Abstract
Animals recognize the availability of nutrients and regulate the intake and storage of these nutrients accordingly. However, the molecular mechanisms underlying nutrient sensing and subsequent changes in behavior and metabolism are not fully understood. Mlx interactor (Mio), the Drosophila homolog of carbohydrate response element binding protein (ChREBP), functions as a transcription factor in the fat body of the fly to control triglyceride storage as well as feeding, suggesting that Mio may act in a nutrient-sensing pathway to coordinate food consumption and metabolism. Here, we show that Mio functions in neurons in Drosophila to regulate feeding and nutrient storage. Pan-neuronal disruption of Mio function leads to increased triglyceride and glycogen storage, and this phenotype is not due to increased food consumption. Interestingly, targeted disruption of Mio specifically in the insulin-producing cells (IPCs) has little effect on nutrient storage, but increases food consumption suggesting that Mio acts in these neurons to control feeding behavior. Since Mio is a transcription factor, one possible way Mio may act in the IPCs to control feeding is through regulating the expression of Drosophila insulin-like peptides (dilps) or drosulfakinin (dsk), neuropeptides produced in the IPCs. Consistent with this hypothesis, IPC-specific knockdown of Mio leads to an increase in dilp3 expression, while not affecting dilp2, 5 or dsk levels. Together, this study indicates a new function for Mio in the Drosophila brain and specifically in the IPCs, controlling neuropeptide gene expression, feeding and metabolism in accordance with nutrient availability.
Keywords: brain, Drosophila, metabolism, feeding, insulin-like peptide
1. Introduction
All animals consume food and metabolize dietary nutrients to produce energy. The regulation of food consumption and maintenance of metabolic homeostasis involves a complex system of hormones that act on multiple tissues. The action of these hormones leads to changes in appetite as well as the activation of intracellular pathways that result in energy production and storage [1]. Organisms store excess nutrients as glycogen and fat for future use, traditionally during times of low food availability [2]. Thus, disruptions to this network will affect food consumption and nutrient storage, consequently leading to obesity, diabetes, and other metabolic diseases.
Two mammalian nutrient-responsive hormones that regulate feeding and metabolism and are often disrupted in metabolic diseases are leptin, an anorexigenic signal made in fat cells, and ghrelin, an orexigenic signal made in the stomach. After secretion, both hormones circulate through the bloodstream, and activate specific receptors in the hypothalamus; this activation produces signals that coordinate feeding as well as energy metabolism [2–4]. Another hormone whose secretion is regulated by nutrient conditions is insulin. Insulin acts on peripheral tissues such as the liver, muscle and adipose to promote glucose uptake and fat and glycogen storage [5]. Insulin also acts centrally to regulate feeding and metabolism. Chronic infusion of insulin into the brains of mammals results in decreased food consumption and fat storage [6,7]. Consistent with this, ablation of the insulin receptor specifically in neurons results in hyperphagia and weight gain [8]. However, the manner by which the expression of these mammalian hormones is regulated in response to changes in nutrient availability is not fully understood.
A mammalian transcription factor that responds to changes in glucose levels is carbohydrate response element binding protein (ChREBP) [9]. ChREBP is highly expressed in the liver, pancreas, and adipose tissue, but is less abundant in the brain, skeletal muscle and small intestine [10]. During times of elevated cellular glucose, ChREBP translocates into the nucleus and dimerizes with the ubiquitously expressed Max-like protein X (Mlx); this complex then binds to DNA to induce target gene expression [11,12]. Specifically, ChREBP functions in the liver by regulating glycogen storage and the expression of lipogenic genes [10]. ChREBP is also important in the development of β-cells of the pancreas and has been implicated in glucose-stimulated insulin secretion in cultured MIN6 β-cells [13,14].
The function of ChREBP is not only limited to peripheral metabolic tissues; ChREBP has also been shown to act centrally to regulate feeding and metabolism. Leptin and ChREBP double knockout mice eat less and have reduced triglyceride stores compared to leptin-deficient mice [15]. Also, these leptin and ChREBP double knockout mice have increased NPY, but lower AgRP mRNA levels (both orexigenic neuropeptides that promote feeding), indicating a role for ChREBP in controlling neuropeptide expression [15]. These results suggest a novel function for ChREBP in the brain, specifically in leptin-responsive hypothalamic neurons to regulate metabolism. Although storage and feeding defects have been previously described in leptin/ChREBP knock-out mice [15], it is largely unknown how ChREBP functions in the brain including how it regulates the expression of neuropeptides.
In order to understand the tissue-specific functions of ChREBP, we study the Drosophila homolog of ChREBP, Mlx interactor (Mio/Mondo), and its binding partner bigmax [16,17]. Mio is expressed predominantly in the fat body, a liver- and adipose-like organ in the fly, and in the malphighian tubules, an excretory organ [18]. Mio responds to glucose in order to regulate lipogenic enzyme expression, similar to ChREBP, defining a conserved function of these genes [19,20]. Additionally, depletion of Mio in the fat body decreases feeding [19], suggesting a role for Mio in mediating the detection of nutrients and the alteration of behavior accordingly. Another group of nutrient-sensitive molecules in Drosophila is the Drosophila insulin-like peptides (dilps). Three of the eight dilps (dilp2, dilp3, and dilp5) are expressed in a region of the fly brain called the pars intercerebralis and have been shown to affect feeding, reminiscent of the hypothalamus in mammals [21–25]. Loss of dilp2 results in increased development time and lifespan, reduced body weight and fecundity, but has no affect on glycogen or triglyceride storage [22]. Interestingly, despite the similarity of dilps 2, 3 and 5, loss of dilp3 or dilp5 only showed a decrease in fecundity [22] Dilp levels and secretion also change in response to nutrient conditions [24,26,27]; however, whether Mio functions in the brain or interacts with the dilps in response to changes in nutrients is unknown.
In this study, we show that Mio functions in the brain not only to control nutrient storage, but also to regulate the expression of previously unknown target genes, such as the dilp class of neuropeptides. When Mio levels are decreased in the entire brain, we observe elevated triglycerides and glycogen with no effect on food consumption; however, targeting of Mio-RNAi specifically in the insulin-producing cells (IPCs) leads to an increase in feeding, while having little effect on nutrient storage. This suggests that Mio is required in multiple neuronal populations to regulate feeding and metabolism. Additionally, when Mio levels are lowered specifically in IPCs, the expression of dilp3 is altered, suggesting that Mio controls the production of certain IPC neuropeptides. Thus, this study identifies a novel player in the nutrient sensing mechanism whereby the brain can detect internal nutrient levels to control nutrient storage, feeding and overall energy homeostasis.
2. Materials and Methods
2.1 Fly genetics
Flies were grown at 25°C on a 12 h:12 h light:dark cycle on standard cornmeal-sugar-yeast medium (9 g Drosophila agar (USA Scientific), 100 mL Karo Lite Corn Syrup, 65 g cornmeal, 40 g sucrose, and 25 g whole yeast in 1.25 L water). The following fly strains used in this study were obtained from the Bloomington Stock Center: w-; UAS-GFPdsRNA (#9331), w-; dilp2-Gal4 (#37516; [24]), and w-; nSyb-Gal4 (#51635; [28]). The w-; UAS-Mio-IR line (#52606) was obtained from the Vienna Drosophila RNAi Center [29]. The w-; UAS-MiodsRNA line was made as previously described [19].
2.2 Triglyceride, glycogen, and protein measurements
Single 5–8 day old female flies were homogenized in lysis buffer (140mM NaCl, 50mM Tris-HCl, pH 7.4, 0.1% Triton-X, 1X protease inhibitor cocktail (Roche Diagnostics)). Triglyceride and protein concentrations were determined using the Stanbio Liquicolor (Fisher Scientific) and BCA Protein Assay (ThermoScientific) kits, respectively, according to manufacturers protocol. Total glucose levels were determined using Glucose Oxidase Reagent (Pointe Scientific) in samples treated with 8 mg/mL amyloglucosidase (Sigma) in 0.2M sodium citrate buffer, pH 5.0. Free glucose was measured in samples not treated with amyloglucosidase and then glycogen concentrations were determined by subtracting the free glucose from total glucose concentration.
2.3 Gene expression analysis
Total RNA was isolated from thirty heads of 5–8 day old adult flies. Heads were homogenized in Ribozol (Amresco) reagent and chloroform extracted. Then an equal volume of isopropanol was added and incubated for 10 minutes at 4°C, after which samples were centrifuged at 12,000 rpm for 15 minutes at 4°C. Supernatants were decanted and pellets were washed twice with 70% ethanol. Pellets were then resuspended in water. Remnant genomic DNA was removed using the TURBO DNA-free Kit (Ambion) as per manufacturer s protocol. Reverse transcription was carried out on 0.25–0.5 μg total RNA with random decamers using the RETROscript kit (Ambion) or qScript cDNA Supermix (Quanta Biosciences) and quantitative PCR was performed on a StepOnePlus thermocycler (Applied Biosystems) using Power SYBR Green (Applied Biosystems). Primer sequences used for qPCR were: Mio (sense 5′ AGCGAGACGAGCTAAACAATTC 3′ and antisense 5′ GTGTAAGAGGCAAGCAAAGGTT 3′), dilp2 (sense 5′ TCTGCAGTGAAAAGCTCAACGA 3′ and antisense 5′ TCGGCACCGGGCATG 3′) [21], dilp3 (sense 5′ AGAGAACTTTGGACCCCGTGAA 3′ and antisense 5′ TGAACCGAACTATCACTCAACAGTCT 3′) [21], dilp5 (sense 5′ GAGGCACCTTGGGCCTATTC 3′ and antisense 5′ CATGTGGTGAGATTCGGAGCTA 3′) [21], dsk (sense 5′ CCGATCCCAGCGCAGACGAC 3′ and antisense 5′ TGGCACTCTGCGACCGAAGC 3′) [30], rp49 (sense 5′ GACGCTTCAAGGGACAGTATCTG 3′ and antisense 5′ AAACGCGGTTCTGCATGAG 3′). The relative concentration of each experimental mRNA was normalized by dividing by rp49 expression levels in each sample. It has been shown that rp49 expression does not change during different dietary conditions, including high carbohydrate intake [31].
2.4 Feeding assay
Food consumption was measured over a 24 hour period by using a modified version of the Capillary Feeder (CAFE) Assay as previously described [32]. Briefly, three 5–8 day old adult female flies were placed in a Drosophila food vial with 1% agar as the only water source and dyed 5% sucrose solution in a 5 μL glass micropipette (Drummond Scientific) as the sole food source. The amount of sucrose solution consumed by the flies was measured after 24hrs and was corrected for any evaporation that occurred during the experiment, which was measured by using identical vials without any flies.
2.5 Statistics
For each set of experiments, the values for GFPdsRNA controls were set to one and the values for the respective experimental measurements are presented relative to the controls. Sample sizes are provided in the figure legends. One-way, independent, weighted ANOVA with Tukey post-hoc tests were then performed to compare each experimental genotype to the GFPdsRNA controls using SPSS software (IBM). P values < 0.05 were considered statistically significant.
3. Results
3.1 Mio is necessary in the brain to regulate nutrient storage
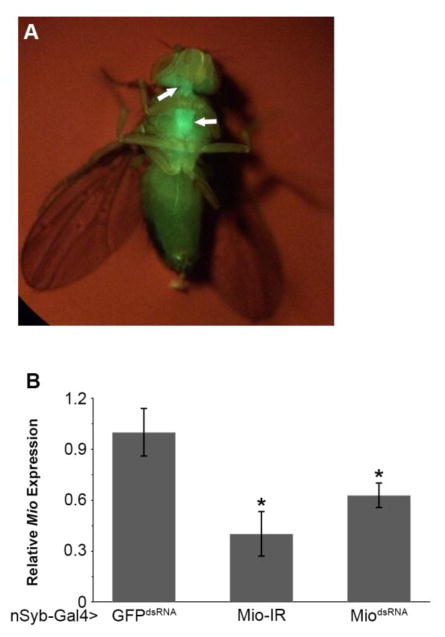
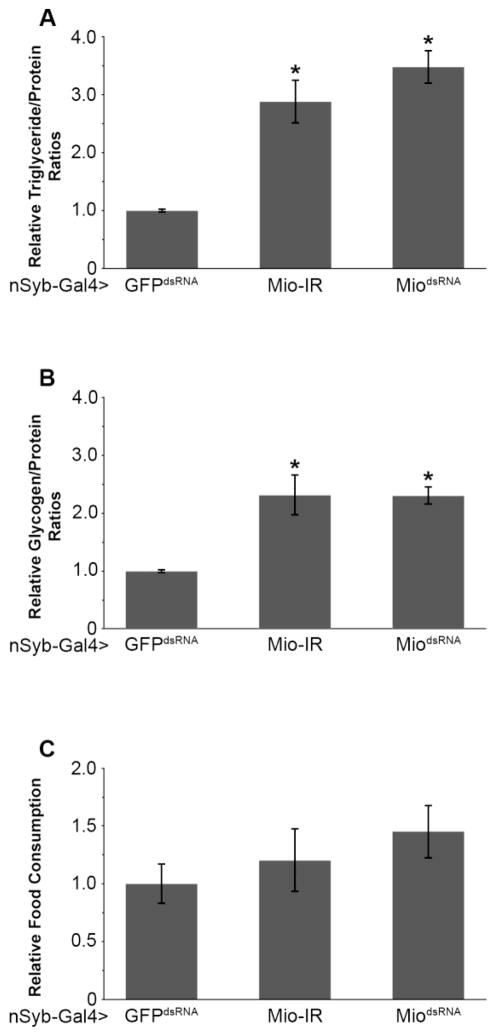
Mio has been shown to control triglyceride storage in the Drosophila fat body [18,19,33]; however, its function in other tissues is unknown. Previously, ChREBP has been shown to be involved in regulating triglyceride storage and neuropeptide expression in mice [15], raising the possibility that Mio also functions in the Drosophila brain to regulate energy storage. To test this hypothesis, we disrupted Mio by targeting RNA interference (RNAi) specifically in neurons using the nSyb-Gal4 driver and two independent Mio-RNAi transgenes. Neural synaptobrevin (nSyb) is a vesicle forming protein essential for synaptic transmission and is present in all neurons [34–36]. To confirm the specificity of the nSyb-Gal4 driver, green fluorescent protein (GFP) was expressed using nSyb-Gal4. As expected, GFP expression was localized specifically to the Drosophila brain as well as the ventral nerve cord (Fig. 1A). nSyb-Gal4 was then used to drive Mio-RNAi in the Drosophila nervous system and knockdown was assessed using quantitative PCR (Fig. 1B). Neuron-specific knockdown of Mio resulted in higher triglycerides and glycogen storage compared to control flies (Fig. 2A,B), suggesting a function for this transcription factor in the brain to limit fat and glycogen accumulation.
Fig. 1. Mio levels are reduced when Mio-RNAi is targeted to the nervous system.
(A) nSyb-Gal4>GFP shows fluorescence expression in neurons. Arrows show GFP fluorescence in the lobes of the brain and the ventral nerve chord. (B) Quantitative PCR was performed for Mio using cDNA from batches of 30 heads from 5–8 day old nSyb-Gal4>Mio-IR (n=12) and nSyb-Gal4>MiodsRNA (n=6) animals compared to nSyb-Gal4>GFPdsRNA controls (n=6). Values represent means ± SEM. *P < 0.05 by one-way independent weighted ANOVA and Tukey post-hoc test comparing each experimental genotype to the GFPdsRNA control.
Fig. 2. Mio acts in the brain to regulate macromolecule storage, but not food consumption.
(A) Triglyceride/protein and (B) glycogen/protein ratios from 5–8 day old nSyb-Gal4>Mio-IR (n=20) and nSyb-Gal4>MiodsRNA (n=45) flies compared to nSyb-Gal4>GFPdsRNA controls (n=34). (C) Total food consumption over 24hrs was measured using the CAFE assay in 5–8 day old nSyb-Gal4>Mio-IR (n=13) and nSyb-Gal4>MiodsRNA (n=27) females compared to nSyb-Gal4>GFPdsRNA controls (n=17). Values represent means ± SEM. *P < 0.05 by one-way, independent, weighted ANOVA and Tukey post-hoc test comparing each experimental genotype to the GFPdsRNA control.
One potential cause for the increase in triglyceride and glycogen storage observed in the Mio-RNAi flies may be higher food consumption. To address this question, expression of Mio was decreased pan-neuronally in flies and feeding was measured using the CAFE assay. Surprisingly, Mio knockdown resulted in no change in feeding behavior (Fig. 2C), suggesting that the increase in triglyceride and glycogen storage is not the result of higher feeding. Together, these data suggest that Mio plays an important role in the Drosophila brain to control whole animal energy storage.
3.2 Mio acts specifically in the IPCs to regulate feeding, but not nutrient storage
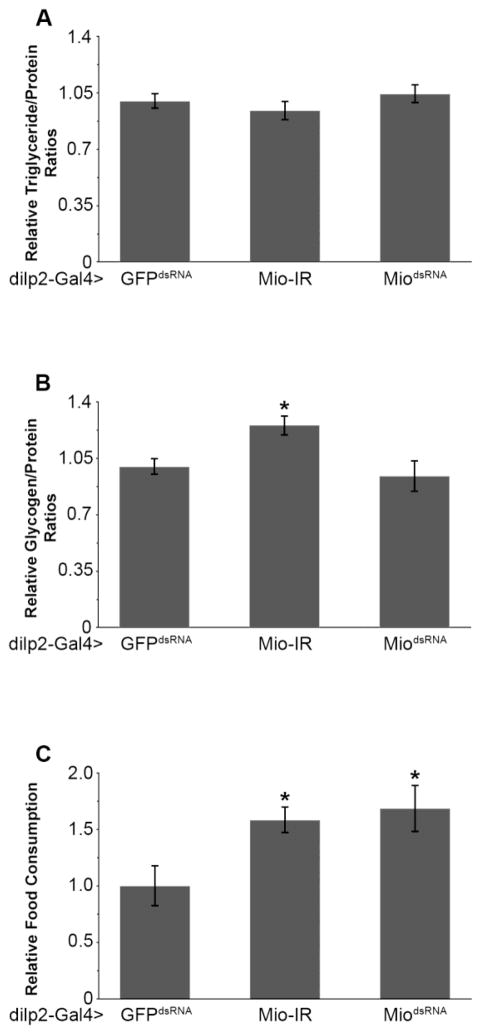
A population of neurons important for the regulation of energy metabolism, longevity and feeding is the collection of insulin producing cells (IPCs) [21,24,37]. To determine whether Mio is required in the IPCs to regulate nutrient storage and metabolism, Mio function was disrupted in the IPCs by inducing RNAi in these cells using the dilp2-Gal4 driver [24]. Decreasing the expression of Mio in the IPCs had no consistently significant effect on triglyceride or glycogen storage (Fig. 3A, B). Interestingly however, IPC-specific knockdown of Mio did result in an increase in food consumption (Fig. 3C). Together, these data suggest that Mio acts in the IPCs to regulate feeding behavior, but not whole body metabolism.
Fig. 3. Mio functions specifically in the IPCs to control feeding, but not nutrient storage.
(A) Triglyceride/protein and (B) glycogen/protein ratios from 5–8 day old dilp2-Gal4>Mio-IR (n=56) and dilp2-Gal4>MiodsRNA (n=40) flies compared to dilp2-Gal4>GFPdsRNA controls (n=36). (C) Total food consumption over 24hrs was measured using the CAFE assay on 5–8 day old dilp2-Gal4>Mio-IR (n=22) and dilp2-Gal4>MiodsRNA (n=15) flies compared to dilp2-Gal4>GFPdsRNA controls (n=12). Values represent means ± SEM. *P < 0.05 by one-way, independent, weighted ANOVA and Tukey post-hoc test comparing each experimental genotype to its respective GFPdsRNA control.
3.3 Mio is essential for normal expression of dilp mRNA
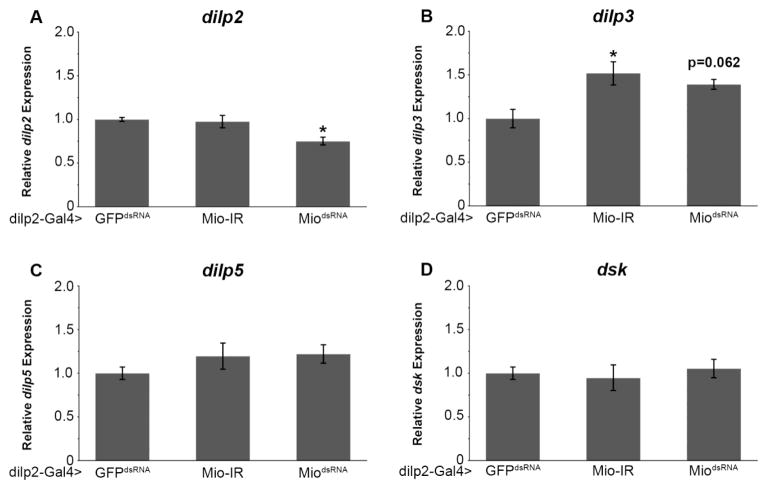
While Mio has been shown to regulate feeding and nutrient storage in the fat body [18,19,33], the full array of target genes of this transcription factor is only beginning to be understood. Many of the metabolic functions of the brain are mediated by peptides that are secreted from IPCs, such as the Drosophila insulin-like peptides 2, 3, and 5 (dilp2, 3, and 5) and the cholecystokinin-like peptide, drosulfakinin (Dsk) [22,30]. Similar to the dilps, Dsk has also been found to be an anorexigenic peptide, limiting food consumption [30]; whether Dsk has any additional metabolic functions is unknown. Since we have shown that Mio acts in the IPCs to control feeding, we hypothesized that Mio may be affecting this process by regulating the expression of the dilp2, 3, 5 or dsk genes. To test this hypothesis, quantitative PCR was used to measure dilp2, 3, 5, and dsk mRNA levels in flies where Mio was decreased in the IPCs. Interestingly, disrupting Mio in IPCs leads to an increase in dilp3 mRNA expression (Fig. 4B), but has no consistent effect on dilp2, dilp5 or dsk expression (Fig. 4A,C,D). Our evaluation of these neuropeptides suggests that Mio acts in the IPCs to regulate the expression of dilp3 to control feeding, but not nutrient storage.
Fig. 4. Mio regulates dilp expression in the IPCs.
Quantitative PCR was performed for (A) dilp2, (B) dilp3, (C) dilp5, and (D) dsk using cDNA from batches of 30 heads from 5–8 day old dilp2-Gal4>Mio-IR (n=4–8) and dilp2-Gal4>MiodsRNA (n=4–8) animals compared to dilp2-Gal4>GFPdsRNA controls (n=6). Values represent means ± SEM. *P < 0.05 by one-way independent, weighted, ANOVA and Tukey post-hoc test comparing each experimental genotype to the GFPdsRNA control.
4. Discussion
In addition to being essential for development [16,18], Mio has also been shown to be important for regulating glucose and lipid metabolism by controlling the expression of many important metabolic genes [18,19,33]. This metabolic function of Mio was previously localized to the fat body, since normal Mio expression in this tissue is necessary for proper feeding and triglyceride storage [19]. Our current findings uncover a novel function of Mio expression in the Drosophila brain, specifically in the insulin-producing cells (IPCs).
Here we show that pan-neuronal depletion of Mio promotes triglyceride and glycogen storage (Fig. 2A,B) while decreasing Mio in the IPCs leads to no change in triglyceride and glycogen levels (Fig. 3A,B). While lowering Mio levels in the entire brain elevates nutrient stores, there is no noticeable change in food consumption (Fig. 2C). This result suggests that the increased nutrient storage phenotypes are not due to increased feeding and caloric intake. Therefore it is possible that the macromolecule storage phenotype shown here is due to altered neural or endocrine actions of specific populations of neurons affecting overall energy expenditure or acting directly on fat body cells to regulate nutrient storage, as seen previously in mammals [8,38–46]. This neuronal function of Mio is different than when Mio functions in the fat body. Decreasing Mio specifically in the fat body tissue results in decreased triglycerides and lipogenic gene expression [19,33] suggesting an autonomous role for Mio in regulating these processes. However, targeted depletion of Mio in the fat body also results in decreased feeding, suggesting a non-autonomous effect on the brain [19]. The fat body has been shown to have endocrine functions producing molecules that act on the brain to regulate metabolism in response to changes in the nutrient conditions. One example is unpaired2 (upd2), the JAK/STAT ligand which is released from the fat body during the fed state and acts on the IPCs in the fly brain to regulate dilp secretion [47]. Therefore, it is possible that Mio may be involved in regulating this process. Further dissection of the fat body and neuron-specific functions of Mio will be essential to understanding how Mio regulates overall metabolic homeostasis in the animal.
The differences in the nutrient storage phenotypes observed when Mio is reduced in all neurons, and specifically in IPCs, suggest that Mio acts in additional non-IPC neurons to regulate energy metabolism. A number of neuronal populations (i.e., the circadian neurons, octopamine-producing neurons and obesity-blocking neurons) have been previously shown to play a role in nutrient storage and metabolism [38,44,48]; whether Mio acts in these cells to regulate energy homeostasis is currently unknown. In addition, Mio may also function in multiple neuronal populations to regulate food consumption as pan-neuronal Mio depletion has no effect on feeding, while IPC-specific Mio-RNAi results in increased food consumption (Fig. 2C, 3C). For example, the neurons that produce neuropeptide F (NPF, a hormone that stimulates feeding) as well as the obesity-blocking neurons have also been shown to have significant effects on feeding behavior [25,38,49,50]. However, whether Mio functions in these groups of neurons is unknown. Therefore, it is possible that decreasing Mio pan-neuronally could affect the NPF-secreting or the obesity-blocking neurons as well as the IPCs, resulting in the differences seen in food consumption when Mio-RNAi is induced in the whole brain and the IPCs.
Additionally, the nSyb driver is active throughout the larval stages of fly development as well as in adult flies (Fig 1A; data not shown). This raises the possibility that Mio could act in the brain throughout development to regulate nutrient storage. As nSyb is a vesicle forming protein found in all neurons [34,35] and dilp2-Gal4 is specific to the IPCs [24], it is possible that the differing feeding and nutrient storage phenotypes between the pan-neuronal and IPC-specific knockdown of Mio could be due to different expression levels in the IPCs in the different drivers. Additional experimentation is necessary to further understand the full localization of Mio action in the brain as well as the temporal requirements of Mio function to control feeding and metabolism.
Interestingly, our data show that decreasing Mio specifically in the IPCs results in an increase in dilp3 mRNA expression; however, whether dilp3 is a direct target of Mio is still unknown. Although exogenous dilp2 expression has been shown to reduce feeding [37], whether dilp3 and dilp5 regulate feeding is still unclear. Additionally, once Drosophila dilps are secreted from IPCs, these peptides function on their target tissues to activate a single insulin-like receptor, dInR, leading to organismal changes in metabolism and feeding behavior [51]. Since all secreted dilps act on a single receptor [51,52], it is possible that the increased dilp3 mRNA found in this study could produce dilp3 protein that acts on similar target cells as dilp2, and act as a compensatory mechanism for increased food consumption. Additionally, dilp3 and 5 expression has also been shown to be controlled by nutrient availability through experiments where expression of dilp3 and 5 is reduced in starved larvae [27]. Since Mio has been shown to coordinate gene expression in response to changes in nutrients [19,33], it is also possible that Mio functions to regulate dilp3 expression in the IPCs in accordance with nutrient availability, as increasing food availability gives rise to increased dilp3 levels [27]. In the future, additional experiments focused on fully understanding the molecular mechanism by which Mio regulates dilp expression in IPCs will help define the tissue-specific roles of metabolic transcription factors.
In summary, the data presented here show that Mio, the Drosophila homolog of the mammalian ChREBP, functions in distinct neuronal populations to regulate lipid and glycogen storage as well as feeding. This study has identified a neuronal site of action for the metabolic transcription factor Mio and furthers our understanding of how the brain coordinates energy metabolism and storage at the organismal level.
Highlights.
Mio/dChREBP is an important metabolic regulator in Drosophila.
Nervous system-specific function of Mio to regulate metabolism is unknown.
Mio acts in the brain to control nutrient storage.
Mio acts in the insulin-producing neurons to regulate dilp3 mRNA levels.
This study furthers our understanding of how the brain controls metabolism.
Acknowledgments
We would like to thank the Bloomington Stock Center (NIH P40OD018537) and the Vienna Drosophila RNAi Center for fly stocks used in this study. This work was supported by NIH grant 1R15NS080155-01A1 and funds from Hofstra University to JRD.
Abbreviations List
- CAFÉ
Capillary feeder assay
- ChREBP
Carbohydrate response element binding protein
- Dilp
Drosophila insulin-like peptide
- Dsk
Drosulfakinin
- IPCs
Insulin-producing cells
- Mio
Mlx interactor
- Mlx
Max-like protein X
- RNAi
RNA interference
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Suzuki K, Simpson KA, Minnion JS, Shillito JC, Bloom SR. The role of gut hormones and the hypothalamus in appetite regulation. Endocr J. 2010;57:359–372. doi: 10.1507/endocrj.k10e-077. [DOI] [PubMed] [Google Scholar]
- 2.Saper CB, Chou TC, Elmquist JK. The Need to Feed: Homeostatic and Hedonic Control of Eating. Neuron. 2002;36:199–211. doi: 10.1016/S0896-6273(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 3.Tschöp M, Smiley DL, Heiman ML. Ghrelin induces adiposity in rodents. Nature. 2000;407:908–913. doi: 10.1038/35038090. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 5.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz MW, Woods SC, Porte D, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 7.Woods SC, Lotter EC, McKay LD, Porte D., Jr Chronic intracerebroventricular infusion of insulin reduces food intake and body weight of baboons. Nature. 1979;282:503–505. doi: 10.1038/282503a0. [DOI] [PubMed] [Google Scholar]
- 8.Brüning JC, Gautam D, Burks DJ, Gillette J, Schubert M, Orban PC, et al. Role of Brain Insulin Receptor in Control of Body Weight and Reproduction. Science. 2000;289:2122–2125. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, et al. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci U S A. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci U S A. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billin AN, Eilers AL, Queva C, Ayer DE. Mlx, a Novel Max-like BHLHZip Protein That Interacts with the Max Network of Transcription Factors. J Biol Chem. 1999;274:36344–36350. doi: 10.1074/jbc.274.51.36344. [DOI] [PubMed] [Google Scholar]
- 12.Filhoulaud G, Guilmeau S, Dentin R, Girard J, Postic C. Novel insights into ChREBP regulation and function. Trends Endocrinol Metab TEM. 2013;24:257–268. doi: 10.1016/j.tem.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Soggia A, Flosseau K, Ravassard P, Szinnai G, Scharfmann R, Guillemain G. Activation of the transcription factor carbohydrate-responsive element-binding protein by glucose leads to increased pancreatic beta cell differentiation in rats. Diabetologia. 2012;55:2713–2722. doi: 10.1007/s00125-012-2623-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamato E, Tashiro F, Miyazaki J. Microarray analysis of novel candidate genes responsible for glucose-stimulated insulin secretion in mouse pancreatic β cell line MIN6. PloS One. 2013;8:e61211. doi: 10.1371/journal.pone.0061211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iizuka K, Miller B, Uyeda K. Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am J Physiol Endocrinol Metab. 2006;291:E358–364. doi: 10.1152/ajpendo.00027.2006. [DOI] [PubMed] [Google Scholar]
- 16.Billin AN, Ayer DE. The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism. Curr Top Microbiol Immunol. 2006;302:255–278. doi: 10.1007/3-540-32952-8_10. [DOI] [PubMed] [Google Scholar]
- 17.Peyrefitte S, Kahn D, Haenlin M. New members of the Drosophila Myc transcription factor subfamily revealed by a genome-wide examination for basic helix-loop-helix genes. Mech Dev. 2001;104:99–104. doi: 10.1016/S0925-4773(01)00360-4. [DOI] [PubMed] [Google Scholar]
- 18.Havula E, Teesalu M, Hyotylainen T, Seppala H, Hasygar K, Auvinen P, et al. Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in Drosophila. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sassu ED, McDermott JE, Keys BJ, Esmaeili M, Keene AC, Birnbaum MJ, et al. Mio/dChREBP coordinately increases fat mass by regulating lipid synthesis and feeding behavior in Drosophila. Biochem Biophys Res Commun. 2012;426:43–48. doi: 10.1016/j.bbrc.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinke I, Schütz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broughton SJ, Piper MDW, Ikeya T, Bass TM, Jacobson J, Driege Y, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grönke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010;6:e1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nässel DR, Winther ÅME. Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol. 2010;92:42–104. doi: 10.1016/j.pneurobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 25.Wu Q, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat Neurosci. 2005b;8:1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
- 26.Géminard C, Rulifson EJ, Léopold P. Remote Control of Insulin Secretion by Fat Cells in Drosophila. Cell Metab. 2009;10:199–207. doi: 10.1016/j.cmet.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-Dependent Expression of Insulin-like Peptides from Neuroendocrine Cells in the CNS Contributes to Growth Regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/S0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 28.Jenett A, Rubin GM, Ngo TTB, Shepherd D, Murphy C, Dionne H, et al. A GAL4-Driver Line Resource for Drosophila Neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 30.Soderberg JAE, Carlsson MA, Nassel DR. Insulin-Producing Cells in the Drosophila Brain also Express Satiety-Inducing Cholecystokinin-Like Peptide, Drosulfakinin. Front Endocrinol. 2012;3 doi: 10.3389/fendo.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lissemore JL, Baumgardner CA, Geer BW, Sullivan DT. Effect of dietary carbohydrates and ethanol on expression of genes encodingsn-glycerol-3-phosphate dehydrogenase, aldolase, and phosphoglycerate kinase inDrosophila larvae. Biochem Genet. 1990;28:615–630. doi: 10.1007/BF00553954. [DOI] [PubMed] [Google Scholar]
- 32.Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musselman LP, Fink JL, Ramachandran PV, Patterson BW, Okunade AL, Maier E, et al. Role of Fat Body Lipogenesis in Protection against the Effects of Caloric Overload in Drosophila. J Biol Chem. 2013;288:8028–8042. doi: 10.1074/jbc.M112.371047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Estes PS, Ho GL, Narayanan R, Ramaswami M. Synaptic localization and restricted diffusion of a Drosophila neuronal synaptobrevin--green fluorescent protein chimera in vivo. J Neurogenet. 2000;13:233–255. doi: 10.3109/01677060009084496. [DOI] [PubMed] [Google Scholar]
- 35.Deitcher DL, Ueda A, Stewart BA, Burgess RW, Kidokoro Y, Schwarz TL. Distinct requirements for evoked and spontaneous release of neurotransmitter are revealed by mutations in the Drosophila gene neuronal-synaptobrevin. J Neurosci Off J Soc Neurosci. 1998;18:2028–2039. doi: 10.1523/JNEUROSCI.18-06-02028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soustelle L, Giangrande A. Novel gcm-dependent lineages in the postembryonic nervous system of Drosophila melanogaster. Dev Dyn. 2007;236:2101–2108. doi: 10.1002/dvdy.21232. [DOI] [PubMed] [Google Scholar]
- 37.Hong SH, Lee KS, Kwak SJ, Kim AK, Bai H, Jung MS, et al. Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in Drosophila and Mammals. PLoS Genet. 2012;8:e1002857. doi: 10.1371/journal.pgen.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Anzi B, Sapin V, Waters C, Zinn K, Wyman RJ, Benzer S. Obesity-blocking neurons in Drosophila. Neuron. 2009;63:329–341. doi: 10.1016/j.neuron.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker KD, Thummel CS. Diabetic larvae and obese flies - emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouret SG, Simerly RB. Minireview: Leptin and development of hypothalamic feeding circuits. Endocrinology. 2004;145:2621–2626. doi: 10.1210/en.2004-0231. [DOI] [PubMed] [Google Scholar]
- 41.Erickson JC, Hollopeter G, Palmiter RD. Attenuation of the obesity syndrome of ob/ob mice by the loss of neuropeptide Y. Science. 1996;274:1704–1707. doi: 10.1126/science.274.5293.1704. [DOI] [PubMed] [Google Scholar]
- 42.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 43.Little TJ, Horowitz M, Feinle-Bisset C. Role of cholecystokinin in appetite control and body weight regulation. Obes Rev. 2005;6:297–306. doi: 10.1111/j.1467-789X.2005.00212.x. [DOI] [PubMed] [Google Scholar]
- 44.DiAngelo JR, Erion R, Crocker A, Sehgal A. The Central Clock Neurons Regulate Lipid Storage in Drosophila. PLoS ONE. 2011;6:e19921. doi: 10.1371/journal.pone.0019921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiAngelo JR, Birnbaum MJ. Regulation of Fat Cell Mass by Insulin in Drosophila melanogaster. Mol Cell Biol. 2009;29:6341–6352. doi: 10.1128/MCB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu K, Zheng X, Sehgal A. Regulation of Feeding and Metabolism by Neuronal and Peripheral Clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajan A, Perrimon N. Drosophila Cytokine Unpaired 2 Regulates Physiological Homeostasis by Remotely Controlling Insulin Secretion. Cell. 2012;151:123–137. doi: 10.1016/j.cell.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erion R, Sehgal A. Regulation of insect behavior via the insulin-signaling pathway. Front Invertebr Physiol. 2013;4:353. doi: 10.3389/fphys.2013.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Q, Wen T, Lee G, Park JH, Cai HN, Shen P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron. 2003;39:147–161. doi: 10.1016/s0896-6273(03)00396-9. [DOI] [PubMed] [Google Scholar]
- 50.Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nässel DR, Kubrak OA, Liu Y, Luo J, Lushchak OV. Factors that regulate insulin producing cells and their output in Drosophila. Invertebr Physiol. 2013;4:252. doi: 10.3389/fphys.2013.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez R, Tabarini D, Azpiazu N, Frasch M, Schlessinger J. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 1995;14:3373–3384. doi: 10.1002/j.1460-2075.1995.tb07343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]