Abstract
The cellular innate immune system plays a critical role in mounting the initial resistance to virus infection. It is comprised of various pattern-recognition receptors that induce type I interferon production, which further shapes the adaptive immunity. However, to overcome this resistance and promote replication, viruses have evolved mechanisms to evade this host innate immune response. Here we discuss a recently described mechanism of boosting the innate immunity by oligoadenylate synthetase-like (OASL) protein, which can potentially be used to overcome viral evasion and enhance innate immunity.
Keywords: Influenza, OASL, interferon, RIG-I
Introduction
Despite remarkable advances in vaccination and treatment, diseases caused by viral infections remain one of the leading causes of death worldwide; as we have seen in recent years, a number of viruses also threaten the global health with pandemic potential. Among the different types of viruses that are human pathogens, RNA viruses pose unique challenges due to their rapid replication kinetics, high mutation rates, and complex evolutionary dynamics. The co-evolution of the virus and the host has resulted in competing strategies to protect and propagate over a long time. As a result, often times the host antiviral immunity that prevents viral infection is targeted by viruses [1]. Therefore, understanding the inner workings of the host innate immune response – the first line of defense against viral infection, and how it is subverted by viruses gives us one of the most promising opportunities to combat diseases caused by RNA viruses.
Host defense against RNA or DNA virus infection is initiated by the innate immune receptors such as RIG-I (Retinoic acid-inducible gene I)-like receptors (RLR), Toll-like receptors (TLR) and specific DNA-sensors through the detection of non-self nucleic acids. This initiates the cellular innate immune response, primarily mediated by type I interferons (IFN), and shapes subsequent adaptive immunity [2–4]. IFN induced by RLR or TLR signaling acts in both autocrine and paracrine manner to induce many IFN-stimulated genes (ISGs), which mediate most of the pleiotropic effects of IFN. Due to common transcriptional elements in their promoters, several ISGs are also directly induced by virus infection (via IRF3/IRF7) without requiring IFN signaling [5,6]. Recently, a large number of ISGs were tested for their antiviral activities against multiple viruses [7–9]. However, among ~ 400 ISGs, the biochemical functions of only a handful have been delineated [10,11]. The best understood mechanism for the antiviral activity of ISG is the generalized inhibition of protein synthesis by dsRNA-activated enzymes (e.g., RNA activated Protein Kinase (PKR), Oligoadenylate Synthetase (OAS) etc.). Recent studies have shed light on the mechanism of antiviral activities of some ISGs, such as Viperin [12], IFIT family proteins [13], IFI16 [14], and cGAS [15]. We have recently described the mechanism of antiviral activity of one such ISG, Oligoadenylate Synthetase-Like (OASL) [16]. Here we discuss some of the unique features of OASL, and the potential for developing broad acting antiviral therapy exploiting this pathway.
OAS family of proteins - Structural features and conservation
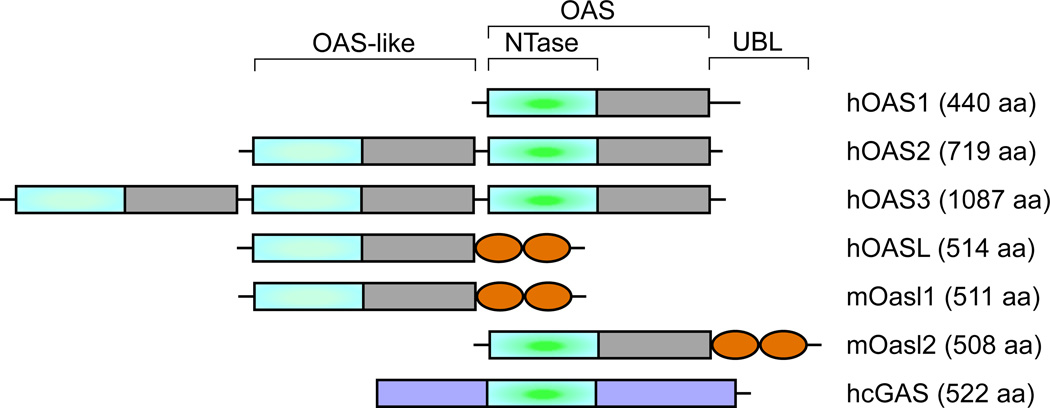
Oligoadenylate synthetases (OAS) belong to a family of ISGs characterized by their ability to synthesize 2’–5’ oligoadenylates, which induce RNA degradation by activating a latent RNase, RNase L [17]. However, the recent identification of the cytoplasmic DNA sensor cyclic GMP-AMP synthetase (cGAS), which is another member of the OAS family, shows potentially diverse functions of this family of proteins [18]. Human oligoadenylate synthetase-like (OASL), is related to the OAS proteins by its N-terminal OAS-like domain, but harbors characteristic changes in the active site, and is thus devoid of 2’–5’ oligoadenylate synthetase activity (Fig. 1). Furthermore, OASL contains two tandem ubiquitin-like domains (UBL) in the C-terminus, which are not present in any of the other members of the OAS family [17]. OASL is directly and rapidly induced by virus infection via interferon regulatory factor (IRF)-3 as well as by IFN signaling and has been shown to have antiviral activities, which requires the UBL domain [8,19]. A number of epidemiological studies have also linked various SNPs in human OASL gene to altered susceptibility to hepatitis C and West Nile virus infections [20–22]. However, in the absence of the catalytic activity to synthesize 2’–5’ oligoadenylates, the mechanism of human OASL antiviral activity remained elusive until recently.
Fig. 1.
Domain organizations of human OAS family and mouse Oasl proteins. Human OAS1, OAS2 and OAS3 (hOAS) contain 1, 2 and 3 OAS-like domains respectively. Among them only one from each has active nucleotidyltransferase (NTase) activity (aligned at the center). Human and mouse OASL proteins have the OAS domain, but the NTase activity is lost in hOASL and mOasl1, whereas mOasl2 has NTase activity. All the OASL proteins contain two repeats of ubiquitin-like domains (UBL) at the C-terminal. Another member of the family, cGAS harbors structurally similar NTase domain, but no other similarity.
The presence of enzymatically active OAS-like proteins was reported in marine sponges [23]. However, as the IFN system is restricted to the jawed vertebrates, the significance of this finding and its contribution in the innate immunity remained largely unappreciated. The discovery of cGAS and its obvious structural similarity with OAS proteins has generated new attention to this family of proteins. It is now clear that these proteins, including the bacterial dinucleotide cyclase (DncV), belong to an ancient nucleotidyltransferase superfamily (NTase domain in Fig. 1) [24,25], and are widely found throughout various forms of life [26]. Interestingly, not all the homologs in this family are predicted to be enzymatically active [26], which indicates divergent functions of these proteins arising during evolution. However, as demonstrated by human OASL, which is devoid of enzyme activity, the absence of enzymatic activity is seldom the defining characteristic of OAS proteins’ involvement in innate immunity. As discussed below, OASL, which seems to have evolved from OAS1 and is confined in vertebrates [27,28], has orthologs in various other vertebrate species with enzyme activity.
Unlike in humans, two OASL orthologs have been identified in mice: Oasl1 and Oasl2, sharing respectively 70% and 48% amino acid sequence identity with human OASL [17]. While mouse Oasl1 is enzymatically inactive, mouse Oasl2 contains two crucial Asp residues in its active site and exhibits OAS enzyme activity [29] (Fig. 1). Similar enzymatically active OASL othologs have also been reported in chickens [30]. The mouse Oasl1 has been recently shown to inhibit IFN induction by binding to the 5’ UTR of IRF7 and inhibiting its translation. Consequently, targeted deletion of Oasl1 led to enhanced IFN induction and diminished viral replication in vivo [31]. Furthermore, Oasl1−/− mice showed better control of viremia and a better virus-specific CD8+ T-cell differentiation upon chronic LCMV infection [32]. In contrast to Oasl1, human OASL and mouse Oasl2 do not bind to the IRF7 5’UTR and are devoid of IRF7 suppression activity. Targeted deletion of Oasl2 in mice showed enhanced viral replication suggesting that Oasl2 acts as the functional equivalent of human OASL [16].
Mechanism of action of OASL proteins - Enhancement of RIG-I activity by OASL
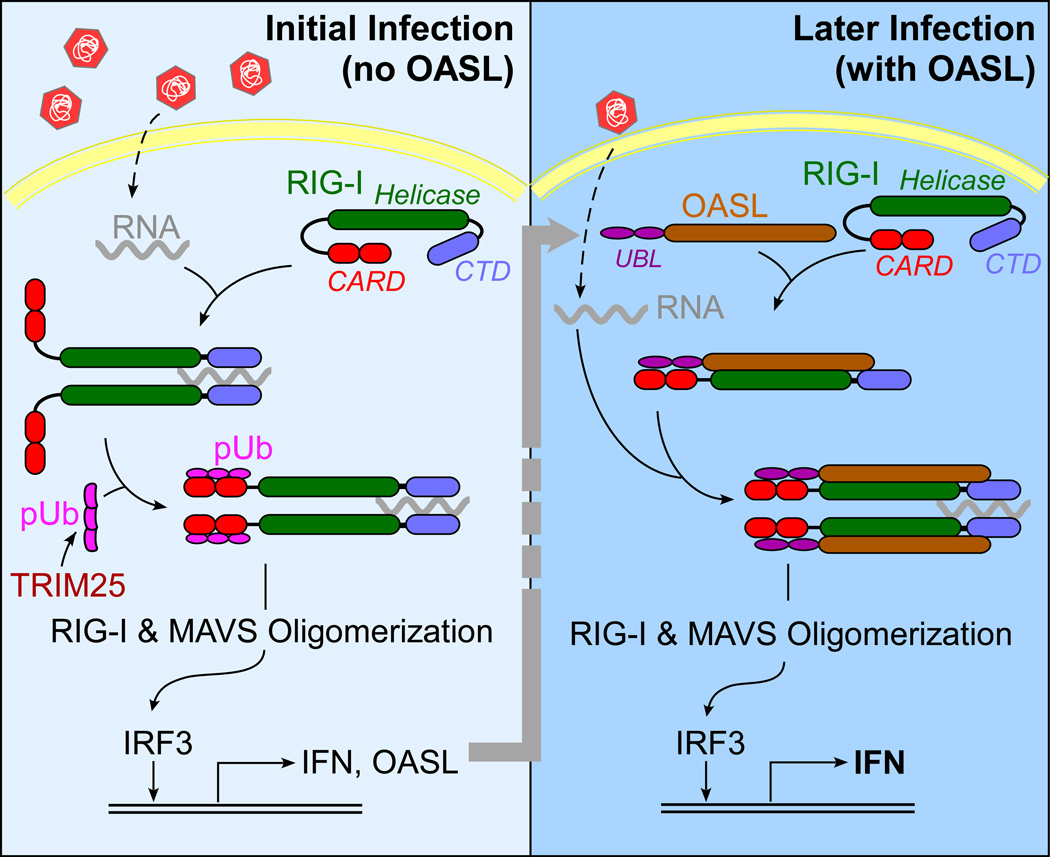
Human OASL promotes antiviral activity by enhancing the sensitivity of RIG-I activation. From a number of biochemical and structural studies [33], a model for RIG-I activation has been proposed where RIG-I adopts a stable auto-inhibited conformation in the absence of RNA. Upon binding to viral RNA through the C-terminal domain (CTD), the helicase domain changes conformation, thereby enabling RIG-I to hydrolyze ATP and further interact with RNA. The N-terminal CARDs (Caspase activation and recruitment domains) then bind to K63-linked polyubiquitin (pUb), converting RIG-I to an active competent state, which is followed by CARD-mediated MAVS aggregation and signaling. Recent observations also suggest that in the case of longer RNA, RIG-I oligomerization occurs without pUb [34]. Although, for larger dsRNA the strict requirement of pUb for RIG-I activation has been a topic of debate, in most cases RIG-I activation is strongly regulated by a two-step mechanism requiring simultaneous binding of two ligands – RNA and pUb. This mechanism allows the RIG-I sensor to avoid aberrant activation of antiviral innate immunity and IFN induction. It has been shown that the synthesis of short K63-linked polyubiquitin chains, or the K63-linked polyubiquitination of RIGI is carried out by the ubiquitin ligase TRIM25 [35,36]. However, we have shown that in presence of OASL, RIG-I can be activated by viral RNA in the absence of TRIM25 [16]. This and additional OASL-RIG-I interaction studies allowed us to propose the following model for the enhancement of RIG-I activity by OASL (Fig. 2). Following the initial viral infection and OASL induction in the infected and the surrounding cells through IFN signaling, OASL binds to RIG-I and mimics pUb. This makes RIG-I activation more sensitive, requiring just one ligand – viral RNA, and leads to enhanced IFN induction. However, it will be premature to conclude that this is the sole mechanism of OASL function, and further investigations are necessary to delineate other possible mechanisms of its activity. For example, OASL expression inhibited HSV-1 replication in HEK293 cells [16]. As HEK293 cells are known to be defective in cGAS-STING-mediated DNA sensor signaling, this antiviral activity was attributed to RIG-I activated through RNA pol III [37]. However, it is not yet clear how OASL affects HSV-1 replication in cells where it is sensed through cGAS pathway.
Fig. 2.
A schematic model of OASL-mediated enhancement of RIG-I signaling.
Viral evasion of innate immunity - Targeting of OASL by viral proteins
As the primary mediators of antiviral innate immunity, the RLR and the IFN pathways are targeted by multiple RNA viruses. Respiratory syncytial virus (RSV), the causative agent for severe bronchiolitis and pneumonia in children and the elderly, accomplishes this with the nonstructural proteins, NS1 and NS2. These proteins specifically promote the degradation of various proteins involved in IFN pathways [38]. Our recent finding that OASL is targeted for degradation by RSV NS1, again supports its physiological importance in providing cellular innate immunity. Interestingly, for the mouse Oasl isoforms, it is only the Oasl2 that inhibits RSV replication, and is degraded in the presence of RSV NS1, while Oasl1 is not (unpublished observation). Similarly, for the influenza virus multiple mechanisms are known to subvert antiviral innate immunity [39]. The influenza non-structural protein, NS1 can directly bind RIG-I and/or limit ligand availability by binding RNA [40,41]. Additionally, NS1 can attenuate activation of RIG-I via inhibition of ubiquitination by the ubiquitin ligase, TRIM25 [35]. As it has been shown that in the presence of OASL, RIG-I activation can be carried out without TRIM25, it is expected that boosting OASL expression may provide strong antiviral activity against all strains of influenza viruses. However, the picorna viruses that are primarily sensed through MDA5 (Melanoma differentiation associated gene 5, another member of the RLR family) are not inhibited by OASL showing specificity. Interestingly, OASL is not targeted by picorna viruses to subvert innate immunity (unpublished observation). These findings together argue in favor of using the OASL-pathway to provide broad antiviral activity against viruses that are primarily sensed through RIG-I, which might help overcome viral subversion of innate immunity.
Conclusions – Unmet needs and how OASL may be useful
Two aspects about OASL-mediated enhancement of RIG-I signaling make it unique for combating viral infection. First, OASL has the potential to overcome the innate immune evasion. According to our results with influenza virus, despite targeting of TRIM25 by NS1, RIG-I can be activated in presence of OASL (unpublished observations). Second, unlike RIG-I expression, which results in IFN induction that can lead to toxicity, expression of OASL by itself does not activate IFN induction. It makes the RIG-I-based RNA detection system much more sensitive to viral RNA, where it can be activated with comparatively sub-threshold levels of virus infection [16]. Therefore, delivering OASL protein or ectopically expressing OASL is less likely to have major toxic side effects and may prove a new mode of combating virus infections. However, the successful delivery of a cytoplasmic protein to obtain therapeutic efficacy is a daunting challenge for the current drug delivery technologies. Thus, we are focusing on the respiratory viruses, which affect the respiratory system that is much more accessible to non-invasive manipulations. In summary, OASL presents a novel molecule that may be able to boost innate host defense, even in the presence of viral inhibition, resulting in improved immunity.
Highlights.
Human OASL provides antiviral activity by enhancing RIG-I signaling.
The presence of OASL allows cells to overcome viral evasion for some viruses.
Targeting the OASL-pathway may be an effective way to combat viral infection.
Acknowledgements
Research in author’s laboratory is supported by NIH funding AI082673 and in part by award P30CA047904 to University of Pittsburgh Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. A comprehensive review describing various types of innate immune sensors involved in virus sensing.
- 3.Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freaney JE, Kim R, Mandhana R, Horvath CM. Extensive cooperation of immune master regulators IRF3 and NFkappaB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 2013;4:959–973. doi: 10.1016/j.celrep.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen GC, Sarkar SN. Transcriptional signaling by double-stranded RNA: role of TLR3. Cytokine Growth Factor Rev. 2005;16:1–14. doi: 10.1016/j.cytogfr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schoggins JW, Macduff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. Functional screening of ISG expression for their antiviral activity against multiple viruses.
- 10.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarkar SN, Sen GC. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther. 2004;103:245–259. doi: 10.1016/j.pharmthera.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Seo JY, Yaneva R, Cresswell P. Viperin: A Multifunctional, Interferon-Inducible Protein that Regulates Virus Replication. Cell Host Microbe. 2011;10:534–539. doi: 10.1016/j.chom.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 15.Xiao TS, Fitzgerald KA. The cGAS-STING pathway for DNA sensing. Mol Cell. 2013;51:135–139. doi: 10.1016/j.molcel.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, Ibsen MS, Schmid-Burgk JL, Schmidt T, Ganapathiraju MK, et al. Antiviral Activity of Human OASL Protein Is Mediated by Enhancing Signaling of the RIG-I RNA Sensor. Immunity. 2014;40:936–948. doi: 10.1016/j.immuni.2014.05.007. Mechanistic description of antiviral activity of human OASL and mouse Oasl2.
- 17.Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 18. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. This and its accompanying reports discovery of the DNA sensor cGAS and cGAMP.
- 19.Marques J, Anwar J, Eskildsen-Larsen S, Rebouillat D, Paludan SR, Sen G, Williams BR, Hartmann R. The p59 oligoadenylate synthetase-like protein possesses antiviral activity that requires the C-terminal ubiquitin-like domain. J Gen Virol. 2008;89:2767–2772. doi: 10.1099/vir.0.2008/003558-0. [DOI] [PubMed] [Google Scholar]
- 20.Ishibashi M, Wakita T, Esumi M. 2',5'-Oligoadenylate synthetase-like gene highly induced by hepatitis C virus infection in human liver is inhibitory to viral replication in vitro. Biochem Biophys Res Commun. 2010;392:397–402. doi: 10.1016/j.bbrc.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 21.Su X, Yee LJ, Im K, Rhodes SL, Tang Y, Tong X, Howell C, Ramcharran D, Rosen HR, Taylor MW, et al. Association of single nucleotide polymorphisms in interferon signaling pathway genes and interferon-stimulated genes with the response to interferon therapy for chronic hepatitis C. J Hepatol. 2008;49:184–191. doi: 10.1016/j.jhep.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yakub I, Lillibridge KM, Moran A, Gonzalez OY, Belmont J, Gibbs RA, Tweardy DJ. Single nucleotide polymorphisms in genes for 2'–5'-oligoadenylate synthetase and RNase L inpatients hospitalized with West Nile virus infection. The Journal of infectious diseases. 2005;192:1741–1748. doi: 10.1086/497340. [DOI] [PubMed] [Google Scholar]
- 23.Wiens M, Kuusksalu A, Kelve M, Muller WE. Origin of the interferon-inducible (2'–5')oligoadenylate synthetases: cloning of the (2'–5')oligoadenylate synthetase from the marine sponge Geodia cydonium. FEBS letters. 1999;462:12–18. doi: 10.1016/s0014-5793(99)01478-7. [DOI] [PubMed] [Google Scholar]
- 24. Hornung V, Hartmann R, Ablasser A, Hopfner KP. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14:521–528. doi: 10.1038/nri3719. A very usefull comprehensive description of structural and functional similarities between OAS proteins and cGAS.
- 25. Wu X, Wu FH, Wang X, Wang L, Siedow JN, Zhang W, Pei ZM. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res. 2014;42:8243–8257. doi: 10.1093/nar/gku569. Molecular evolutionary analysis of the ancient family of NTase.
- 26.Pari M, Kuusksalu A, Lopp A, Kjaer KH, Justesen J, Kelve M. Enzymatically active 2',5'-oligoadenylate synthetases are widely distributed among Metazoa, including protostome lineage. Biochimie. 2014;97:200–209. doi: 10.1016/j.biochi.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Kjaer KH, Poulsen JB, Reintamm T, Saby E, Martensen PM, Kelve M, Justesen J. Evolution of the 2'–5'-oligoadenylate synthetase family in eukaryotes and bacteria. J Mol Evol. 2009;69:612–624. doi: 10.1007/s00239-009-9299-1. [DOI] [PubMed] [Google Scholar]
- 28.Perelygin AA, Zharkikh AA, Scherbik SV, Brinton MA. The mammalian 2'–5' oligoadenylate synthetase gene family: evidence for concerted evolution of paralogous Oas1 genes in Rodentia and Artiodactyla. J Mol Evol. 2006;63:562–576. doi: 10.1007/s00239-006-0073-3. [DOI] [PubMed] [Google Scholar]
- 29.Eskildsen S, Justesen J, Schierup MH, Hartmann R. Characterization of the 2'–5'-oligoadenylate synthetase ubiquitin-like family. Nucleic Acids Res. 2003;31:3166–3173. doi: 10.1093/nar/gkg427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatsumi R, Sekiya S, Nakanishi R, Mizutani M, Kojima S, Sokawa Y. Function of ubiquitin-like domain of chicken 2'–5'-oligoadenylate synthetase in conformational stability. J Interferon Cytokine Res. 2003;23:667–676. doi: 10.1089/107999003322558809. [DOI] [PubMed] [Google Scholar]
- 31. Lee MS, Kim B, Oh GT, Kim YJ. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol. 2013;14:346–355. doi: 10.1038/ni.2535. This paper reports the function of mouse Oasl1.
- 32.Lee MS, Park CH, Jeong YH, Kim YJ, Ha SJ. Negative regulation of type I IFN expression by OASL1 permits chronic viral infection and CD8(+) T-cell exhaustion. PLoS pathogens. 2013;9:e1003478. doi: 10.1371/journal.ppat.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Neill LA, Bowie AG. The powerstroke and camshaft of the RIG-I antiviral RNA detection machine. Cell. 2011;147:259–261. doi: 10.1016/j.cell.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 34.Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 35.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barik S. Respiratory syncytial virus mechanisms to interfere with type 1 interferons. Curr Top Microbiol Immunol. 2013;372:173–191. doi: 10.1007/978-3-642-38919-1_9. [DOI] [PubMed] [Google Scholar]
- 39.Fernandez-Sesma A. The influenza virus NS1 protein: inhibitor of innate and adaptive immunity. Infect Disord Drug Targets. 2007;7:336–343. doi: 10.2174/187152607783018754. [DOI] [PubMed] [Google Scholar]
- 40.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr, Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]