Abstract
Aberrant activation of the Wnt signaling pathway is an important step in the initiation and progression of tumor development in diverse cancers. The central effector of canonical Wnt signaling, β-catenin (CTNNB1), is a multifunctional protein, and has been extensively studied with respect to its roles in cell-cell adhesion and in regulation of Wnt-driven transcription. Here, a novel mass spectrometry-based proteomics technique in colorectal cancer cells expressing stabilized β-catenin, was used to identify a protein-protein interaction between β-catenin and DNA methyltransferase I (Dnmt1) protein, the primary regulator of DNA methylation patterns in mammalian cells. Dnmt1 and β-catenin strongly co-localized in the nuclei of colorectal cancer cells, and the interaction is mediated by the central domain of the Dnmt1 protein. Dnmt1 protein abundance is dependent upon the levels of β-catenin, and is increased in cells expressing stabilized mutant β-catenin. Conversely, the Dnmt1 regulates the levels of nuclear β-catenin and β-catenin/TCF driven transcription. In addition, lysine-specific demethylase 1 (LSD1/KDM1A), a regulator of DNMT1 stability, was identified as a component of the Dnmt1/β-catenin protein complex and perturbation of the Dnmt1/β-catenin interaction altered DNA methylation. In summary, a functional protein-protein interaction was identified between two critically important oncoproteins, in turn revealing a link between Wnt signaling and downstream nuclear functions mediated by Dnmt1.
Keywords: β-catenin, Wnt Pathway, DNA methyltransferase, protein-protein interactions, proteomics, Signal Transduction
INTRODUCTION
Cancer cells typically exhibit complex, multi-layered perturbations of signalling pathways and regulatory mechanisms (1). Dys-regulation of the Wnt signalling pathway is a major factor in the initiation and progression of colorectal cancer. Canonical or β-catenin-dependent Wnt signalling is the best-defined branch of Wnt signalling, and is activated by binding of a Wnt ligand with specific cell surface receptor complexes. The subsequent signalling cascade leads to accumulation and nuclear translocation of β-catenin, the central effecter of canonical Wnt signalling. Activating mutations of β-catenin, that stabilize the protein, or loss of function of tumour suppressors such as Adenomatous Polyposis Coli (APC), are causal events in the initiation of colorectal cancer (2,3). β-catenin is one of the best studied oncoproteins and diverse β-catenin protein-protein interactions have been identified. Recent studies, for example, have revealed the association of β-catenin with chromatin and epigenetic modifying complexes (4,5), and it is clear that our knowledge of β-catenin protein function is quite incomplete.
In this study we identify a protein-protein interaction between β-catenin and Dnmt1, the primary maintenance DNA methyltransferase in mammalian cells. Dnmt1 (Dnmt1 refers to the protein product of the DNMT1 gene) expression is altered in many different tumours (6–10), although somatic mutations of DNMT1 are relatively rare events in human cancers. Although best known for its role in DNA methylation, other functions have been defined for Dnmt1 protein, including the formation of transcriptional repressor complexes with HDAC and DMAP1 proteins (11) and with the LSD1 histone demethylase (12). In addition, many protein-protein interactions between Dnmt1 and transcription factors have been identified, principally mediated via the N-terminal regulatory region of the protein (13). DNMT1 itself is subject to multiple layers of regulation, including transcriptional (14) and post-translational regulation through control of Dnmt1 protein stability (13,15,16). How these different mechanisms contribute to regulation of Dnmt1 (and DNA methyltransferase activities in general) in cancer cells is only partially understood. In addition, in many cancer cells aberrant expression of DNA methyltransferases occurs alongside dys-regulated signal transduction pathways, but the extent to which signalling pathways regulate Dnmt1 and associated functions remains to be determined.
Here we use mass-spectrometry, co-immunoprecipitation and confocal microscopy to identify and characterize the Dnmt1-β-catenin interaction. We show that the interaction is mediated by the central portion of the Dnmt1 protein and that Dnmt1 and β-catenin protein levels are mutually dependent. We show that Dnmt1 protein levels are responsive to exogenous Wnt activation that the response is not mediated via transcriptional mechanisms. Finally, to investigate the functional consequences of the Dnmt1-β-catenin interaction, we show that the interaction with Dnmt1 protein regulates both the level of β-catenin and β-catenin/TCF-driven transcriptional activity, and that CpG methylation of an imprinted locus is significantly reduced in cells lacking β-catenin. In summary, our study identifies a novel mechanism by which the levels of these two important oncoproteins are regulated in cancer cells and points to a regulatory link between Wnt signalling and epigenetic functions of Dnmt1.
MATERIALS AND METHODS
Cell culture
Colorectal cancer cell lines RKO and HCT116 were maintained in McCoy-5A media (Life Technologies, 16600-108, Carlsbad, CA) containing 10% fetal bovine serum (Life Technologies, 10438-026, Carlsbad, CA) and 1% streptomycin-penicillin (Life Technologies, 15140-148, Carlsbad, CA) at 37°C in CO2 incubator (5% CO2, 100% H2O). Human Embryonic Kidney cell line HEK293T was maintained in DMEM media (Life Technologies, 11965-092, Carlsbad, CA) containing 10% fetal bovine serum and 1% streptomycin-penicillin under the same conditions. For Wnt activation, media were removed and the cells were washed twice with serum-free McCoy5A media, and 1ml serum-free McCoy5A media, added in each well of 6-well cell culture plate with purified Wnt3a protein (R&D Systems, Inc. 5036- WNP-010/CF, Minneapolis, MN) at the required final concentration of 30ng/ml (17) and the cells were cultured for an additional 0.5, 1, 2, 3, 6, 12, or 24 hours before harvesting. Cells were harvested by scraping the cells off plates and then washed with cold PBS twice for immediate use or storage (-80°C). Knock-out cell-lines were provided by the respective laboratories in which they were generated and cultured under the same conditions as the parent cell-lines (20, 21).
Protein extraction and quantification
Harvested cells were lysed in buffer (25 mM Tris-HCl, pH7.4, 1 mM EDTA, 150mM NaCl, 1% NP-40, 50% glycerol, Protease inhibitor cocktail) by homogenization and incubated on ice for 30 minutes followed by centrifugation at 13,000rpm for 30 minutes. Benzonase nuclease (Sigma E1014, Saint Louis, MO) was added to the lysis buffer as required and the supernatant (soluble fraction) kept for further analysis. Proteins were quantified by Bio-Rad protein assay dye (Bio-Rad 500-0006, Hercules, CA) at 595nm.
RNA extraction and RT-PCR
Total RNA was extracted from Wnt3a stimulated HEK293T time course cells using RNeasy Mini Kit (Qiagen, 74104, Valencia, CA) and 1μg RNA was used in one step RT-PCRs using SuperScript™ One-Step RT-PCR with Platinum® Taq (Life Technologies 10928-034, Carlsbad, CA) on DNMT1 with forward primer 5’-GTGGGGGACTGTGTCTCTGT-3’and reverse primer 5’- TGCTGCCTTTGATGTAGTCG-3’, CTNNB1 with forward primer 5’-AAGCCTCTCGGTCTGTGG-3’ and reverse primer 5’-TGATGGTTCAGCCAAACGCT-3’, and GAPDH with forward primer 5’-CCGTCTAGAAAAACCTGCC -3’ and reverse primer 5’-GCCAAATTCGTTGTCATACC -3’ as loading control. one-step RT-PCR conditions were set according to manufactory's protocol (Life Technologies 10928-034, Carlsbad, CA) with annealing temperature at 55°C. The amplified DNA products were then electrophoresed on 1.5% agarose gel containing SYBR® Safe DNA gel stain (Life Technologies, S33102, Carlsbad, CA).
Flow cytometry analysis
HEK293T cells at different time points (0hr, 0.5hrs, 1hrs, 2hrs, 3hrs, 6hrs, 12hrs and 24hrs) after stimulation were collected, washed twice with ice cold PBS buffer (137mMNaCl, 2.7mMKCl, 8mM Na2HPO4, 1.46mM KH2PO4), resuspended in 50 μl PBS buffer and fixed with 450 μl 100% Methanol in -20°C for at least 20 minutes. The cells were then treated with RNase (Life Technologies, 12091, Carlsbad, CA) in for 30 minutes before stained with 100 μg/ml propidium iodide and subsequently subjected to flow cytometry analyser (Beckman Coulter Epics XL flow cytometer). A minimum of ten thousand cells within the gated region were analysed and data were captured and presented by Expo32 ADC Cytometry List Mode Data Acquisition & Analysis Software.
SDS-PAGE and Immunoblotting
Equal amounts (20 μg) of proteins from different samples were loaded on precast 4−12% Bis-Tris gel (Life Technologies NP-0335, Carlsbad, CA). Following electrophoresis, gels were either stained with Coomassie Brilliant Blue (Pierce 20278, Rockford, IL) or transferred to nitrocellulose membrane (Whatman 10402594, Dassel, Germany). Western blotting was used to detect the protein with super signal ELISA Pico chemiluminescent substrate. Primary antibodies anti-β-catenin (Cell Signaling Technology 9581, Danvers, MA), anti-β-catenin (active) (Signaling Technology8814, Danvers, MA), anti-Dnmt1 (Cell Signaling Technology 5119, Danvers, MA) and anti-α-tubulin (Cell Signaling Technology, Inc., 2144, Danvers, MA) as loading control were applied at 1:1000 and secondary antibodies horseradish peroxidase (HRP)-conjugated anti-mouse (Promega W4011, Madison, WI) and HRP- conjugated anti-rabbit (Cell Signaling Technology 7074, Danvers, MA) were added at 1:20,000. Chemiluminescence detection using SuperSignal* ELISA Pico Chemiluminescent Substrate (Thermo Scientific PI-37070, Rockford, IL) was applied to all westerns. Bands were quantified by ImageJ (http://rsbweb.nih.gov/ij/) (20,21) and the mean values and standard deviations computed for three replicates.
Proteomic Analysis
Standard in-gel tryptic digestion was performed according to the published method (22). The combined elution fractions were lyophilized in a SpeedVac Concentrator (Thermo Electron Corporation, Milford, MA), resuspended in 100μl of 0.1% formic acid and further cleaned up by reverse phase chromatography using C18 column (Harvard, Southborough, MA). The final volume was reduced to 10μl by vacuum centrifugation and addition of 0.1% formic acid. Tryptic peptides were separated by online reverse phase nanoscale capillary liquid chromatography (nano-LC, Dionex Ultimate 3000 series HPLC system) coupled to electoral spray injection (ESI) tandem mass spectrometer (MS−MS) with octopole collision cell (Thermo-Finnegan LTQ Orbitrap). Loaded peptides were eluted on nano-LC with 90 min gradients ranging from 6 to 73% acetonitrile in 0.5% formic acid with a flow rate of 300 nl/min. Data dependent acquisition was performed on the LTQ-Orbitrap using Xcalibur software (version2.0.6, Thermo Scientific) in the positive ion mode with a resolution of 60 000 at m/z range of 325.0−1800.0, and using 35% normalized collision energy, up to five most intensive multiple charged ions were sequentially isolated, fragmented and further analysed.
Mass-spectrometry data processing
Raw LC-MS/MS data were processed using Mascot version 2.2.0 (Matrix Science, Boston, MA). The sequence database was searched with a fragment ion mass tolerance of 0.8Da and a parent ion tolerance of 15 PPM. The raw data were searched against the human International Protein Index database (released in 2009 and containing 74,017 protein sequences) with fixed modification carbamidomethyl (C) and variable modification oxidation (M). The five fractions for each sample were combined as a single search in Mascot. Peptides were filtered at a significance threshold of P < 0.05 (Mascot). Scaffold (Proteome Software Inc., Portland, OR, USA; version 3.00.04) was used to analyse LC-MS/MS-based peptide and protein identifications (23).Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (24). Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides (24).
Immunofluorescence
Cells were grown to 50% confluence on cover slip slides (12-545-80, Fisherbrand* Cover Glasses) overnight and fixed first with 4% formaldehyde in PBS (phosphate Buffered Saline) for 15 min at room temperature. Fixed cells were blocked with blocking buffer composed of 1xPBS with 5% goat serum (Cell Signaling Technology 5425, Danvers, MA) and 0.3% Triton X-100 for 60 minutes at room temperature. Cells were then probed with an anti-FLAG (Sigma F1804, Saint Louis, MO) sera at dilution of 1:200 and β-catenin (D10A8) XP Rabbit antibody (Cell Signaling Technology 8480, Danvers, MA) at dilution of 1:100 for 2 hours at room temperature, washed with PBS buffer three times of 5 min each, then incubated with secondary Alexa Fluor® 594 Goat Anti-Mouse IgG, highly cross-adsorbed antibody (Life Technologies A31624, Carlsbad, CA) and Alexa Fluor® 488 Goat Anti-Rabbit IgG (H+L), highly cross-adsorbed antibody (Life Technologies A11034, Carlsbad, CA) at a dilution of 1:1000 for 30 min at room temperature in dark. Nuclei were stained with DAPI (Cell Signaling Technology 8961, Danvers, MA). Next, the Slides were rinsed with PBS and Prolong Gold Anti-Fade Reagent (Cell Signaling Technology 9071, Danvers, MA) was applied. Slides were then analysed and images were taken on Leica TCS SP2 AOBS filter-free UV/spectral confocal laser scanner on an inverted DM IRE2 microscope.
RNAi
HCT116 and RKO cells were transfected with targeted siRNAs against DNMT1 and CTNNB1 using Lipofectamine 2000 transfection reagent (Life Technologies 11668, Carlsbad, CA) and the cells were then incubated at 37°C in CO2 incubator and harvested 48 hours after transfection. Three gene unique 27mer siRNAs for each target were synthesized by Origene (Rockville, MD). The three target sequences of DNMT1are ACCAAAUUACGUAAAGAAGAAUUAT (SR301244A), AGCACAGAAGUCAACCCAAAGAUCT (SR301244B) and UGAGUGGAAAUUAAGACUUUAUGTA (SR301244C). The three target sequences of CTNNB1 are GGAUCACAAGAUGGAAUUUAUCAAA (SR301063A), CGCAUGGAAGAAAUAGUUGAAGGTT (SR301063B) and AGAAUUGAGUAAUGGUGUAGAACAC (SR301063C). Control siRNA (SR30004) is designed and provided by Origene.
LEF/TCF reporter assays
Targeted cells (HCT116, RKO, DNMT1KO-HCT116 and CTNNB1KO-HCT116) were seeded onto multiwall plates one day before transfection and grown to 70-90% confluence. Vectors of LEF/TCF reporter, negative control and positive control were then individually premixed with transfection reagent Lipofectamine2000 (Life Technologies 11668, Carlsbad, CA) and Opti-MEM serum-free culture medium and incubated at room temperature for 20 minutes. For co-transfection with siRNA, three target sequences of either DNMT1 or CTNNB1 (see previous description in RNAi) were added into the premix and incubated at room temperature for 20 minutes before use. Luciferase assay was performed using Dual-Luciferase reporter assay system (Promega 1910, Madison, WI) 48 hours after transfection. The cells were lysed in plate using passive lysis buffer (Promega 1941, Madison, WI) for 15 minutes at room temperature with gentle shaking. Cell lysates were then transferred into a 96-well plate and development substrates were added according to the instructions (Promega 1910, Madison, WI). Firefly luciferase and Renilla luciferase activity were measure by microplate reader premium Quad4 Monochromators (Tecan Group Inc. Infinite M1000 pro, Switzerland). The promoter activity values were expressed as arbitrary units using a Renilla reporter for internal normalization according to manufactory's protocol (Promega 1910, Madison, WI). For experiments using TOPFlash/FOPFlash plasmids used were M50 and M51 Super 8x TopFLASH (25) (Addgene). Experiments were done in triplicates for all biological cell cultures/ transfections and luminescent measurements. Mean (bar height) of relative luciferase units for each sample plus standard deviation (error bar) were then calculated and plotted.
Quantification of Protein Half-Life
HCT116, DNMT1KO-HCT116 and CTNNB1KO-HCT116 were grown in separate 6-well plates to log phase. Cycloheximide (Cell Signaling 2112, Danvers, MA) was added to a final concentration of 50 μg/ml to terminate protein synthesis. After cycloheximide treatment, equal numbers of cells were collected at 0, 3, and 6 hours, and cell lysates were prepared as described. The lysates were analysed by SDS-PAGE and Western blotting. Bands corresponding to each protein were detected by using chemiluminescence and the intensity of bands was quantified using ImageJ. Half-life (T1/2) was calculated as previously described (26).
Methylation analysis of H19 CpG islands
Genomic DNA was extracted with the QIAamp DNA Mini Kit (Qiagen 51804, Valencia, CA). Bisulfite treatment of the genomic DNA samples was carried out with the Qiagen EpiTect kit (Qiagen 59104, Valencia, CA) according to the manufacturer's instructions, followed by PCR amplification with specific primers for H19 promoter region (forward: 5′- GGTCCCA/ideoxyU/ATGTAAGATTTTGGTGGAATAT-3′; reverse: 5′- GGCATAG/ideoxyU/ACAAACTCACACATCACAACC-3′). The PCR products were gel-purified, inserted into USER cloning vector zw102 (courtesy of Wang lab), and sequenced with T3 universal primer.
RESULTS
A Dnmt1-β-catenin protein-protein interaction
We previously performed a large-scale proteomics study to map the Wnt-responsive proteome (17). Proteins were identified whose expression levels responded to exogenous Wnt3A stimulation and we observed that Dnmt1 protein levels exhibited a robust response to Wnt activation in HEK293T cells and in the colorectal cancer cell-lines, RKO and HCT116. In the current study, we used a novel affinity-purification mass-spectrometry (AP-MS) technique previously applied by us to identify Dnmt1 protein-protein interactions (15,27). We introduced a 3xFLAG tag into the DNMT1 locus in HCT116 cells (DNMT1KI-HCT116), expressing a stabilized mutant β-catenin (i.e. with constitutive activation of Wnt signalling). The DNMT1KI-HCT116 cells were then used in AP-MS experiments as previously described (27). We filtered the dataset to remove contaminants and non-specific binding proteins (28,29) and then analysed the identified peptides and proteins (Supplementary Table 1). Peptides corresponding to β-catenin (2 distinct peptides) were identified suggesting that Dnmt1 protein and β-catenin interact in HCT116 cells.
To validate the association between β-catenin and Dnmt1 proteins, co-immunoprecipitation experiments were performed. First, anti-FLAG immunoprecipitates from DNMT1KI-HCT116 cells were analysed as shown in Figure 1A. To assess whether the β-catenin – Dnmt1 interaction is chromatin-dependent, we performed the co-IP experiments with and without the Benzonase nuclease (Supplementary Figure 2). No differences were observed in the Dnmt1 signal indicating a soluble β-catenin-Dnmt1 protein complex. Western blots were also analysed with anti-HAUSP/USP7 antibodies as a positive control, since USP7 has been shown previously to be associated with Dnmt1 protein complexes (15,30). A co-IP experiment using native anti-Dnmt1 antibodies (instead of anti-FLAG) in parent HCT116 cells was also performed, showing the same result (Supplementary Figure 3) as the anti-FLAG experiments. We next performed the reciprocal co-IP experiment from DNMT1KI HCT116 cells using native anti-β-catenin antibodies and observed Dnmt1 proteins in these samples (Figure 1B), confirming the association of β-catenin and Dnmt1.
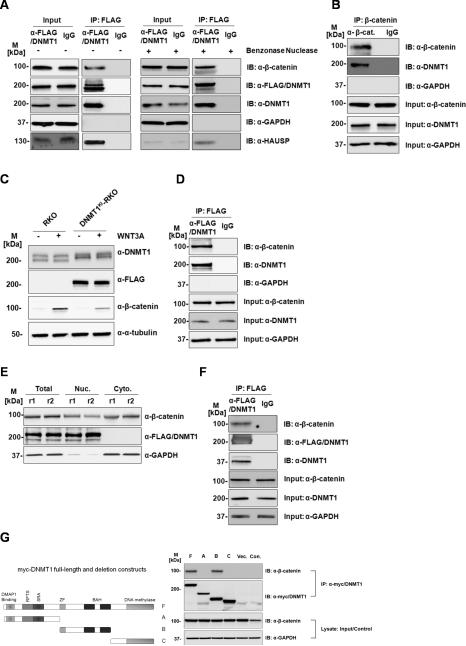
Figure 1. Dnmt1 protein interacts with β-catenin in colorectal cancer cells.
(A) DNMT1 (FLAG) immunoprecipitates β-catenin in HCT116 cells. Total protein lysate from HCT116 with Benzonase untreated (-) or treated (+) was used in the immunoprecipitation and elution fractions were loaded on gel and blotted (B) β-catenin immunoprecipitates DNMT1 in HCT116 cells (C) β-catenin is induced by Wnt3a stimulation in both RKO and DNMT1KI-RKO cells. (D) Dnmt1 (anti-FLAG) immunoprecipitates β-catenin in DNMT1KI-RKO cells. (E) Sub-cellular fractionation and protein expression levels inDNMT1KI-HCT116 cells. (F) Dnmt1 (anti-FLAG) immunoprecipitates β-catenin in nuclear fraction of DNMT1KI-HCT116 cells. Twenty micrograms of proteins from each fraction was loaded on SDS-PAGE followed by Western blot analyses with anti-Dnmt1, anti-FLAG, anti-β-catenin, anti-HAUSP, anti-α-tubulin, and anti-GAPDH. (M - standard protein marker; r - biological replicate; Nuc - nuclear fractions; Cyto - cytoplasmic fractions; IP – immunoprecipitation; IB – immunoblotting; Input indicates equal loading for IP experiments). (G) The central portion of Dnmt1 interacts with β-catenin. HEK293 cells were transfected with plasmids expressing full length Myc-tagged Dnmt1 (F), three Myc-tagged Dnmt1 deletion constructs (A, B, C) as shown in the diagram of Dnmt1 protein as well as empty plasmid vector (Vec.). Cell lysates were immunoprecipitated with anti-Myc antibodies and cell lysate from non-transfected HEK293 cells (Con.) were used as immunoprecipitation control. Elution fractions were then loaded on SDS-PAGE followed by Western blot analyses with anti-β-catenin antibodies. M refers to standard protein marker
We previously performed AP-MS experiments on RKO cells with FLAG-tagged DNMT1 (DNMT1KI-RKO) (15,27) but did not identify β-catenin peptides in these experiments. We reasoned however, that since endogenous β-catenin levels are significantly lower in RKO cells than in HCT116 cells, exogenous activation of Wnt signalling in RKO cells might allow detection of the β-catenin-Dnmt1 interaction in RKO cells. We therefore treated DNMT1KI-RKO cells with Wnt3a prior to Western and co-IP analysis as shown in Figures 1C and 1D. Wnt3a treatment of DNMT1KI-RKO cells increases β-catenin levels and β-catenin is detected in anti-FLAG immunoprecipitates from DNMT1KI-RKO cells. We also analysed nuclear and cytosolic sub-cellular fractions from DNMT1KI-HCT116 cells. Dnmt1 protein is strongly localized to the nucleus of DNMT1KI-HCT116 cells (Figure 1E) and anti-FLAG IP from nuclear fractions identifies β-catenin (Figure 1F).
To map which regions of Dnmt1 protein are necessary for the interaction with β-catenin, Myc-tagged deletion constructs of Dnmt1 protein were constructed as shown in Figure 1G. These were expressed in HEK293 cells and immunoprecipitated for Western analysis with anti-β-catenin antibodies. As shown in Figure 1G, β-catenin is only detected in association with full length Dnmt1 protein or with the central region of the protein encompassing the zinc finger and two BAH (Bromo-adjacent homology) domains. Thus neither the N-terminal region nor the C-terminal DNA methylase catalytic domains are necessary for the interaction of Dnmt1 protein with β-catenin.
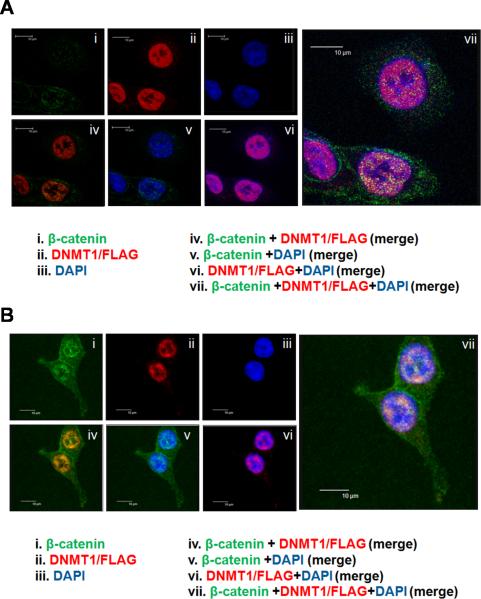
We next studied the sub-cellular localization of β-catenin-Dnmt1 association using confocal microscopy and immunofluorescence to visualize the proteins in vivo (Figure 2). DNMT1KI-HCT116 cells were either untreated (Figure 2A) or treated with Wnt3a (Figure 2B) and analysed using anti-β-catenin and anti-FLAG antibodies. As is clearly shown, β-catenin signal is detected in both the cytosolic and nuclear compartments, whereas Dnmt1 signal is confined to the nucleus. The merged β-catenin and Dnmt1 signal shows strong co-localization of the two proteins in the nucleus, and this is most evident in cells treated with Wnt3a (Figure 2B, panel iv).
Figure 2. Nuclear co-localization of Dnmt1 and β-catenin proteins.
(A) Dnmt1 protein colocalizes with β-catenin protein in HCT116 cells. (B) Dnmt1 protein colocalizes with β-catenin protein in Wnt3a stimulated HCT116 cells. Images shown were captured by confocal microscopy. Dnmt1 is shown in red (anti-mouse-AlexaFluor594 secondary antibody); β-catenin is shown in green (anti-rabbit-AlexaFluor488 secondary antibody) and co-localization of Dnmt1 and β-catenin yellow (overlay images iv show all three colours). DAPI was used for cell nuclear staining (Blue). Scale bar represents 10 μm.
In summary, mass-spectrometry, co-immunoprecipitation and immunofluorescence results indicate that β-catenin and Dnmt1 proteins are co-complexed in the nucleus, that this interaction increases in response to Wnt3a, and that the interaction occurs in multiple different cell-lines.
Levels of Dnmt1 and β-catenin proteins are mutually dependent
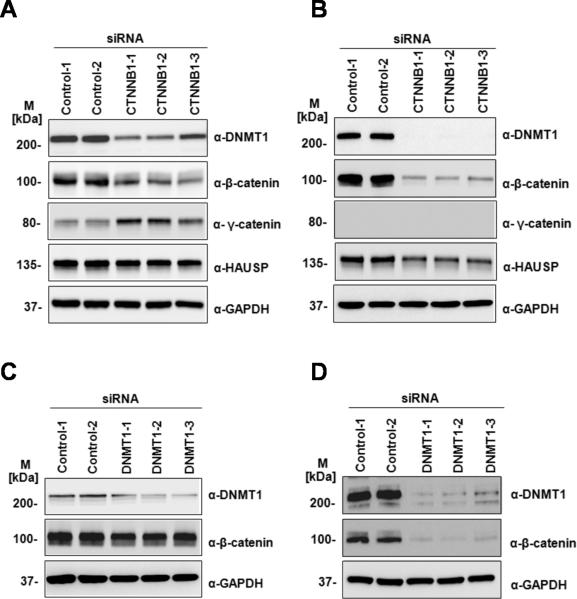
We next determined how the association between Dnmt1 protein and β-catenin affects the levels of these two proteins. Two previously generated knock-out cell-lines, DNMT1−/− (DNMT1KO-HCT116) (18), CTNNB1−/− (CTNNB1KO-HCT116) (19) and were compared to parent HCT116 cells. Figure 3A shows Western and RT-PCR analysis of parent HCT116 and CTNNB1KO-HCT116 cells. The levels of Dnmt1 protein are substantially reduced in the CTNNB1KO-HCT116 cells as compared to HCT116 parent cells. Notably, however, RT-PCR analysis reveals no difference between DNMT1 transcript levels in CTNNB1KO-HCT116 and HCT116 cell-lines, indicating that the lack of β-catenin does not affect DNMT1 transcript levels. We also immunoblotted these samples using anti-gamma-catenin (plakoglobin) antibodies, and showed that the levels of plakoglobin are elevated in CTNNB1−/− cells, consistent with the previously described observations that plakoglobin can independently promote Wnt/TCF signalling in β-catenin-deficient cells (31). Figure 3B shows similar analysis in DNMT1−/− (DNMT1KO-HCT116) cells. β-catenin levels are substantially reduced in DNMT1KO-HCT116 as compared to HCT116 cells, although no difference in CTNNB1 transcript levels is apparent. We re-introduced β-catenin into CTNNB1KO-HCT116 by transient transfection of a full-length β-catenin expression construct (Figure 3C). As shown clearly in the Western analysis of these cells, re-expression of β-catenin rescues Dnmt1 protein expression in the CTNNB1−/−cells, indicating the dependence of Dnmt1 protein levels on β-catenin, although in the reciprocal experiment (Figure 3D) in which Dnmt1 was expressed in DNMT1KO-HCT116 cells, significant restoration of β-catenin protein levels was not observed.
Figure 3. The Dnmt1- β-catenin association is mutually stabilizing.
(A) Knock-out of CTNNB1 leads to reduced abundance of endogenous Dnmt1. Twenty micrograms of total protein lysate from CTNNB1KO-HCT116 or HCT116 cells were loaded on SDS-PAGE followed by Western blot analyses with anti-Dnmt1, anti-β-catenin, anti-ɣ -catenin, anti-HAUSP, and anti-GAPDH. One step RT-PCR was performed on the total RNA extracted from both HCT116 and CTNNB1KO HCT116 cells with specific primer pairs for Dnmt1 and GAPDH (as loading control).(B) Knock-out of DNMT1 leads to reduced abundance of endogenous β-catenin. Twenty micrograms of total protein lysate from DNMT1KO-HCT116or HCT116 cells were loaded on SDS-PAGE followed by Western blot analyses with anti-Dnmt1, anti-β-catenin and anti-GAPDH. One step RT-PCR was performed on the total RNA extracted from both HCT116 and DNMT1KO-HCT116 cells with specific primer pairs for CTNNB1 and GAPDH (as loading control). M refers to standard protein marker. (C) Dnmt1 protein expression is rescued in CTNNB1KO-HCT116 cells by addition of β-catenin. β-catenin protein expression level was restored by making transient transfection in CTNNB1KO-HCT116 cells with plasmid vector containing full length β-catenin. Total protein lysate (20 μg) from HCT116, CTNNB1KO-HCT116 or CTNNB1KO-HCT116 with ectopic expressed β-catenin cells were loaded on SDS-PAGE followed by Western blot analyses with anti-Dnmt1, anti-β-catenin, anti-β-catenin (active) and anti-GAPDH (loading control). M refers to standard protein marker. (D) β-catenin expression is not rescued in DNMT1KO-HCT116 cells by addition of Dnmt1 protein. Transient transfection of DNMT1KO-HCT116 cells with plasmid vector containing full length Dnmt1 does not restore β-catenin expression. Total protein lysate (20 μg) from HCT116, DNMT1KO-HCT116 or DNMT1KO-HCT116 with ectopically expressed Dnmt1 were loaded on SDS-PAGE followed by Western blot analyses with anti-Dnmt1, anti-β-catenin and anti-GAPDH (loading control). M refers to standard protein marker. (E) Degradation profiles for β-catenin and Dnmt1 in HCT116, DNMT1KO-HCT116 or CTNNB1KO-HCT116 cells following cycloheximide treatment. Degradation rate constants were quantified by measuring the relative intensity of each protein by quantitative Western blotting at 0, 3, and 6 hours after cycloheximide treatment. The intensity data were fit to a first-order decay function to estimate the degradation rate constant, which then was used to calculate the half-life.
To investigate how Dnmt1 and β-catenin affect one another's stability, we measured protein half lives in the presence or absence of each protein. CTNNB1KO-HCT116 and DNMT1KO-HCT116 cells were treated with cycloheximide to block translation and then the protein degradation profiles observed. As shown in Figure 3E, we found that Dnmt1 has a significantly shorter half-life than β-catenin, and that in CTNNB1KO-HCT116 cells the half-life is reduced by ~30%. β-catenin has a longer half-life that is reduced in the absence of Dnmt1. In DNMT1KO-HCT116 cells, β-catenin half-life is reduced by ~40% as compared to parent HCT116 cells. DNMT1KO-HCT116 and CTNNB1KO-HCT116 cells were also treated with MG-132 proteasome inhibitor, and levels of β-catenin and Dnmt1 analysed by Western blot (Supplementary Figure 6). In DNMT1KO-HCT116 cells, levels of β-catenin are markedly increased by the addition of MG-132 whereas in CTNNB1KO-HCT116 cells, levels of Dnmt1 increase in response to MG-132 suggesting that the destabilization of Dnmt1 and β-catenin is mediated via the proteasome and can be inhibited through inhibition of proteasomal activity.
To further study the interdependence of β-catenin and Dnmt1 protein levels we performed siRNA-mediated knock-down of DNMT1 or CTNNB1 in HCT116 and RKO cells. Knock-down of CTNNB1 in HCT116 (Figure 4A) and in RKO cells (Figure 4B) shows decreased levels of Dnmt1 protein in both cell-lines. The effect is more marked in RKO cells than in HCT116 cells, and we noted that in HCT116 cells, siRNA-mediated CTNNB1 knock-down only partially reduced the levels of CTNNB1 (Figure 4A), possibly due to the substantially higher levels of endogenous β-catenin in HCT116 cells. Similarly, knock-down of DNMT1 in HCT116 (Figure 4C) and RKO (Figure 4D) reduced levels of β-catenin proteins. The effect is also more marked in RKO cells, where the siRNA-mediated knock-down of DNMT1 dramatically reduced levels of Dnmt1 and β-catenin.
Figure 4. siRNA analysis of the Dnmt1- β-catenin association.
Knock-down of CTNNB1 leads to reduced abundance of endogenous Dnmt1 in (A) HCT116 and (B) RKO cells. HCT116 or RKO cells were transfected with control siRNA or three independent siRNAs against CTNNB1. Immunoblots were used to quantify Dnmt1, β-catenin, ɣ -catenin and HAUSP with anti-Dnmt1, anti-β-catenin, anti-ɣ -catenin, anti-HAUSP sera. GAPDH was used as loading control. Knock-down of Dnmt1 leads to reduced abundance of endogenous CTNNB1in (C) HCT116 and (D) RKO cells. HCT116 or RKO cells were transfected with control siRNA or three independent siRNAs against Dnmt1. Immunoblots were used to quantify Dnmt1and β-catenin with antibodies against Dnmt1 and β-catenin, respectively. GAPDH was used as loading control. M refers to standard protein marker.
Regulatory mechanisms of the Dnmt1-β-catenin protein-protein interaction
We next tested two HCT116 derivative cell-lines in which either the wild-type CTNNB1 allele (CTNNB1−/Δ45-HCT116) or mutant CTNNB1 allele (CTNNB1WT/−-HCT116) was disrupted (19). Using two different clones for each cell-line, we analysed expression of β-catenin and Dnmt1. As shown in Figure 5A, the levels of Dnmt1 protein are decreased in CTNNB1WT/−-HCT116 cells, in line with the decreased levels of β-catenin in CTNNB1WT/−-HCT116 as compared to CTNNB1−/Δ45-HCT116 cells. Nuclear and cytosolic sub-cellular fractionations show a substantial reduction of Dnmt1 protein in the nucleus of the cells expressing only wild-type β-catenin (Figure 5B). We next stimulated each of these cell-lines with exogenous Wnt3A (Figure 5C-D). The levels of β-catenin show a more robust response in the wild-type β-catenin cells as compared to the mutant β-catenin cells, and this response is reflected in the levels of Dnmt1 protein which show a more marked increase in response to Wnt3A in the wild-type β-catenin cells than in the mutant β-catenin cells. We also tested whether the Dnmt1 response was responsive to exogenous Wnt stimulation in CTNNB1KO-HCT116 cells. HCT116 and CTNNB1KO-HCT116 cells were treated with Wnt3a and Western analysis performed (Supplementary Figure 4). As expected, Wnt3a only stabilizes Dnmt1 in the HCT116 cells, showing that the stabilization of Dnmt1 in response to Wnt3A requires β-catenin. Indeed, in these experiments, we observed a slight decrease of Dnmt1 abundance in CTNNB1KO-HCT116 cells treated with Wnt3A when compared to CTNNB1KO-HCT116 cells not treated with Wnt3A, suggesting a β-catenin-independent Wnt-mediated mechanism may also regulate Dnmt1 expression.
Figure 5. Immunoblot analysis of Dnmt1 expression in cells expressing mutant or wild-type β-catenin.
(A) Immunoblot analysis of cells expressing wild-type β-catenin (CTNNB1WT/−-HCT116) and mutant β-catenin (CTNNB1−/Δ45 -HCT116). Total soluble proteins were extracted and compared in two separate clones for each cell-line, 20ug of total protein was loaded on SDS-PAGE and western blots were performed using α-β-catenin, α-Dnmt1 and α-α-tubulin (loading control). (B) Immunoblot analysis of sub-cellular fractions of cells expressing wild-type β-catenin (CTNNB1WT/−-HCT116) and mutant β-catenin (CTNNB1−/Δ45-HCT116) (two clones for each cell type). For each fraction, a total protein of 20ug was loaded on SDS-PAGE and western blots were performed using α-β-catenin and α-Dnmt1. (C) Wnt3A time course stimulation of CTNNB1WT/−-HCT116 (clone D075) and CTNNB1−/Δ45-HCT116 cells (clone K058). Cells were collected at 0, 1, 3, 6 and 24 hours post Wnt3A (30ng/ml) stimulation on each cell line followed by total protein extraction. Total protein of 20ug was loaded on SDS-PAGE and western blots were performed using α-β-catenin and α-Dnmt1.
Since Dnmt1 protein plays a central role in DNA methylation during cell division and DNA-replication (32,33), we also tested whether Wnt driven modulation of Dnmt1 protein levels might be attributed to cell-cycle regulation following Wnt3a stimulation. Flow cytometry was performed on cells harvested across the 24hr time course following Wnt3a treatment. No gross alterations of relative numbers of cells at each time point were apparent (Supplementary Figure 1) and we therefore concluded that the response of Dnmt1 protein levels to Wnt3a treatment is not cell-cycle related.
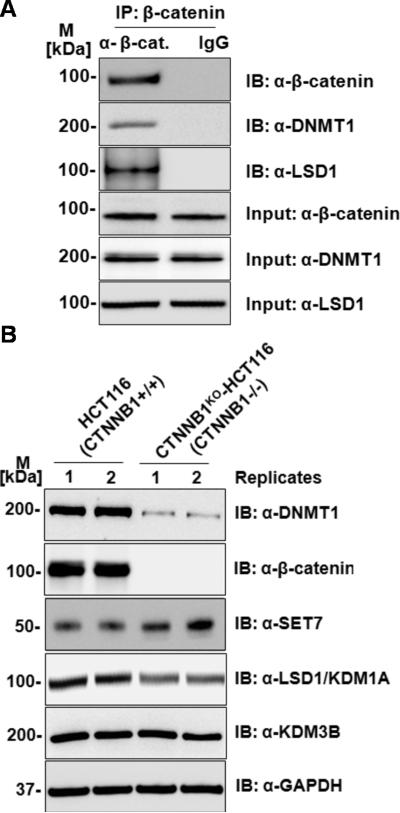
To investigate mechanisms by which β-catenin regulates Dnmt1 protein levels, we analysed the expression of several Dnmt1 interacting proteins including proteins that regulate Dnmt1 stability through methylation (SET7, LSD1) (16,34). Immunoblots of these proteins were performed on lysates from HCT116 and CTNNB1KO-HCT116 cells as shown in Figures 6A and 6B. We were particularly interested by the reduction in levels of LSD1 protein in CTNNB1KO-HCT116 cells. LSD1 (KDM1A) encodes a lysine-specific demethylase which has been shown to stabilize Dnmt1 through demethylation (35). As a negative control, we also immunoblotted a related lysine-specific demethylase, KDM3B, and found no difference in the levels of KDM3B protein between HCT116 and CTNNB1KO-HCT116 cells (Figure 6A). It may be that the reduction in LSD1 protein levels is attributable to destabilization of Dnmt1-LSD1 protein complexes by the reduction of Dnmt1 in and CTNNB1KO-HCT116 cells. We next tested whether LSD1 protein was present in anti-β-catenin immunoprecipitates. Coimmunoprecipitation using anti-β-catenin showed that both Dnmt1 and LSD1 proteins were present (Figure 6B) indicating that LSD1 is present in Dnmt1- β-catenin protein complexes, and indicating that LSD1 may function in the β-catenin-dependent regulation of Dnmt1.
Figure 6. Analysis of known Dnmt1 interacting and regulatory proteins.
(A) Anti-β-catenin immunoprecipitates both Dnmt1 and LSD1 proteins in HCT116 cells. Total protein lysate from HCT116 or immunoprecipitates were blotted with β-catenin, Dnmt1 and LSD1 antibodies. (B) Immunoblot analysis of HCT116 and CTNNB1 knock-out (CTNNB1KO-HCT116) cells (2 replicates each) of Dnmt1 interacting and regulatory proteins. Total soluble proteins were extracted and compared in two separate clones for each cell-line (Clones are indicated as 1, 2), 20ug of total protein was loaded on SDS-PAGE and western blots were performed using anti-SET7, anti-LSD1 (lysine demethylase regulating Dnmt1 stability) antibody and related lysine demethylase anti-KDM3B
Functional consequences of the Dnmt1-β-catenin protein-protein interaction
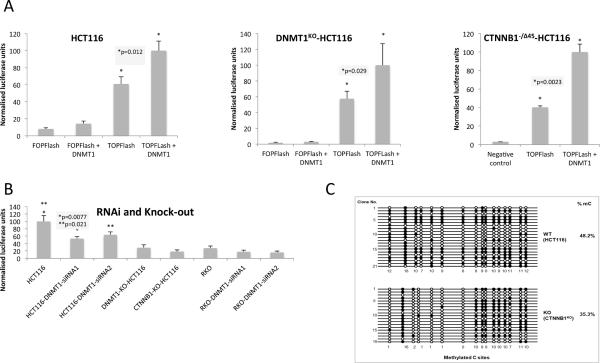
Our results predict that Wnt/β-catenin signalling should be altered according to Dnmt1-dependent regulation of β-catenin protein levels. We first compared TCF-reporter (TOPFlash) activity in cells transfected with DNMT1 expression vector in HCT116 and and CTNNB1−/Δ45-HCT116 (Figure 7A). Next, we compared β-catenin/TCF signalling in DNMT1KO-HCT116 and HCT116 cells treated with siRNA to reduce Dnmt1 levels as shown in Figure 7B. Over-expression of DNMT1 increases TCF-reporter activity approximately 2-fold and targeting of DNMT1 via siRNA or knock-out reduces β-catenin signalling (~ 2-fold and ~4-fold respectively; all p<0.05 Student's T-test). In RKO cells and CTNNB1−/WT-HCT116 cells, overall levels of TCF-reporter activity are much lower (~10 fold), as expected (lower β-catenin levels) and statistically significant changes in TCF-reporter were not detected in either over-expression or RNAi experiments (supplementary information). Although we cannot rule out that knock-down or knock-out of DNMT1 affects TCF signalling through a β-catenin-independent mechanism, these results suggest that Dnmt1 protein regulates Wnt/TCF signalling by stabilizing β-catenin in the nucleus.
Figure 7. Functional consequences of the Dnmt1- β-catenin interaction.
(A) TCF (luciferase) reporter activity in HCT116 and CTNNB1−/Δ45-HCT116 (mutant β-catenin) cells(B) TCF (luciferase) reporter activity in HCT116 and RKO cells (treated with DNMT1 siRNAs) as well as DNMT1KO-HCT116 and CTNNB1KO-HCT116 cells. All cells were transfected with LEF/TCF reporter. Negative and positive controls were also used for signal normalization and transfection efficiency monitoring. Dual Luciferase assay was performed 48 hours after transfection and promoter activity values were expressed as arbitrary units using a Renilla reporter for internal normalization. Experiments were done in triplicates for biological cell cultures/ transfections and luminescent measurements. Average numbers (bar height) of relative luciferase units for each sample plus standard deviation (error bar) were plotted relative to maximum. (C) Methylated CpG sites are decreased in CTNNB1 KO cells at the H19 locus, especially on the first six (1, 3, 4, 5, 6, 7) of seven CpG sites. Genomic DNAs from wild-type HCT116 (WT) and CTNNB1 knockout HCT116 (KO) clones were bisulfite treated. The CpG island of H19 locus was PCR amplified, cloned, and sequenced with the Sanger sequencing method. Twenty one and nineteen clones were sequenced for WT and KO, respectively (methylated CpG sites are black spots, non-methylated CpG sites are white spots and DNA sequences are represented as black lines).
Finally, since DNMT1 is the primary mammalian maintenance DNA methyltransferase, we sought to determine whether destabilization of Dnmt1 in the absence of β-catenin also alters CpG methylation patterns. The H19 imprinted locus encodes a non-protein-coding RNA and functions as a tumour suppressor (35). We also previously found that the H19 imprinted locus was a sensitive marker for DNMT1 methylation activity in HCT116 cells (15). We therefore compared the CpG methylation status of ~20 clones each of HCT116 and CTNNB1KO-HCT116 cells. As shown in Figure 7C, there is significantly reduced methylation in the CTNNB1KO-HCT116 cells at several CpG loci, indicating that the lower levels of Dnmt1 protein in CTNNB1KO-HCT116 cells has a knock-on effect on DNA methylation activity. In concordance with this finding, we also observed increased levels of H19 mRNA transcripts in CTNNB1KO-HCT116 as compared to HCT116 cells (Supplementary Figure 7).
DISCUSSION
Here we analyse an interaction between Dnmt1 and β-catenin proteins that mutually regulates the levels of each protein in colorectal cancer cells. The regulation is not mediated via transcriptional activation of DNMT1, but instead through the interaction of Dnmt1 protein with β-catenin in the nucleus. Given the critical roles of both β-catenin and Dnmt1 cells in normal as well as cancer cells, our results point to an important mechanism by which the two proteins cross-regulate. We found that the lysine demethylase, LSD1, which regulates Dnmt1 stability, is present in Dnmt1-β-catenin protein complexes, suggesting that the stabilization of Dnmt1 in these protein complexes is mediated via lysine methylation. We investigated the downstream functional consequences of the β-catenin-Dnmt1 association, and showed that Dnmt1 protein levels regulate β-catenin/TCF signalling and that the absence of β-catenin impacts CpG methylation status, pointing to a regulatory link between Wnt/β-catenin signalling and the downstream functions of Dnmt1.
Whilst protein stability of Dnmt1 and β-catenin is an important outcome of the interaction between the proteins, the precise molecular mechanisms that result in increased β-catenin/TCF signalling activity remain to be determined. Although β-catenin expression rescues Dnmt1 expression in CTNNB1KO-HCT116 cells, we did not observe rescue of β-catenin protein levels in DNMT1KO-HCT116 cells following expression of Dnmt1 (Figure 3). This may imply that the increased β-catenin/TCF signalling activity induced by Dnmt1 is not due to stabilisation and increased levels of β-catenin but from an alternative mechanism by which Dnmt1 increases β-catenin transcriptional activity. Other studies have shown that Dnmt1 can regulate transcription through protein interactions, in a methylation-independent manner. The Dnmt1 N-terminus was shown to mediate the interaction of Dnmt1 with HDAC and DMAP1 proteins, to form transcriptional repressor complexes (11), and many interactions between Dnmt1 and transcription factors have been identified (13). E-cadherin expression (in HCT116 cells) is regulated by an interaction between Dnmt1 and the transcriptional repressor Snail1, and in the same study that loss of the Dnmt1 N-terminal domains promoted β-catenin translocation to the nucleus (36). Stabilization of Dnmt1 protein, but not alterations of DNMT1 transcript levels are the cause of DNMT1 dys-regulation in human mammary epithelial cells, and this stabilization is mediated via the Dnmt1 protein N-terminus (37). How LSD1 functions in the Dnmt1-β-catenin interaction remains to be determined, since LSD1 is known to regulate Dnmt1 stability via demethylation (34). Other modifying proteins may also be important; interactions of Dnmt1 protein with HAUSP/USP7, UHRF1 and Tip60 proteins regulate Dnmt1 by ubiquitination and acetylation (15,38). Tip60 interacts with both Dnmt1 and β-catenin (39), and further mass- spectrometry analysis will determine whether Tip60 is present in β-catenin-Dnmt1 protein complexes. Although we previously showed that Tip60 acetylates Dnmt1 protein which promotes Dnmt1 degradation (15), the significance of the β-catenin-Tip60 interaction is as yet undetermined.
Our study points to a mechanism of cross-talk between Wnt signalling and DNA methylation mediated via a protein-protein interaction between β-catenin and Dnmt1. How this cross-talk manifests itself in tumours is as yet unknown, although in mouse, hypomorphic Dnmt1 alleles in the Apc Min background substantially reduce the number of polyps observed (40). Although the suppression of polyp formation was attributed to decreased CpG island methylation in the Dnmt1 hypomorphic mice, reduced Dnmt1 protein levels may also suppress polyp formation by destabilizing β-catenin and reducing Wnt/β-catenin driven transcriptional activity via the mechanism that we have described. Transcriptional regulation of DNMT1 by Wnt has also been shown (41). Using colorectal HT-29 cells (expressing truncated APC) the authors showed that transfection of full length APC reduces DNMT1 mRNA and DNMT1 promoter-reporter activity. In addition the authors also inhibited TCF-driven transcription using a dominant-negative TCF and found that this also reduces DNMT1 mRNA levels, indicating transcriptional control of DNMT1 expression. In our study we do not observe transcriptional effects of Wnt/β-catenin on DNMT1 (as measured by steady-state RNA levels), but instead observed dramatic effects of Wnt and β-catenin on Dnmt1 protein levels. These differences may imply that activation of Wnt/β-catenin by APC mutations is not entirely functionally equivalent to activation by stabilizing CTNNB1/β-catenin mutations. We also note, as did the authors of the previous study, that there are no apparent TCF binding sites in the DNMT1 promoter and so the transcriptional activation of DNMT1 is unlikely to be mediated via direct binding of β-catenin/TCF in the DNMT1 promoter. Taken together, our study and these previous studies show that multi-layered communication exists between DNMT1 and Wnt/β-catenin signalling. Although these and previous studies have been performed in cancer cell models, future work should establish the possible importance of cross-talk between DNMT1 and Wnt/β-catenin signalling in clinical samples. In addition, whilst our study has identified that an interaction between Wnt/ β-catenin and Dnmt1 may alter DNA methylation patterns on a specific locus, it would be interesting to measure global patterns of DNA methylation to see how widespread this effect is.
Interestingly, several studies have demonstrated methylation-independent functions for Dnmt1. In HCT116 cells, Dnmt1 proteins lacking the C-terminal catalytic domain function as transcriptional repressors at specific loci (12). The authors speculated that Dnmt1 serves as a scaffold for recruitment of transcriptional repressive complexes including interaction with the LSD1 histone demethylase. Intriguingly, knock-down of Dnmt1 protein was shown to regulate gene-expression via a methylation-independent and histone deacetylation – independent mechanism (42). Our analysis showed that knock-down of Dnmt1 protein can regulate TCF/β-catenin transcriptional activity. Future studies will establish whether Dnmt1 protein levels also regulate endogenous Wnt/β-catenin targets. The role played by chromatin should also be considered. The interplay between soluble nuclear Dnmt1 proteins and chromatin-associated Dnmt1 is complex. For example, Dnmt1 protein forms soluble complexes with USP7 that then form trimeric complexes with chromatin-bound UHRF1 (30). In addition, in contrast to DNMT3A/B, a substantial portion of Dnmt1 protein appears to exist as a soluble pool in the nucleus rather than being associated with chromatin (43,44). Finally, whilst the primary nuclear function of β-catenin is to regulate transcription via association with TCF transcription factors, our study shows that regulation of other proteins via stabilizing protein-protein interactions may also be an important function of nuclear β-catenin.
In summary, our work identifies a novel mechanism by which the levels of two key oncoproteins, β-catenin and Dnmt1 are regulated in cancer cells. The β-catenin-Dnmt1 interaction stabilizes each protein, and in turn regulates downstream β-catenin and Dnmt1 functions. Our study indicates a possible cancer-relevant mechanism of cross-regulation between Wnt signalling and DNA methylation. Given that the Wnt pathway plays a critical role in development and tissue differentiation, the mechanism revealed in this study could also explain how Wnt signalling drives tissue differentiation.
Supplementary Material
ACKNOWLEDGEMENTS
R.M.E and Z.W. acknowledge NCI award 1R21CA16006 that supported in part the work described here. R.M.E acknowledges an EU Marie Curie FP7-PEOPLE-2012-CIG award. Z.W. acknowledges support from NCI award P50CA150964. J.S. acknowledges support from the Cancer Pharmacology Training Program (R25CA148052) at Case Western Reserve University. J.S. acknowledges support via NIH S10RR017980 for use of the Imaging Facility, Dept. of Genetics and Genome Sciences.
Footnotes
Conflict of Interest Disclosure: The authors declare that there are no known conflicts of interest
Implications: Two critical oncoproteins, Dnmt1 and β-catenin mutually regulate one another's levels and activities in colorectal cancer cells.
REFERENCES
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 4.Yakulov T, Raggioli A, Franz H, Kemler R. Wnt3a-dependent and -independent protein interaction networks of chromatin-bound β-catenin in mouse embryonic stem cells. Mol Cell Proteomics MCP. 2013 doi: 10.1074/mcp.M112.026914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–86. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 6.Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito Y, Kanai Y, Nakagawa T, Sakamoto M, Saito H, Ishii H, et al. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer J Int Cancer. 2003;105:527–32. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 8.Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res Off J Am Assoc Cancer Res. 2003;9:4415–22. [PubMed] [Google Scholar]
- 9.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, et al. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–9. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 10.De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855–60. [PubMed] [Google Scholar]
- 11.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–77. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 12.Clements EG, Mohammad HP, Leadem BR, Easwaran H, Cai Y, Van Neste L, et al. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012;40:4334–46. doi: 10.1093/nar/gks031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin W, Leonhardt H, Pichler G. Regulation of DNA methyltransferase 1 by interactions and modifications. Nucl Austin Tex. 2011;2:392–402. doi: 10.4161/nucl.2.5.17928. [DOI] [PubMed] [Google Scholar]
- 14.Kinney SRM, Pradhan S. Regulation of expression and activity of DNA (cytosine-5) methyltransferases in mammalian cells. Prog Mol Biol Transl Sci. 2011;101:311–33. doi: 10.1016/B978-0-12-387685-0.00009-3. [DOI] [PubMed] [Google Scholar]
- 15.Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estève P-O, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci U S A. 2009;106:5076–81. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song J, Wang Z, Ewing RM. Integrated analysis of the Wnt responsive proteome in human cells reveals diverse and cell-type specific networks. Mol Biosyst. 2013 doi: 10.1039/c3mb70417c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, et al. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–7. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 19.Chan TA, Wang Z, Dang LH, Vogelstein B, Kinzler KW. Targeted inactivation of CTNNB1 reveals unexpected effects of beta-catenin mutation. Proc Natl Acad Sci U A. 2002;99:8265–70. doi: 10.1073/pnas.082240999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying Western blots: pitfalls of densitometry. Electrophoresis. 2009;30:1845–55. doi: 10.1002/elps.200800720. [DOI] [PubMed] [Google Scholar]
- 21.Tan HY, Ng TW. Accurate step wedge calibration for densitometry of electrophoresis gels. Opt Commun. 2008;281:3013–7. [Google Scholar]
- 22.Jiménez CR, Huang L, Qiu Y, Burlingame AL. In-gel digestion of proteins for MALDI-MS fingerprint mapping. Curr Protoc Protein Sci Editor Board John E Coligan Al. 2001 doi: 10.1002/0471140864.ps1604s14. Chapter 16:Unit 16.4. [DOI] [PubMed] [Google Scholar]
- 23.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–9. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 24.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 25.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13:680–5. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 26.Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci U S A. 2006;103:13004–9. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J, Hao Y, Du Z, Wang Z, Ewing RM. Identifying novel protein complexes in cancer cells using epitope-tagging of endogenous human genes and affinity-purification mass spectrometry. J Proteome Res. American Chemical Society. 2012;11:5630–41. doi: 10.1021/pr300598t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ewing RM, Chu P, Li H, Taylor P, Climie S, McBroom L, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dazard J-EJ, Saha S, Ewing RM. ROCS: A reproducibility index and confidence score for interaction proteomics. BMC Bioinformatics. 2012;13:128. doi: 10.1186/1471-2105-13-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felle M, Joppien S, Németh A, Diermeier S, Thalhammer V, Dobner T, et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39:8355–65. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda O, Usami N, Kondo M, Takahashi M, Goto H, Shimokata K, et al. Plakoglobin (gamma-catenin) has TCF/LEF family-dependent transcriptional activity in beta-catenin-deficient cell line. Oncogene. 2004;23:964–72. doi: 10.1038/sj.onc.1207254. [DOI] [PubMed] [Google Scholar]
- 32.Unterberger A, Andrews SD, Weaver ICG, Szyf M. DNA methyltransferase 1 knockdown activates a replication stress checkpoint. Mol Cell Biol. 2006;26:7575–86. doi: 10.1128/MCB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–6. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–9. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 35.Yoshimizu T, Miroglio A, Ripoche M-A, Gabory A, Vernucci M, Riccio A, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci. 2008;105:12417–22. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espada J, Peinado H, Lopez-Serra L, Setién F, Lopez-Serra P, Portela A, et al. Regulation of SNAIL1 and E-cadherin function by DNMT1 in a DNA methylation-independent context. Nucleic Acids Res. 2011;39:9194–205. doi: 10.1093/nar/gkr658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agoston AT, Argani P, Yegnasubramanian S, Marzo AM De, Ansari-Lari MA, Hicks JL, et al. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280:18302–10. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 38.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112:439–44. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 39.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eads CA, Nickel AE, Laird PW. Complete Genetic Suppression of Polyp Formation and Reduction of CpG-Island Hypermethylation in ApcMin/+Dnmt1-Hypomorphic Mice. Cancer Res. 2002;62:1296–9. [PubMed] [Google Scholar]
- 41.Campbell PM, Szyf M. Human DNA methyltransferase gene DNMT1 is regulated by the APC pathway. Carcinogenesis. 2003;24:17–24. doi: 10.1093/carcin/24.1.17. [DOI] [PubMed] [Google Scholar]
- 42.Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem. 2004;279:27915–27. doi: 10.1074/jbc.M312823200. [DOI] [PubMed] [Google Scholar]
- 43.Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–76. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. Nature Publishing Group. 2012;19:1155–60. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.