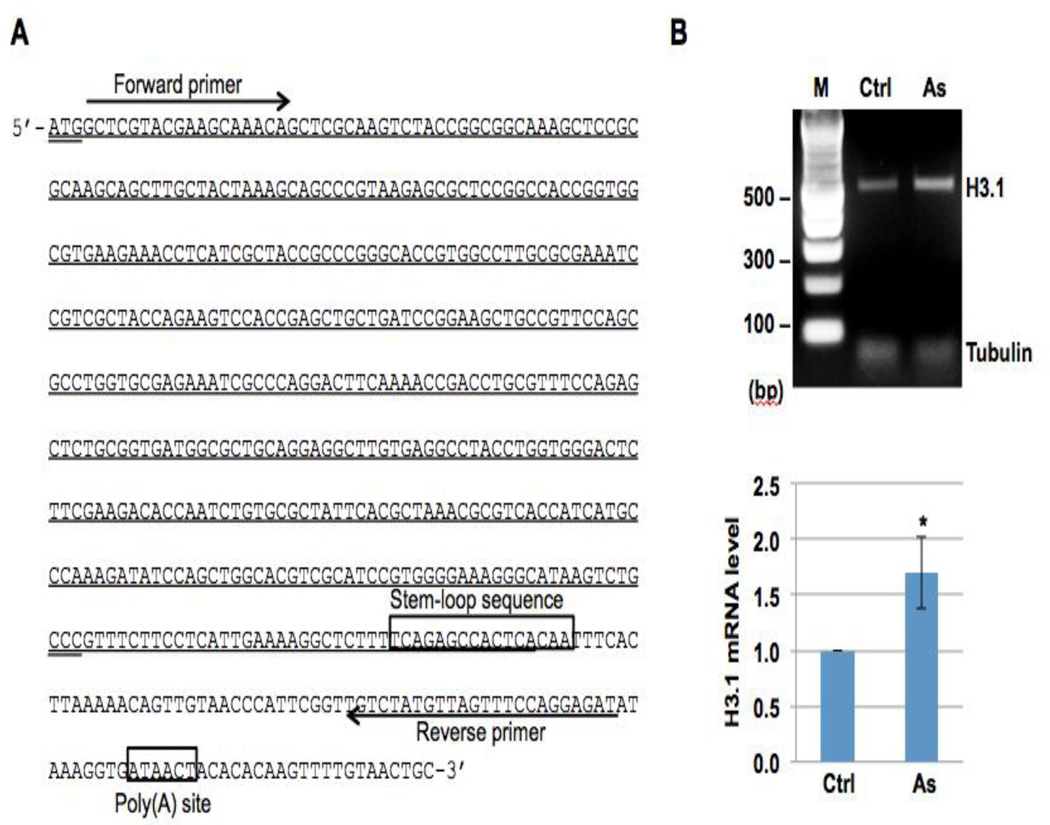
It is likely that the aberrant polyadenylation of H3.1 observed after arsenic exposure is due to arsenic’s effect on SLBP; however, the mechanisms behind the polyadenylation were still unclear. A poly(A) signal or a histone stem-loop sequence is required for termination of transcription. It was hypothesized that the loss of SLBP prevents it from binding to the stem-loop sequence and the transcription can not be terminated near the stem-loop sequence. In this case, the histone mRNA acquires a poly(A) tail via a downstream poly(A) signal [17]. To investigate this theory, we analyzed the nucleotide sequence of H3.1 gene 3’ end and found a potential poly(A) site, TATAAA, 63 nt downstream of the stem-loop sequence (Figure 1A). RT-PCR of H3.1 mRNA using primers upstream of the stem-loop sequence and near the poly(A) site will determine how the transcription is terminated - by the stem-loop sequence or by the poly(A) signal. We treated BEAS-2B cells with 1 µM of arsenic for 48 hours. Total RNA was extracted and then converted to cDNA. H3.1 cDNA downstream of the stem-loop sequence was amplified by PCR with specific primers as shown in Figure 1 A. The results of the RT-PCR shown in Figure 1B demonstrate that arsenic exposure increases the levels of H3.1 mRNA containing downstream of the stem-loop sequence. These data support the idea that lack of SLBP allows the H3.1 mRNA to be polyadenylated using the downstream poly(A) signal.
Figure 1. Arsenic treatment increases the levels of H3.1 mRNA containing the sequence downstream of the stem-loop sequence.
(A) The nucleotide sequence of the histone H3.1 gene (HIST1H3C). The start codon and stop codon are underlined twice. The stem-loop sequence and potential poly(A) site are boxed. The region transcribed by the stem-loop mechanism is underlined. The primers used for amplifying H3.1 cDNA containing the downstream sequence of the stem-loop sequence are underlined by an arrow. (B) RT-PCR results. Upper panel: BEAS-2B cells were treated with (As) or without (Ctrl) 1 µM arsenic for 48 hours. Total RNA was extracted and then converted to cDNA. H3.1 cDNA with the downstream sequence of the stem-loop sequence was then amplified by PCR with specific primers as shown in (A). g-tubulin was used as an internal control. Lower panel: The bar graphs show relative quantification of H3.1 mRNA levels normalized to g-tubulin. The data shown are the means ± S.D. from experiments performed in triplicate. * p<0.05.
3. Polyadenylated H3.1 mRNA leads to cell transformation
BEAS-2B cells overexpressed with H3.1 poly(A) mRNA promote anchorage-independent growth
Aberrant polyadenylation of H3.1 mRNA was increased in BEAS-2B cells transformed by arsenic [37]. Disruption of histone variant deposition into nucleosomes has been linked to cancer and increased presence of canonical histones outside of S-phase may promote this dysfunction. These observations allude to the notion that increased H3.1 levels may play a role in arsenic-induced carcinogenesis. Here, for the first time, we report the ability of increased H3.1 polyadenylated mRNA to induce cell transformation.
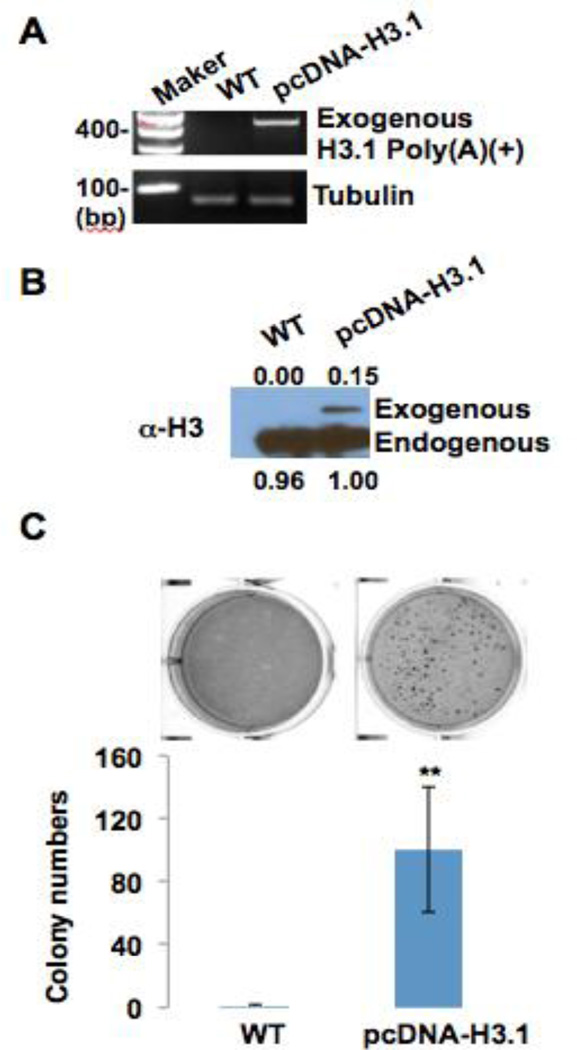
We inserted canonical histone H3.1 cDNA into a pcDNA vector. The pcDNA vector contained a poly(A) signal which produced polyadenylated H3.1 mRNA. The pcDNA-H3.1 vector was stably transfected into BEAS-2B cells. RT-PCR analysis displayed that ectopically expressed poly(A) H3.1 mRNA was significantly increased in cells with the pcDNA-H3.1 vector (Figure 2A). A Western blot displayed an increase in ectopically expressed H3.1 protein levels generated from H3.1 polyadenylated mRNA in cells containing the vector (Figure 2B). In order to investigate if the stably transfected BEAS-2B cells would undergo cell transformation, we plated wild-type BEAS2B cells and pcDNA-H3.1-expressing cells in 0.35% agar and cultured them for 4 weeks. Interestingly, the cells overexpressing polyadenylated H3.1 formed significantly more colonies in soft agar than wild-type cells. About 100 colonies formed in the pcDNA-H3.1-expressing cells, while very little to no colonies formed in the wild-type (Figure 2C). These data suggest that polyadenylated H3.1 mRNA holds potential as a mechanism to facilitate carcinogenesis by toxicants such as arsenic that deplete SLBP.
Figure 2. Polyadenylation of H3.1 facilitates colony formation of BEAS2B cells on soft agar.
(A) RT-PCR results. Canonical histone H3.1 cDNA was inserted into pcDNA vector, which contains poly(A) signal and thus generates polyadenylated H3.1 mRNA. pcDNA-H3.1 vector was stably transfected into BEAS2B cells. Total RNA was extracted from each cell line and then converted to cDNA using oligo dT primers. Exogenous polyadenylated H3.1 mRNA was then amplified by PCR with specific primers for H3.1 and vector. (B) Western blot shows exogenous H3.1 protein generated from ectopically expressed polyadenylated H3.1 mRNA. (C) BEAS2B cells (WT) and pcDNA-H3.1-expressing cells were plated in 0.35% soft agar, and cultured for 4 weeks. The data shown are the means ± S.D. from experiments performed in triplicate. ** p<0.01.
Increased polyadenylated H3.1 mRNA in carcinogenesis
This is the first report to describe overexpression of a canonical histone to induce cell transformation. It still remains unclear how polyadenylated H3.1 is inducing anchorage-independent growth and if this mechanism is involved in arsenic-induced carcinogenesis. Disruption of H3 has been demonstrated in many cancer types and much of the dysfunction is attributed to alterations in histone tail modifications [46–48]. To our knowledge, no investigations have reported on the upregulation of H3 protein levels themselves and their possible involvement in cancer. Earlier, we demonstrated the presence of H3.1 poly(A) mRNA in mitosis. This datum may have significant impact on cellular processes since a failure to repress histone expression following DNA replication is highly toxic due to abnormal chromosome segregation in mitosis [49]. If large amounts of canonical histones are present outside of S phase, they may compete with histone variants such as H3.3 for nucleosome assembly at specific genomic loci such as promoters, enhancers, and pericentric heterochromatin regions. Since canonical and variant histones possess distinctive properties [50], this substitution could disrupt appropriate chromatin structure and may affect functions relevant to histone variants including transcription, and DNA damage repair, and eventually cause genomic instability [51, 52].
The most likely theory regarding polyadenylated H3.1 mRNA’s role in carcinogenesis involves increased levels of H3.1 competing with the H3.3 variant for nucleosomal deposition. Despite only a 5 amino acid difference between H3.1 and H3.3, the two histones play different roles in a variety of cellular processes, such as development, germline integrity, and gene expression [53–55]. As stated earlier, H3.3 is found in the promoters of active genes. H3.3-containing nucleosomes promote a less stable nucleosome accompanied by an opened chromatin structure. These features create an environment that fosters gene activation [55]. H3.1 gets evicted from the nucleosome and replaced by H3.3 [54, 55]. Should H3.1 levels increase, H3.1 may outcompete H3.3 for deposition into the nucleosome. H3.3 eviction and subsequent replacement with H3.1 has been shown to increase the deactivating histone mark, H3K27me3, and decrease the activating histone mark, H3K4me3 [56]. Thus, presence of H3.1 in nucleosomes in promoters creates a more repressive environment than H3.3-containing nucleosomes. H3.1 outcompeting H3.3 for deposition in nucleosomes found in the promoters of tumor suppressor genes will silence these genes, which will facilitate carcinogenesis. A study by Harada et al. has demonstrated that increasing H3.1 to alter the ratio of H3.1 to H3.3 in the cell leads to H3.1 replacement of H3.3 in regulatory regions of skeletal muscle cells. This replacement was accompanied by an increase in H3K27me3 and a decrease in H3K4me3 [56]. Another study by Wen et al. found that replacement of H3.1 by H3.3 is required for the activation of key pluripotency genes in embryos by removing H3.1 repressive histone modifications [57].
In contrast to the theories stated above, there is evidence that H3.3 likely replaces H3.1 at the promoters of cell cycle-promoting genes during senescence, contributing to the transcriptional suppression of these genes by removing H3K4me3 through H3.3 cleavage [58]. Increased levels of H3.1 may outcompete H3.3 deposition and prevent the H3.3 tail cleavage by cathepsin and the removal of the active modification H3K4me3, resulting in the proliferation genes being left on. Continued expression of genes involved in proliferation and the cell cycle may be one way overexpressed H3.1 drives carcinogenesis. It is likely that more than one of these mechanisms may participate in tumorigenesis, depending on the location of the nucleosome assembly dysfunction.
To further support the hypothesis that upregulation of H3.1 facilitates cancer, several studies have reported increased levels of CAF-1 (chromatin assembly factor 1) in various cancer types [59–64]. CAF-1 is a chaperone protein that guides histone H3.1 into the nucleosome at S-phase in a replication-dependent manner (1). Increased levels of CAF-1 alongside upregulation of H3.1would work together to interfere with proper nucleosome assembly further fostering carcinogenesis. Future studies involving the overexpression of polyadenylated H3.1 should examine whether the role of CAF-1in cancer development is dependent on the increase in H3.1 deposition. It would also be interesting to see if arsenic induces an increase in CAF-1 levels, which coincide with the arsenic-induced increase in H3.1.
H3.3 is incorporated into chromatin by HIRA (histone regulator A) in a replication-independent manner (1–3). Unlike CAF-1 upregulation, increased HIRA levels work to prevent carcinogenic events by incorporating histone H3.3 and other canonical core histones (histone H4) into a dynamic chromatin landscape [65]. These studies reporting the different carcinogenic endpoints observed between upregulation of CAF-1 or upregulation of HIRA support the notion that interfering with proper nucleosome assembly of H3.1 and H3.3 plays a role in carcinogenesis. Few investigations have been conducted on HIRA and future studies should examine its function in nucleosome assembly. It would be interesting to see if arsenic or overexpression of H3.1 would affect HIRA levels directly or indirectly.
H3.3 deposition may be alternatively mediated by another pathway involving ATP-dependent helicase (ATRX) and death associated protein 6 (DAXX). Dysfunctions in these proteins may also affect nucleosome assembly and contribute to carcinogenesis. A study by Jiao et al. demonstrated that 40% of human pancreatic neuroendocrine tumors (PanNETs) contained loss-of-function mutations in either the ATRX or DAXX genes [66]. In addition, somatic mutations in the H3.3–ATRX– DAXX chromatin assembly pathway have been identified in 44% of pediatric glioblastoma tumors. These data further support the hypothesis that disrupting nucleosome deposition of H3.3 is an oncogenic pathway.
4. Knockdown of SLBP induces cell transformation
Knockdown of SLBP expression by shRNA promotes anchorage-independent growth in BEAS-2B cells
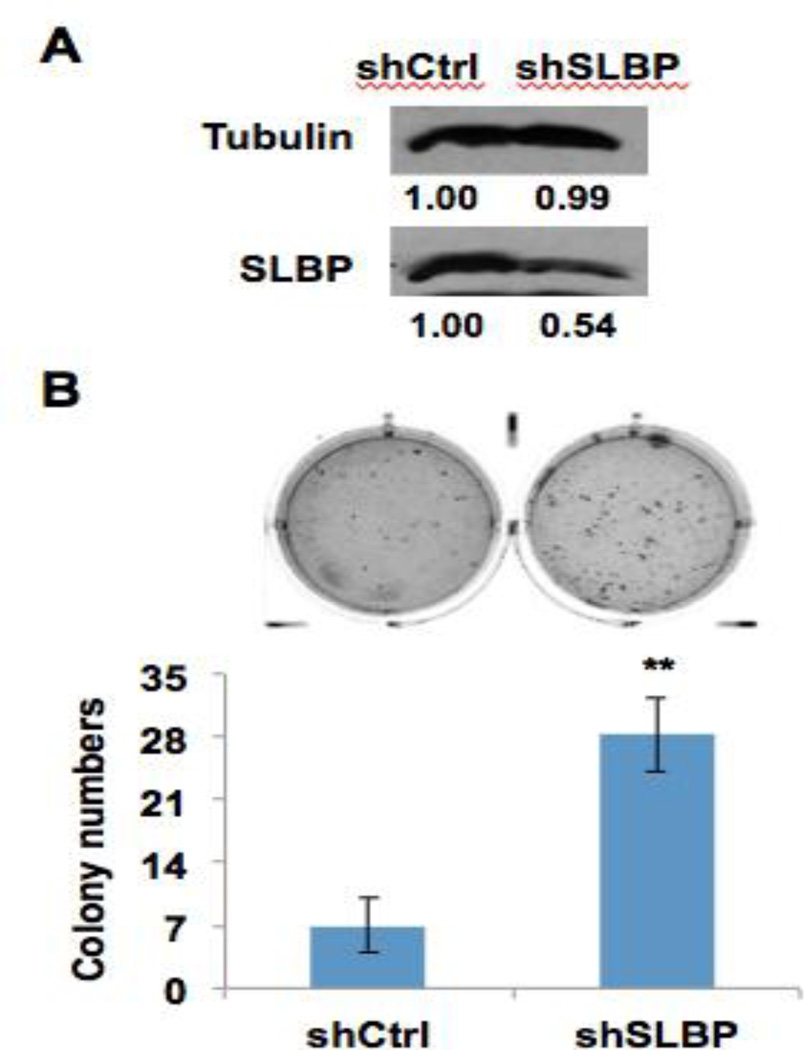
SLBP was depleted in arsenic-transformed clones. To provide further insight into the role that SLBP depletion plays in carcinogenesis, we knocked down the level of SLBP gene expression and observed its affect on cell transformation. BEAS-2B cells were stably transfected with an SLBP small-hairpin RNA (shRNA) vector. A Western blot analysis demonstrated reduction of SLBP levels in SLBP shRNA transfected cells (shSLBP) compared to control cells (shCtrl) (Fig. 3A). The shSLBP and shCtrl cells were plated in 0.35% soft agar and cultured for four weeks. The shSLBP cells formed significantly more colonies at the end of the four weeks than shCtrl cells (Fig. 3B).
Figure 3. Figure 1. Reduction of SLBP expression facilitates colony formation of BEAS2B cells on soft agar.
(A) BEAS2B cells were stably transfected with SLBP shRNA vector. Western blot shows reduction of SLBP protein levels in SLBP shRNA-transfected cells (shSLBP) as compared with control cells (shCtrl). (B) shCtrl and shSLBP cells were plated in 0.35% soft agar, and cultured for 4 weeks. The data shown are the means ± S.D. from experiments performed in triplicate. ** p<0.01.
Loss of SLBP and its role in carcinogenesis
These data suggest that loss of SLBP alone is sufficient to induce cell transformation and is playing a key role in driving arsenic-induced carcinogenesis. The mechanisms behind SLBP reduction and cancer likely involve upregulation of canonical histone levels since our previous data showed that SLBP loss leads to increased histone H3 levels and increased H3.1 promotes anchorage-independent cell growth. Many of the hypothesized mechanisms presented in section 3 concerning why induction of H3.1 promotes anchorage-independent cell growth are pertinent to the loss of SLBP in cell transformation.
Dysfunction in nucleosome assembly and its affects on gene expression is likely one of the primary mechanisms at play in Figure 3. In contrast to Figure 2, the increase in colony formation likely involves dysregulation of other canonical histones besides H3.1. Our previous data [37] show that arsenic exposure affects the expression of 90% of all the histone genes and expression of histone H4 mRNA was significantly induced. It is likely that loss of SLBP is increasing the levels of other canonical histones and this may affect the nucleosomal assembly of other histone variants besides H3.3. For example, H2A gets replaced by the histone variant H2AZ. Nucleosomes containing H2AZ act in a similar fashion as H3.3-containing nucleosomes at certain genomic regions. H2AZ in combination with H3.3 decreases nucleosome stability and creates an open chromatin structure that promotes gene activation. H3.3 and H2AZ co-occupy nucleosomes flanking promoters and the nucleosome at the transcription start site [55]. Increased level of canonical H2A also holds potential to disrupt DNA repair since the histone variant H2AX is involved in DNA double-strand break repair [55].
5. Conclusion
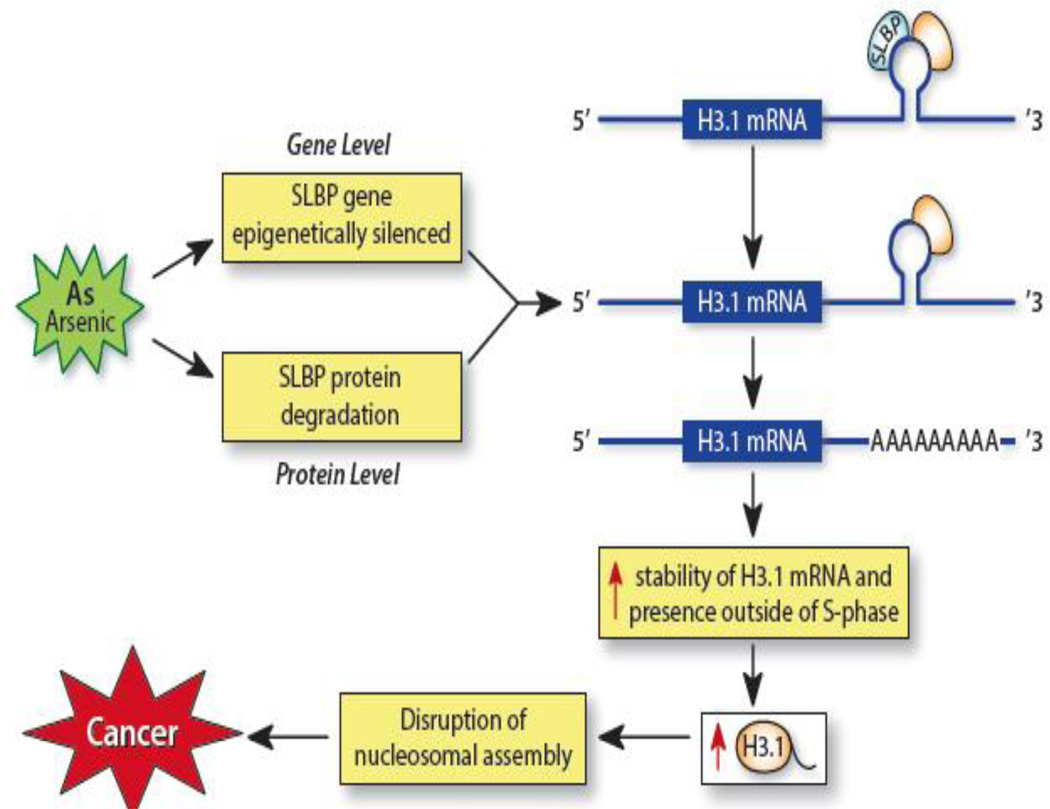
Mechanisms surrounding arsenic-induced carcinogenesis are vague. Although there are multiple known mechanisms in which arsenic exerts its carcinogenic potential such as altering DNA methylation levels and inducing oxidative stress, our data reveals a new pathway in which arsenic exerts its tumorigenic effects. Figure 4 summarizes these findings. Arsenic induces depletion of SLBP by affecting both the protein levels and the mRNA levels of SLBP and this reduction subsequently results in aberrant polyadenylation of H3.1 mRNA, which continues to exist outside of S-phase. Other carcinogenic metals such as nickel and cadmium may also facilitate tumorigenesis via SLBP depletion and future investigations should examine the effects of these metals on canonical histone mRNA structure. In vivo studies also are necessary to confirm these results as a route of arsenic-induced carcinogenesis.
Figure 4. Arsenic-induced depletion of SLBP and subsequent aberrant polyadenylation of H3.1 mRNA and its effects.
Arsenic exposure induces depletion of the stem-loop binding protein (SLBP) by affecting both its protein and mRNA levels. Arsenic epigenetically silences the SLBP gene reducing SLBP mRNA levels and enhances SLBP’s proteasomal degradation. The loss of SLBP prevents it from binding to the stem-loop structure on H3.1 mRNA and the mRNA acquires a poly(A) tail via a downstream poly(A) signal sequence.. The poly(A) tail provides the H3.1 mRNA with increased stability and translation, which allow for presence of H3.1 mRNA outside of S-phase and increased H3.1 levels. Increased polyadenylated H3.1 mRNA may disrupt nucleosome assembly and interfere with H3.3 deposition, which can influence gene expression and facilitate carcinogenesis.
Dysfunction in both canonical and variant histones have been linked to cancer; however, the actual levels of the proteins have not been fully investigated in carcinogenic studies. Here, for the first time, we demonstrate the involvement of increased levels of H3.1 in cell transformation. Polyadenylated H3.1 mRNA and exogenous H3.1 protein levels were significantly increased in transfected cells and these cells displayed increased colony formation in soft agar. These data suggest that increased levels of H3.1 due to polyadenylation of its mRNA holds potential as a mechanism to facilitate carcinogenesis by toxicants such as arsenic that deplete SLBP. This hypothesis was further supported by data showing the enhanced ability of SLBP knockdown cells to undergo cell transformation.
Currently, our studies have focused on histone H3; however, overexpression of other canonical histone mRNAs with a poly(A) tail may also promote cell transformation. Future investigations should check if arsenic-induced upregulation of other canonical histones are due to aberrant polyadenylation of their mRNA. Future studies will focus on the overexpression of polyadenylated H4, H2A, and H2B to see if induction of these canonical histones will also facilitate cell transformation.
Acknowledgements
This work was supported by National Institute of Health (NIH) grants, ES010344, ES014454, ES000260, ES022935, ES023174 and ES005512.
Footnotes
Conflict of Interest Statement
None of the authors of this manuscript have a conflict of interest.
References
- 1.Daban JR. High concentration of DNA in condensed chromatin. Biochem Cell Biol. 2003;81(3):91–99. doi: 10.1139/o03-037. [DOI] [PubMed] [Google Scholar]
- 2.Strunnikov AV. Condensin and biological role of chromosome condensation. Prog Cell Cycle Res. 2003;5:361–367. [PubMed] [Google Scholar]
- 3.Davey CA, et al. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 4.Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 5.Hacques MF, et al. Use of an immobilized enzyme and specific antibodies to analyse the accessibility and role of histone tails in chromatin structure. Biochem Biophys Res Commun. 1990;168(2):637–643. doi: 10.1016/0006-291x(90)92368-a. [DOI] [PubMed] [Google Scholar]
- 6.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28(7):672–682. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominski Z, Marzluff WF. Formation of the 3' end of histone mRNA. Gene. 1999;239(1):1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- 8.Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17(3):274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Tan D, et al. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3'hExo ternary complex. Science. 2013;339(6117):318–321. doi: 10.1126/science.1228705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitfield ML, et al. SLBP is associated with histone mRNA on polyribosomes as a component of the histone mRNP. Nucleic Acids Res. 2004;32(16):4833–4842. doi: 10.1093/nar/gkh798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal N, et al. Assembly of the SLIP1-SLBP complex on histone mRNA requires heterodimerization and sequential binding of SLBP followed by SLIP1. Biochemistry. 2013;52(3):520–536. doi: 10.1021/bi301074r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iampietro C BJ, Wang X, Cody NA, Chin A, Lefebvre FA, Douziech M, Krause HM, Lécuyer E. Developmentally regulated elimination of damaged nuclei involves a Chk2-dependent mechanism of mRNA nuclear retention. Dev Cell. 2014;29(4):468–481. doi: 10.1016/j.devcel.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 13.Whitfield ML, et al. Stem-loop binding protein, the protein that binds the 3' end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20(12):4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng L, et al. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol Cell Biol. 2003;23(5):1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koseoglu MM, Graves LM, Marzluff WF. Phosphorylation of threonine 61 by cyclin a/Cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Mol Cell Biol. 2008;28(14):4469–4479. doi: 10.1128/MCB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan N, et al. The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol Cell Biol. 2012;32(21):4306–4322. doi: 10.1128/MCB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanzotti DJ, et al. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3' end processing in vivo. Mol Cell Biol. 2002;22(7):2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan E, et al. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 2001;15(2):173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan KD, Steiniger M, Marzluff WF. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell. 2009;34(3):322–332. doi: 10.1016/j.molcel.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, in Some Drinking-water Disinfectants and Contaminants, including Arsenic. Lyon: France; 2004. [PMC free article] [PubMed] [Google Scholar]
- 21.Schuhmacher-Wolz U, et al. Oral exposure to inorganic arsenic: evaluation of its carcinogenic and non-carcinogenic effects. Crit Rev Toxicol. 2009;39(4):271–298. doi: 10.1080/10408440802291505. [DOI] [PubMed] [Google Scholar]
- 22.Chiu H, et al. Does arsenic exposure increase the risk for liver cancer? J Toxicol Environ Health A. 2004;67(19):1491–1500. doi: 10.1080/15287390490486806. [DOI] [PubMed] [Google Scholar]
- 23.Benbrahim-Tallaa L, Waalkes M. Inorganic arsenic and human prostate cancer. Environ Health Perspect. 2008;116(2):158–164. doi: 10.1289/ehp.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman M, Ng J, Naidu R. Chronic exposure of arsenic via drinking water and its adverse health impacts on humans. Environ Geochem Health. 2009;31(Suppl 1):189–200. doi: 10.1007/s10653-008-9235-0. [DOI] [PubMed] [Google Scholar]
- 25.Wasserman GA, et al. Water arsenic exposure and intellectual function in 6-year-old children in Araihazar, Bangladesh. Environ Health Perspect. 2007;115(2):285–289. doi: 10.1289/ehp.9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, et al. Arsenic exposure at low-to-moderate levels and skin lesions, arsenic metabolism, neurological functions, and biomarkers for respiratory and cardiovascular diseases: review of recent findings from the Health Effects of Arsenic Longitudinal Study (HEALS) in Bangladesh. Toxicol Appl Pharmacol. 2009;239(2):184–192. doi: 10.1016/j.taap.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng CH, et al. Long-term arsenic exposure and ischemic heart disease in arseniasis-hyperendemic villages in Taiwan. Toxicol Lett. 2003;137(1–2):15–21. doi: 10.1016/s0378-4274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- 28.Wasserman GA, et al. Water arsenic exposure and children's intellectual function in Araihazar, Bangladesh. Environ Health Perspect. 2004;112(13):1329–1333. doi: 10.1289/ehp.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CJ, et al. Increased prevalence of hypertension and long-term arsenic exposure. Hypertension. 1995;25(1):53–60. [PubMed] [Google Scholar]
- 30.Kinniburgh DG, Kosmus W. Arsenic contamination in groundwater: some analytical considerations. Talanta. 2002;58(1):165–180. doi: 10.1016/s0039-9140(02)00265-5. [DOI] [PubMed] [Google Scholar]
- 31.Flora SJ. Arsenic-induced oxidative stress and its reversibility. Free Radic Biol Med. 2011;51(2):257–281. doi: 10.1016/j.freeradbiomed.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Reuter S, et al. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott M, et al. Mitochondria, oxidative stress and cell death. Apoptosis. 2007;12(5):913–922. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- 34.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12(10):1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 35.Valko M, et al. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39(1):44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Kile ML, et al. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics. 2014;9(5):774–782. doi: 10.4161/epi.28153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brocato J, et al. Arsenic induces polyadenylation of canonical histone mRNA by down-regulating stem-loop-binding protein gene expression. J Biol Chem. 2014;289(46):31751–31764. doi: 10.1074/jbc.M114.591883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith AH, et al. Increased lung cancer risks are similar whether arsenic is ingested or inhaled. J Expo Sci Environ Epidemiol. 2009;19(4):343–348. doi: 10.1038/jes.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arita A, Costa M. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics. 2009;1(3):222–228. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brocato J, Costa M. Basic mechanics of DNA methylation and the unique landscape of the DNA methylome in metal-induced carcinogenesis. Crit Rev Toxicol. 2013;43(6):493–514. doi: 10.3109/10408444.2013.794769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passantino L, Munoz AB, Costa M. Sodium metavanadate exhibits carcinogenic tendencies in vitro in immortalized human bronchial epithelial cells. Metallomics. 2013;5(10):1357–1367. doi: 10.1039/c3mt00149k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun H, et al. Comparison of gene expression profiles in chromate transformed BEAS-2B cells. PLoS One. 2011;6(3):e17982. doi: 10.1371/journal.pone.0017982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chervona Y, et al. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol Biomarkers Prev. 2012;21(12):2252–2260. doi: 10.1158/1055-9965.EPI-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392(6675):516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 45.Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44(1):43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 46.Funato K, et al. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346(6216):1529–1533. doi: 10.1126/science.1253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan F, Shi Y. Histone H3.3 and cancer: A potential reader connection. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1418996111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah MA, et al. A global assessment of cancer genomic alterations in epigenetic mechanisms. Epigenetics Chromatin. 2014;7(1):29. doi: 10.1186/1756-8935-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meeks-Wagner DHL. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44(1):43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 50.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9(1):15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 51.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11(4):264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 52.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46(6):722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Hum Genet. 1997;101(3):284–294. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- 54.Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Curr Opin Genet Dev. 2010;20(2):110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.HenikoffS SSM. Histone Variants and Epigenetics. Cold Spring Harb Perspect Biol. 2015;7(1) doi: 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harada A, et al. Incorporation of histone H3.1 suppresses the lineage potential of skeletal muscle. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen D, et al. H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos. Nucleus. 2014;5(5):369–375. doi: 10.4161/nucl.36231. [DOI] [PubMed] [Google Scholar]
- 58.Duarte LF, et al. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat Commun. 2014;5:5210. doi: 10.1038/ncomms6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barbieri E, et al. Histone chaperone CHAF1A inhibits differentiation and promotes aggressive neuroblastoma. Cancer Res. 2014;74(3):765–774. doi: 10.1158/0008-5472.CAN-13-1315. [DOI] [PubMed] [Google Scholar]
- 60.Barbieri E, et al. A p53 drug response signature identifies prognostic genes in high-risk neuroblastoma. PLoS One. 2013;8(11):e79843. doi: 10.1371/journal.pone.0079843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bethke L, et al. Comprehensive analysis of the role of DNA repair gene polymorphisms on risk of glioma. Hum Mol Genet. 2008;17(6):800–805. doi: 10.1093/hmg/ddm351. [DOI] [PubMed] [Google Scholar]
- 62.Glinsky GV, et al. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113(6):913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polo SE, et al. Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res. 2004;64(7):2371–2381. doi: 10.1158/0008-5472.can-03-2893. [DOI] [PubMed] [Google Scholar]
- 64.Staibano S, et al. Chromatin assembly factor-1 (CAF-1)-mediated regulation of cell proliferation and DNA repair: a link with the biological behaviour of squamous cell carcinoma of the tongue? Histopathology. 2007;50(7):911–919. doi: 10.1111/j.1365-2559.2007.02698.x. [DOI] [PubMed] [Google Scholar]
- 65.Rai TS, et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 2014;28(24):2712–2725. doi: 10.1101/gad.247528.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–11203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]