Abstract
The recent years have been characterized by a surge of studies on the role of transcription factors and histone modifications in regulating the progression of progenitors into oligodendrocytes. This review summarizes this body of evidence and presents an integrated view of transcriptional networks and epigenetic regulators defining proliferating progenitors and their differentiation along the oligodendrocyte lineage. We suggest that transcription factors in proliferating progenitors have direct access to DNA, due to predominantly euchromatic nuclei. As progenitors differentiate, however, transcriptional competence is modulated by the formation of heterochromatin, which modifies the association of DNA with nucleosomal histones and renders the access of transcription factor dependent on the activity of epigenetic modulators. These concepts are delineated within the context of development and the potential functional implications are discussed.
Keywords: development, brain, myelin, epigenetic, histone
1. INTRODUCTION
Myelinating cells provide metabolic support (Funfschilling et al. 2012) and insulation to axons and help to adjust neuronal activity to the level of environmental stimulation (Fields 2008; Liu et al. 2012 ; Makinodan et al. 2012). Oligodendrocytes are the myelin-forming cells in the brain and spinal cord and are the last cells to differentiate during development. The word “oligo-dendro-cyte” derives from greek (ολιγοσ=few, δενδρον=tree/branch and κυτοσ=hollow vessel/cell) and was originally used to define the distinctive morphological features of this highly branched cell with membrane extensions. The definition has then evolved through the years to include the immunophenotypic characterization of stage-specific antigens and the identification of a functional output (i.e. myelination; metabolic support to axon). Underlying the “oligodendrocyte” identity lays the activation of a transcriptional repertoire, resulting from the interplay of extracellular signals with intrinsic molecular components. This review addresses oligodendrocyte differentiation in response to extrinsic signals, by focusing on transcription and epigenetics.
From an historical perspective, the word “epigenetics” is the likely result of the fusion between “genetics” (the study of inheritable traits) and “epigenesis” (the study of developmental events) and it was introduced to define developmental changes in gene expression (Waddington 1959; Waddington 2012). This initial concept was followed by a series of seminal discoveries on the molecular basis of inheritance (Avery et al. 1944) and by the resolution of DNA structure (Watson and Crick 1953). The definition of chromatin as repeating units of proteins and DNA (Kornberg 1974), the proposed role of histones in transcription (Allfrey et al. 1964), the resolution of nucleosomal structure (Luger et al. 1997) and the discovery of histone modifying enzymes(Jenuwein and Allis 2001; Strahl and Allis 2000), broadened the concept of epigenetics to include temporal and spatial control of developmental gene expression (Holliday 1990), mechanisms of environmentally-induced plasticity, trans-generational transmission, imprinting and X-chromosome inactivation (Feinberg 2007; Heard et al. 1997; Heard and Disteche 2006; Youngson and Whitelaw 2008). In simple terms, “epigenetics” refers to “changes in gene expression that persist through cell division and are independent of DNA sequence” (Youngson and Whitelaw 2008), including all the mechanisms of transcription regulation that result from DNA and histone modifications, histone variants, ATP-dependent chromatin remodeling and microRNAs.
This review integrates the current knowledge of transcriptional networks and post-translational modifications of nucleosomal histones during the differentiation of oligodendrocyte progenitors (OP) into myelinating oligodendrocytes.
2. EPIGENETIC MARKS AND REGULATORS
As discussed in the introduction, “epigenetic regulation” is a term that refers to the changes in gene expression occurring independently of changes in the DNA sequence. It provides a link between genotype and phenotype, and it is the basis of cellular diversity and differentiation in a multicellular organism where different cell types share the same genome (Goldberg et al. 2007). Chromatin, the complex of DNA and histone proteins, has been widely shown to shape the epigenetic landscape of the cell and its responsiveness to extracellular stimuli (Jaenisch and Bird 2003; Riccio 2010; Wu et al. 2012). Nucleosomes are the basic unit of chromatin and are composed by a complex of four histone proteins (H2A, H2B, H3 and H4) forming an octamer around which 147 base pairs of DNA is wrapped. Multiple molecular players regulate the accessibility of the DNA in this context, including ATP-dependent remodeling complexes and chromatin-modifying enzymes. Post-translational modifications of nucleosomal histones are dynamic and play an essential role in chromatin structure and transcriptional regulation. The most well studied histone modifications that have been shown to regulate chromatin structure and transcription include lysine acetylation, methylation and ubiquitination, arginine methylation and threonine/serine phosphorylation.
Acetylation of lysine residues at the N terminal domain of nucleosomal histones is catalyzed by enzymes called histone acetyl transferases (i.e. HATs), which include three large families: the MYST, the GNAT and CBP/p300 families (Table I). Acetylated lysine residues at position K9, K14, K18, and K36 in histone H3 and at position K5, K8, K12, K16, and K91 of histone H4 have been reported in transcriptionally active promoters (Feller et al. 2015; Karmodiya et al. 2012; Morris et al. 2007; Pokholok et al. 2005; Shogren-Knaak et al. 2006; Sternglanz 1996; Wolffe and Pruss 1996; Zhao et al. 2011), while acetylation of H3K27 has been associated to enhancer regions (Shlyueva et al. 2014; Zentner et al. 2011).
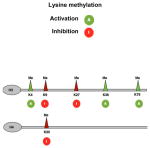
Table I. Enzymatic activities that regulate histone epigenetic marks.
Represented here is a synopsis of activating (green) and repressive (red) post-translational modifications of lysine residues in histone H3 and H4. Note that the scheme does not distinguish between mono-, di- and tri-methylation of lysines and the reader is invited to refer to the text for their functional significance. A list of the enzymes responsible for the deposition of the mark (i.e. addition of acetyl or methyl groups) is presented with the relative residue specificity. Erasers define enzymes catalyzing the removal of the mark. Please note that Sirtuins can be distinguished by HDACs due to their ability to respond to redox states. Also note that mono, di and trimethylation of lysine residues at specific positions is mediated by distinct classes of enzymes with EHMT1 and EHMT2 modulating dimethylation and SUV39H1/2 modulating trimethylation of K9 histone H3.
| Type | Writers | Erasers |
|---|---|---|
|
HATs ATF2 CREBBP ELP3 EP300 HAT1 KAT2A, KAT2B KAT7 KAT8 NCOA1 |
HDACs and SIRTUINS HDAC1 HDAC2 HDAC3 HDAC8 HDAC9 HDAC11 SIRT1 SIRT2 |
|
HMTs H3K4 MLL1, MLL2, MLL3, MLL4 SETD1A SETD1B SETD7 SMYD2 SMYD3 H3K9 EHMT1 EHMT2 PRDM2 SETDB1 SUV39H1 SUV39H2 H3K27 EZH2 H3K36 NSD1 ASH1L SETD2 SMYD2 H3K79 DOT1L H4K20 NSD1 SUV420H1 SUV420H2 SETD8 |
KDMs H3K4 KDM1A KDM1B KDM2B KDM5A KDM5B KDM5C KDM4D H3K9 KDM1A KDM3A KDM4A KDM4B KDM4C KDM4D PHF8 H3K27 KDM6A KDM6B PHF8 H3K36 KDM2A KDM2B KDM4A KDM4B KDM4C KDM4D KDM8 H4K20 PHF8 |
The removal of acetyl groups from nucleosomal histones is carried out by histone deacetylases (i.e HDACs), and is mostly associated with transcriptional repression (Braunstein et al. 1993; Braunstein et al. 1996; Gregoretti et al. 2004). HDACs have also the ability to deacetylate transcription factors (i.e. YY1, GLI, E2F) and thereby enhance or decrease their transcriptional activity (Canettieri et al. 2010; Martinez-Balbas et al. 2000; Yao et al. 2001). The HDAC protein family is diverse and is composed of 4 different classes, based on subcellular localization and structural features. The HDACs with nuclear localization (and thereby able to deacetylate nucleosomal histones) include: class I (HDAC 1,2,3,8), class IIb (HDAC4), and class IV (HDAC11) (Table I) (Seto and Yoshida 2014). Sirtuins are NAD-dependent deacetylases that have also been shown to act on nucleosomal histones, although their substrate specificity is quite broad and their subcellular localization varies in different cell types (Brachmann et al. 1995; Hecht et al. 1995; Imai et al. 2000).
The transcriptional role of lysine and arginine methylation is more complex than acetylation, as it can be associated with different functional outcomes depending on the number of methyl groups and their localization. Lysine residues can be mono (me1), di (me2), or tri (me3) methylated on their ε amino-group. The most widely studied histone modifications include H3K4, H3K9, H3K27, H3K36, H3K79 and H4K20. Monomethylation of H3K4 (H3K4me1) has been associated with enhancers while trimethylation of H3K4 (H3K4me3) has been correlated with transcriptionally active promoters, often in combination with additional marks (i.e. H3K9Ac, H3K27Ac)(Karmodiya et al. 2012; Shlyueva et al. 2014; Vermeulen and Timmers 2010; Zentner et al. 2011). In contrast, trimethylation of H3K9 and H3K27 has been associated with transcription repression(Barski et al. 2007; Boggs et al. 2002; Kim and Kim 2012).
The family of histone methyl-transferases (i.e HMTs) catalyzing the removal of methyl groups is quite diverse and specific for individual residues (Table I). They use S-adenosyl methionine as a cofactor to donate the methyl group to specific lysine residues. The MLL subfamily is responsible for the trimethylation of H3K4, while EHMT1 and EHMT2 catalize dimethylation of H3K9; SETDB2 or SUV39H1 carry out trimethylation of K9 (Greer and Shi 2012) and EZH2 is responsible for the trimethylation of H3K27 (Feng et al. 2002; Rea et al. 2000).
The removal of methyl groups from lysine residues is also quite specific (Table I). It is carried out by lysine demethylases (i.e KDMs), iron-dependent dioxygenases composed by Jumonji C (JmjC) domain-containing enzymes and by flavin adenine dinucleotide (FAD)-dependent amine oxidases (i.e. KDM1A and KDM1B) (Kooistra and Helin 2012; Shi et al. 2004; Tsukada et al. 2006).
The modification of arginine residues in nucleosomal histones is also complex and involves: mono-methylation (me1) and either symmetric (me2s) or asymmetric (me2a) dimethylation. The functional output of these modifications varies, with symmetric arginine methylation implicated in transcriptional repression and asymmetric arginine methylation associated with transcriptional activation (Di Lorenzo and Bedford 2011; Migliori et al. 2010; Strahl et al. 2001; Wysocka et al. 2006). Arginine methylation is catalyzed by protein arginine methyl-transferases (i.e PRMTs), a family of enzymes composed by 11 members (PRMT1-11) in mammalian cells(Bedford and Clarke 2009). While no arginine demethylase has been identified, a well-studied modification of arginine residues that precludes methylation is deamination, which is catalyzed by peptidylarginine deiminases (i.e PADIs), enzymes that convert methylated arginine into citrulline (Cuthbert et al. 2004; Migliori et al. 2010; Wang et al. 2004).
This vast and expanding literature reveals the level of complexity of epigenetic regulation, which includes the action of multiple enzymes and post-translational modifications on specific histone residues, thereby defining complex histone code modulating the transcriptional output at particular gene loci (Table I).
3. TRANSCRIPTION FACTORS INVOLVED IN OLIGODENDROCYTE LINEAGE DEVELOPMENT
The literature on the transcriptional networks modulating oligodendrocyte lineage progression encompasses a vast number of studies, ranging from factors modulating cell specification to those preventing the expression of differentiation markers in progenitor cells (Liu and Casaccia 2010; Liu et al. 2015), and from those repressing genes involved in pluripotency or proliferation (Sher et al. 2008), to those regulating the expression of myelin components. In this chapter we shall review the major family of transcription factors regulating oligodendrocyte progenitors and their differentiation into myelin-forming oligodendrocytes.
3.1. BASIC HELIX-LOOP-HELIX (bHLH) FAMILY
The bHLH class includes a family of proteins characterized by the presence of an amino-terminal basic domain, that binds the consensus DNA sequence called E-box, and a C-terminal helix-loop-helix domain called HLH, that facilitates the homo- or hetero-dimerization with other protein partners. The function of these factors as transcriptional repressors or activators depends largely on the interaction with specific protein partners and on the presence or absence of DNA binding domains. Members of the HLH family include: the ubiquitously expressed E-proteins (e.g. E2-2, HEB, E12 and E47) and their tissue-specific dimerization factors (e.g. NEUROD, NGN, MYOD, MASH1/Ascl, OLIG1, OLIG2, HES1, HES5) with the ability to either activate or repress transcription by binding to E-box sites at promoters or regulatory regions of target genes. The ID family members lack DNA binding activity and act as repressors by sequestration (Benezra et al. 1990b). Much work in the past recent history was dedicated to the elucidation of the molecular mechanisms regulating the sequential fate restriction of oligodendrocyte lineage cells from undifferentiated multipotential precursors (Jones 2004; Ledent and Vervoort 2001). Inductive theories were introduced in the 1990s, when the transcription field focused largely on the role of lineage-specific factors, as lineage determinants in response to morphogens and growth factors. The discovery of MyoD, a basic helix-loop-helix (bHLH) transcription factor that was both necessary and sufficient for the generation of muscle cells (Weintraub et al. 1991), followed by the identification of Myogenin, another bHLH protein synergizing with MYOD, spurred a search for similar “master regulatory genes” in other lineages. NeuroD in Xenopus laevis (Lee et al. 1995) and Neurogenin in mammals(Ma et al. 1996) were discovered to be neuronal determination genes and Olig1 and Olig2 were discovered to be oligodendrocyte-specific genes (Lu et al. 2000; Zhou et al. 2000) in avians and in mammals.
Despite their similarity to other bHLH transcription factors, expression of Olig2 or Olig1 was not sufficient to induce oligodendrocyte differentiation in unrelated cell types (Lu et al. 2000; Sun et al. 2001; Zhou et al. 2001; Zhou et al. 2000). Both induced by SHH and expressed in ventral domains of the spinal cord, these two transcription factors were shown to be critical for oligodendrogliogenesis, as no oligodendrocyte progenitors were formed when both genes were ablated (Lu et al. 2002; Zhou and Anderson 2002). However, OLIG1 1 and OLIG2 appear to play distinct roles in oligodendrocyte development. The expression of Olig2 in the embryonic spinal cord, for instance, preceded that of Olig1 (Zhou et al. 2000) and only Olig2 was shown to play a role in the establishment of clear boundaries of cellular domains leading to the generation of motorneurons and oligodendrocytes. It is recognized that the definition of domains in the spinal cord is regulated by transcriptional cross-repression between OLIG2, the homeodomain transcription factors NKX2.2 and NKX6.1 and another transcription factor implicated in interneuron fate called IRX3 (Lu et al. 2002; Zhou and Anderson 2002; Zhou et al. 2001). Overexpression studies further highlighted differences between the role of OLIG1 and OLIG2 in oligodendrocyte development and the importance of protein-protein interactions. Injection into the chick neural tube of Olig2 alone, promoted motor neuron development. Injection of Olig2 and NKx2.2 in the same system, promoted the development of oligodendrocytes and interneuron’s (Zhou et al. 2001). Mis-expression of Olig1 in the murine embryonic forebrain, in contrast, induced the formation of clones enriched in glial cells without neuronal cell types but not myelinating oligodendrocytes(Lu et al. 2001). It was proposed that the ability of OLIG2+ cells to generate motor neurons or oligodendrocytes was based on the relative amounts of OLIG2 and neurogenin 2 (NGN2) in neural stem cells (Novitch et al. 2001; Zhou et al. 2001). Recent studies have refined this concept and led to the idea that OLIG2 dephosphorylation is critical for oligodendrocyte specification (Li et al.) due to the sequestration of NGN2. Post-translational modifications of OLIG2 have also been shown to favor its interaction with the homeodomain protein NKX2.2 (Sun et al. 2003), with the nuclear factor I/A (NFIA)(Deneen et al. 2006) and with MASH1, another bHLH protein whose ablation in mice (Guillemot et al. 1993), resulted in severely impaired spinal cord myelination at birth (Sugimori et al. 2008). Based on the multitude of transcription factors and their ability to form complexes with other proteins either enhancing or repressing their ability to modulate transcription, it is likely that future studies might elucidate additional transcriptional networks regulating oligodendrocyte differentiation in dorsal areas of the brain and spinal.
The critical function of OLIG2 for oligodendrogliogenesis was clearly supported by the analysis of Olig2 mutant mice, which revealed severely impaired development of motor neurons and oligodendrocytes in the ventral spinal cord (Lu et al. 2002) but not in the hindbrain, thereby suggesting that residual Olig1 function could be sufficient to rescue OP generation in a region-specific fashion. However the role of OLIG1 in developmental myelination remained the subject of an active debate through the years. While the original study on Olig1 ablation (obtained by insertion of a Cre-Neo cassette within the Olig1 locus) reported a very mild phenotype (Lu et al. 2002), a subsequent study revealed a severe hypomyelinated phenotype after deletion of the Neo cassette (Xin et al. 2005), and raised the possibility that OLIG1 is an important determinant of myelination. Based on the conclusions of these two studies the evidence that OLIG1 plays a role in developmental myelination can be defined - at best - as controversial, while its role in adult remyelination is well accepted (Arnett et al. 2004). A recent study brought additional evidence on the dispensable role of OLIG1 in developmental myelination by generating two novel Olig1 null lines either by homologous recombination in ES cells followed by blastocyst injection or by rescue of the Olig1;Olig2 double knockout line (Paes de Faria et al. 2014). The results of the published study unequivocally supported the conclusions of the original study by Lu et al., 2002 since the absence of Olig1 only minimally impaired developmental myelination. The importance of OLIG1 in developmental myelination in the hindbrain (Xin et al. 2005) and in remyelination (Arnett et al. 2004) prompted a series of studies that identified SIP1 and ZFP488 as downstream effectors of OLIG1 as discussed below (Wang et al. 2006; Weng et al. 2012).
HES1 and HES5 are transcriptional repressors, initially identified as NOTCH-dependent inhibitors of neurogenesis (Ohtsuka et al. 1999) with the ability to bind to N-boxes on pro-neural genes (Akazawa et al. 1992) and to recruit complexes containing the co-repressors TLE/Groucho and histone deacetylases (Grbavec and Stifani 1996; Paroush et al. 1994). HES5 is an important inhibitor of oligodendrocyte differentiation in cultured cells (Kondo and Raff 2000) and a regulator of timing of myelination in vivo (Liu et al. 2006). As well as binding directly to DNA consensus sequences on myelin genes and recruiting HDAC-containing repressive complexes, HES proteins also form non-functional protein complexes with the bHLH proteins E47 and MASH1(Akazawa et al. 1992) and sequester positive regulators of myelin gene expression, such as SOX10 (Liu et al. 2006).
ID2 and ID4 are also transcriptional repressors and members of the HLH proteins, lacking the basic DNA binding motif. They are induced by BMP signaling (Ying et al. 2003) and inhibit transcriptional activation by sequestration of OLIG1 and OLIG2, thereby preventing DNA binding (Benezra et al. 1990a; Benezra et al. 1990b; Samanta and Kessler 2004). Id4 expression declines during differentiation and forced expression of either Id4 (Kondo and Raff 2000; Samanta and Kessler 2004) or Id2 (Wang et al. 2001) prevented differentiation of OPs in vitro, while gene ablation of Id4 (Marin-Husstege et al. 2006) resulted in ectopic and heterochronic myelin gene expression.
More recently, new controversies have risen regarding the role of OLIG2 in the oligodendrocyte lineage. The first one relates to the role of this transcription factor in regulating the decision of NG2 cells to become oligodendrocytes or astrocytes. While the initial studies on OLIG2 function focused on the importance of this transcription factor in the ventral spinal cord and forebrain (Lu et al. 2002; Zhou and Anderson 2002), and highlighted the importance of this transcription factor for the generation of motor neurons and oligodendrocytes, the ablation of Olig2 in NG2 cells revealed a potential new role for this transcription factor in repressing astroglial cell fate in specific regions, such as the neocortex and the corpus callosum (Zhu et al. 2012). This suggests that the function of OLIG2 could be region-specific and stage specific and possibly affected by post-translational modifications and protein-protein interactions.
The second controversy relates to the role of OLIG2 as recruiter of the chromatin remodeling complex SMARCA4/BRG1 and activating a program of oligodendrocyte differentiation (Yu et al. 2013). It was proposed OLIG2 acts as key transcription factor initiating oligodendrocyte differentiation by targeting the SMARCA4/BRG11 ATPase remodeling complex to enhancer elements on key modulators of myelination (such as Myrf, Sox10, Zfp191, described below). This model was proposed on the basis of identified target genes by ChIP seq analysis and on the reported severe hypomyelination in the spinal cord of Smarca4/Brg1 null mice using the Olig1cre driver line to achieve conditional ablation (Yu et al. 2013).
However, subsequent experiments from other groups using the CnpCre driver (Bischof et al. 2015; Mei et al. 2013) to ablate either Olig2 (Mei et al. 2013) or Smarca4/Brg1 (Bischof et al. 2015), have not confirmed the originally proposed model. Therefore, despite the general consensus related to the role of OLIG2 in the early stages of oligodendrocyte development, its role in differentiating oligodendrocytes remains controversial. The report that late Olig2 ablation resulted in increased- rather than the predicted decreased – myelination, questioned the model of OLIG2 as key recruiter of the ATP remodeling complexes to genomic loci necessary for myelination (Yu et al. 2013). A potential explanation for the stage-specific effects of Olig2 ablation could be the compensatory role of OLIG1 in mice with Olig2 ablation at later stages of development. Additional questions to the originally proposed model were posed by a careful analysis of mice with ablation of Smarca4/Brg1 in progenitors (using an NG2Cre line) and at later stages of differentiation (using a CnpCre line). In both cases the phenotype was relatively mild and thorough investigation of the potential underlying mechanism identified BRG1 as early inducer of Sox10 expression, although not necessary for its expression at later stages. Together, these manuscript define a controversy which might be explained by the possibility that ChipSeq data in the Yu et al. 2013 study were performed on immature progenitor cells rather than differentiating oligodendrocytes, by the fact that an independent validation of the ChipSeq data using quantitative immunoprecipitation of chromatin samples isolated at multiple times of oligodendrocyte development (as carefully analyzed by Bischof et al. 2015 was not included in the original study.
3.2 HIGH MOBILITY GROUP OR HMG FAMILY
The high mobility group family of differentiation inhibitors includes the SOX family members as well as other HMG molecules with a bimodal expression profile (such as Tcf7l2/Tcf4) and a more complex functional role (Table II), including the ability to counteract inhibitory Wnt signaling in progenitor cells (Ye et al. 2009) while inhibiting myelin gene expression in differentiating cultured oligodendrocytes (He et al. 2007). The members of this family of transcription factors are characterized by the presence of the HMG (high mobility group) domain, a 79 amino-acid protein motif that binds to specific non-B conformations of DNA(Leslie et al. 1980). B-DNA is the most typical conformation of DNA found in cells (Leslie et al. 1980), whereas non-B DNA is either characterized by a wider minor groove and detected in cells when DNA forms complexes with either RNA or with enzymes (A-DNA) or is characterized by methylation (Z-DNA) (Nickol et al. 1982; Shaw and Arya 2008).
Table II. Synopsis of transcription factors relevant to oligodendrocyte lineage biology.
This table summarizes the main classes and subgroups of transcription factors implicated in the regulation of distinct stages of oligodendrocyte biology. Please see text for detailed description and complete reference list.
| Transcription factor Class | Protein | Gene ID | Extracellular Signal | Function | References | |
|---|---|---|---|---|---|---|
| bHLH | MASH1 | Ascl1 (Mash1) | SHH | Specification. Activation of myelin gene expression | (Gokhan et al. 2005; Guillemot et al. 1993; Liu et al. ; Sugimori et al. 2008) | |
| OLIG1 | Olig1 | SHH | Differentiation. Remyelination. | (Arnett et al. 2004 ; Gokhan et al. 2005; Li et al. 2007; Lu et al. 2001; Lu et al. 2002; Lu et al. 2000; Sun et al. 2001; Zhou and Anderson 2002; Zhou et al. 2001; Zhou et al. 2000) | ||
| OLIG2 | Olig2 | SHH | Specification. Differentiation. | (Zhou and Anderson 2002; Zhou et al. 2000), (Lu et al. 2002) (Li et al. 2011; Lu et al. 2001; Lu et al. 2002; Lu et al. 2000; Novitch et al. 2001; Sun et al. 2001; Zhou and Anderson 2002; Zhou et al. 2001; Zhou et al. 2000) | ||
| HES1 | Hes1 | NOTCH, BMP4 | Maintenance of the progenitor stage. Inhibition of differentiation. | (Akazawa et al. 1992; Grbavec and Stifani 1996; Kondo and Raff 2000; Ogata et al. ; Ohtsuka et al. 1999; Paroush et al. 1994; Wu et al.) | ||
| HES5 | Hes5 | NOTCH, BMP4 | Maintenance of the progenitor stage. Inhibition of differentiation. | (Kondo and Raff 2000; Liu et al. 2006; Wu et al.) | ||
| HEY1 | Hey1 | NOTCH, BMP4 | Maintenance of the progenitor stage. Inhibition of differentiation. | (Wu et al.) | ||
| HLH | ID2, ID4 | Id2, Id4 | BMPs | Inhibition of differentiation. | (Benezra et al. 1990a; Benezra et al. 1990b; Samanta and Kessler 2004; Wang et al. 2001; Ying et al. 2003) | |
| Homeodomain | NKX2.2 | Nkx2.2 | SHH | Specification. Differentiation (?). | (Lu et al. 2002; Qi et al. 2001; Sun et al. 2003; Wei et al. 2005; Zhou and Anderson 2002) | |
| NKX6.1, NKX6.2 | Nkx6.1, Nkx6.2 | SHH | Specification. Regulation of paranodal gene expression. | (Cai et al. 2005 ) | ||
| Homeodomain/Zinc finger | SIP1 | Zeb2 (Sip1/ Zfhx1b) | SHH (?) | Inhibition of BMP4 signaling. Myelination. | (Weng et al.) | |
| Kruppel-like | KLF9 | Klf9 | T3 | Differentiation. Remyelination. | (Dugas et al.) | |
| HMG (High Mobility Group) | Sox | SOX2 | B1: Sox 2 | N.D | Maintenance of neural progenitor stage. | (Bylund et al. 2003; Ellis et al. 2004; Graham et al. 2003; Hoffmann et al. 2014; Hutton and Pevny 2011; Thier et al.) |
| SOX11 | C: Sox11 | N.D | Inhibition of differentiation and myelin gene expression | (Swiss et al.) | ||
| SOX5, SOX6 | D: Sox5, Sox6 | N.D | Inhibition of differentiation. | (Stolt et al. 2006) | ||
| SOX9, SOX10 | E: Sox9, Sox10 | N.D | Lineage progression. Activation myelin gene expression. Survival. | (Finzsch et al. 2008; Pozniak et al. ; Sock et al. 2001; Stolt et al. 2004; Stolt et al. 2003; Stolt et al. 2002; Stolt et al. 2006; Stolt et al. 2005) | ||
| SOX17 | F: Sox17 | N.D | Lineage progression Activation myelin gene expression | (Chew et al. ; Ming et al. 2013; Moll et al. 2013; Sohn et al. 2006) | ||
| TCF7L2 | Tcf7l2(Tcf4) | WNTs | Inhibition myelin gene expression. | (Fancy et al. 2009; He et al. 2007; Ye et al. 2009) | ||
| Zinc Fingers | ZNF24 | Znf24 (Zfp191) | N.D | Myelination. | (Howng et al.) | |
| MYT1 | Myt1 | N.D | Modulation of Plp expression | (Nielsen et al. 2004) | ||
| ZFP488 | Zfp488 | N.D | Differentiation | (Soundarapandian et al. ; Wang et al. 2006) | ||
| ZFP191 | Zfp191 | N.D | Myelination | (Howng et al. 2010) | ||
| YY1 | Yy1 | Differentiation. Inhibition of myelin genes repressors. | (Berndt et al. 2001; He et al. 2007; He et al. 2010; Liu et al. 2015; Liu J. 2014; Zolova and Wight 2011) | |||
| MYRF | Myrf(Mrf) | N.D | Myelination and myelin maintenance. | (Emery et al. 2009; Koenning et al.) (Bujalka et al. 2013; Cahoy et al. 2008) | ||
| Nuclear Receptor | THRα | Thra | T3 | Timing of differentiation, Remyelination. | (Barres et al. 1994; Billon et al. 2002) | |
| PPARδ, PPARγ | Pparg, Ppard | 15-Deoxy-Δ-12,14-prostaglandin J2 (15-d-PGJ2) | Differentiation. | (De Nuccio et al. ; Feinstein et al. 2002; Saluja et al. 2001) | ||
| RXRγ | Rxrg | RA (9-cis) | Differentiation. Remyelination. | (Huang et al.) | ||
| NFIA | Nfia | Notch | Specification | (Deneen et al. 2006; Okano and Temple 2009) | ||
The SOX (Sry-related HMG box) family of transcription factors includes more than 20 members sharing the homology for the HMG box of the Sry gene, which is involved in sex determination. They can be further classified in subgroups based on homology, structure and function (Table II), and they are characterized by DNA sequence specific recognition and consequent transcriptional activation or inhibition, depending on the interacting partners. The subgroup A identifies the SRY factor, which gives the name to the family and is present on the Y chromosome. The subgroup B of the Sox family includes Sox1, 2 and 3, transcriptional activators which are expressed in proliferating neural and oligodendrocyte progenitor cells. Among these family members, SOX2 (Ellis et al. 2004; Graham et al. 2003; Hutton and Pevny) has been shown to increase proliferation and inhibit neuronal differentiation (Bylund et al. 2003) and to support re-programming of OPs into neural stem cells(Kondo and Raff 2004). SOX2 is down-regulated during the late stages of oligodendrocyte maturation (Shen et al. 2008) due to the activity of HDACs. However, in contrast with neural progenitors where they are down-regulated, Sox2 and Sox3 remain expressed during the early stages of differentiation and exert a pro-oligodendrogliogenic effect that has been in part attributed to their ability to down-regulate the microRNA miR145(Hoffmann et al. 2014).
The subgroup C of Sox family members includes Sox 4 and Sox11 which was identified as one of the target genes repressed by HDAC1 during OP differentiation (Swiss et al. 2011). The repressive activity of SOX11 was confirmed by the evidence that its forced overexpression in oligodendrocyte lineage cells reduced the expression of myelin genes during the process of differentiation (Swiss et al. 2011).
The subgroup D of the Sox family includes SOX5 and SOX6 that are both expressed in OPs and have been shown to inhibit differentiation (Stolt et al. 2006; Swiss et al. 2011). Genetic ablation of Sox5 and Sox6 in mice induced precocious specification and differentiation of oligodendrocytes (Stolt et al. 2006).
The subgroup E includes SOX8, SOX9 and SOX10, which have a crucial role in peripheral and central nervous system myelination. Sox8 and Sox9 are expressed in OPs, with Sox9 being down-regulated during differentiation (Stolt et al. 2005) and Sox8 still detected in oligodendrocytes, albeit at lower levels (Sock et al. 2001; Stolt et al. 2004). The redundancy of SOX8 and SOX9 function was inferred by the reduced number of OPs in Sox 9 null mice (Stolt et al. 2003) and their complete absence in Sox8/Sox9 double knockouts (Stolt et al. 2005). Sox10 is expressed later than Olig2 in OPs, and its expression persists at high levels throughout lineage progression (Stolt et al. 2002). Genetic ablation of Sox10 in mice did not affect OPs, possibly owing to compensation by Sox8 and Sox9, but caused severe hypomyelination (Britsch et al. 2001) (Finzsch et al. 2008; Stolt et al. 2002), thereby suggesting a role for this transcription factor in promoting the late stages of differentiation. The current view is that the ability of SOX10 to promote oligodendrocyte differentiation is mediated by the transcriptional activation of genes associated with the myelinating stage including Mbp, Plp1, and Cnx47 (Schlierf et al. 2006; Stolt et al. 2002), which can be attributed to the formation of protein complexes with other transcription factors such as OLIG1 (Li et al. 2007), MYRF (Hornig et al., 2013) and direct binding to the Mediator subunit12 of the transcriptional machinery (Vogl et al. 2013). At the progenitor stage the activity of SOX10 would be inhibited by sequestration into complexes containing transcriptional inhibitors (Liu et al. 2006) and by competition with Sox family members of the subgroup C and D, which have the ability to compete for binding at the same DNA consensus sequence. In this respect, an interesting area of future investigation will be the identification of the post-translational modifications of SOX10, which might underlie its ability to form protein complexes with distinct partners.
The subgroup F includes SOX17, a factor that was originally identified in a microarray screen for transcription factors that were enriched in sorted oligodendrocyte lineage cells and implicated in cell cycle exit and differentiation of OP (Sohn et al. 2006). It was shown that SOX17 plays an essential role in cell cycle exit, by down-regulating genes related to cell proliferation (e.g. cyclin D1) and beta-catenin signaling pathway (e.g. axin 2, beta-catenin) (Chew et al. 2011). Overexpression of Sox17 also increased OP differentiation as observed in transgenic overexpressing lines (Ming et al. 2013), possibly due to sequestration of myelin gene transcriptional inhibitors (e.g. Tcf7l2/Tcf4) and Wnt-signaling related genes (Chew et al. 2011).
3.3 ZINC FINGER FAMILY
Zinc fingers transcription factors are a large protein family able to recognize specific DNA sequences through their Zinc finger domain, which is structurally stabilized by a zinc ion bound to conserved Cys and His residues within the motif (Klug and Schwabe 1995; Mackay and Crossley 1998). In oligodendrocyte biology, zinc fingers transcription factors have been shown to be critically involved in transcription regulation at different stages of differentiation.
YY1 is a zinc-finger molecule with the ability to activate or repress genes in a cell-specific fashion, depending on the post-translational modifications occurring in response to different signals. In Schwann cells, for instance, YY1 is phosphorylated and acts as positive regulator of myelin genes (He et al. 2010), while in oligodendrocyte progenitors it is acetylated and preferentially acts as repressor of genes characteristic of the progenitor state. In differentiating OP, YY1 was detected in protein complexes containing the repressive enzymes HDAC1 (He et al. 2007) as well as K9 histone methyltransferases (Liu et al. 2015). Conditional ablation of Yy1 in the oligodendrocyte lineage did not compromise cell cycle exit, but arrested progenitors at an undifferentiated state characterized by high levels of transcriptional inhibitors of differentiation such as Id4, Sox11 and Tcf7l2/Tcf4. Together these data reinforce the complexity of the transcriptional activity of YY1, whose function is modulated in a context-specific fashion. The cell and developmental stage-specific post-transcriptional modifications of YY1 could potentially serve as interpretative key to reconciliate previous reports of YY1 as repressor (Zolova and Wight 2011) or activator (Berndt et al. 2001) of myelin genes in distinct cellular contexts.
ZFP191 is a zinc finger transcription factor implicated at later stages of differentiation. It was discovered by screening C3H/HeJ background mice for the occurrence of spontaneous resulting in a hypomyelinating phenotype characterized by tremor and death by the third postnatal week (Howng et al.). These mutants showed a significant decrease in myelin gene expression and were characterized by oligodendrocytes that extended processes to contact axons but failed to wrap them, a phenotype reminiscent of Olig1 null mice (Howng et al. 2010). Interestingly, morphological studies revealed no difference in the number of precursor cells or late-stage differentiated oligodendrocyte, despite the severe hypomyelination of mutant mice (Howng et al. 2010). This suggested that ZFP191 is critical for later stages of myelination, probably by direct or indirect transcriptional regulation of myelin gene expression.
ZFP488, on the other hand is a transcriptional repressor that cooperates with OLIG2 to promote differentiation. In vivo experiments have shown that ectopic overexpression of Zfp488 and Olig2 produced precocious differentiation in chick embryos (Wang et al. 2006). Additional studies have shown that ZFP488 promoted oligodendrogliogenesis upon overexpression in the subventricular zone, in the cuprizone model of demyelination (Soundarapandian et al. 2011).
MYT1 (myelin transcription factor 1) is a zinc finger transcription factor that binds the Plp promoter, the most abundant of oligodendrocytes myelin genes (Kim et al. 1997). In proliferating oligodendrocyte precursors MYT1 localizes to the nuclear speckles, where it plays an important role in proliferation and differentiation (Nielsen et al. 2004). In demyelination lesions from MHV-induced demyelinated mouse samples and MS human samples, Myt1 is highly expressed in both demyelinated and remyelinated lesions, suggesting a potential role for MYT1 in remyelination (Vana et al. 2007). However, it is not clear whether Myt1 act as a repressor and/or an activator in transcription regulation. Evidences of MYT1 and SIN3B interaction suggest its role as a repressor in progenitor cells (Romm et al. 2005).
SIP1 is a transcription factor that is structurally characterized by the presence of a homeodomain and a zinc finger motif and recognizes E-boxes on the DNA sequence. This factor is a SMAD-interacting protein (Verschueren et al. 1999) and acts as a repressor of BMP signaling pathway (van Grunsven et al. 2007). Interestingly it has been shown that SIP1 acts as master regulator of cell fate determination by integrating BMP4 and WNT signaling pathways (Weng et al. 2012). Studies on Sip1 conditional knockout mouse have shown its role in myelination, since the knockout exhibits severe demyelinating defects in the central nervous system (Weng et al. 2012). Sip1, initially identified as a target gene for OLIG1 and OLIG2, can act as a transcriptional activator or repressor depending on the interacting cofactors. It has been shown that SIP1 negatively regulates BMP4 signaling, either by interacting with receptor-activated SMADs (i.e. R-SMADs), or by repressing Id2/Id4 and Hes1 expression or by inducing the expression of Smad7, a negative regulator of BMP pathway. In addition to its role as a negative regulator of BMP signaling, SIP1 can also activate the expression of Mbp and myelin genes activator like Sox10 and Myrf (Weng et al. 2012).
The Krüppel-like family of transcription factors (KLF) is characterized by the presence of zinc finger domains, localized at the C-terminus of the protein sequence and binding to GC-rich DNA sequences. Recent studies have shown that KLF9 is downstream of T3 signaling and is sufficient to induce differentiation of oligodendrocytes in vitro, though the Klf9 knockout mouse appears to develop normally (Dugas et al. 2012).
3.4 ADDITIONAL TRANSCRIPTION FACTORS MODULATING DIFFERENTIATION
Myrf was identified on the basis of its expression profile, which is enriched in differentiated and myelinating oligodendrocytes (Cahoy et al. 2008). MYRF is a membrane-bound transcription factor that undergoes an activating proteolitic cleavage, allowing the N-terminal domain to directly bind enhancer regions specific to oligodendrocyte and myelin genes to regulate transcription during differentiation (Bujalka et al. 2013). Genetic ablation of Myrf in mice resulted in profound hypomyelination and death at the third postnatal week, and silencing of Myrf in cultured OPCs also impaired differentiation (Emery et al. 2009).
Among the most potent inducers of myelination in rodents are thyroid hormone (Barres et al. 1994), and retinoic acid (Huang et al. 2011) which bind to receptors with the ability to bind DNA and modulate transcription. Together with the PPAR family members PPARγ and PPARδ (De Nuccio et al. 2011; Saluja et al. 2001), these nuclear receptors have been implicated in myelin synthesis and future studies will need to determined how this intricate network of transcription factor family cross talks, especially in response to extracellular signals in the developing brain and in pathological conditions.
4. MICRO-RNAs
The role of microRNAs during development is critical to mediate cell differentiation and maintenance of cell identity. In the oligodendrocyte lineage, multiple microRNAs have been discovered in the last decade as modulators of differentiation. The role of microRNAs in early stages of oligodendrogliogenesis was suggested by in vivo studies on Dicer ablation in neural precursor cells, which revealed a significant reduction of oligodendrocyte lineage cells (Kawase-Koga et al. 2009). An important microRNA for oligodendrocyte specification is miR7a (Zhao et al. 2010), which is widely expressed but shows highest levels in oligodendrocyte precursors. Electroporation of miR7a in mouse embryos and overexpression studies revealed an expanding population of oligodendrocyte progenitors and up-regulation of Pdgfra, Olig1 and Olig2. Consistently, miR7 silencing inhibited oligodendrocyte specification and differentiation. Targets of this microRNA include neuronal (i.e. Pax6 and NeuroD4), astrocytic (i.e. Nfib and Nfic), and oligodendrocytic (i.e. Cnp, Sp1) transcripts. Additional microRNAs identified in oligodendrocyte precursors include the miR-17-92 cluster, which affects the proliferation of progenitors in vivo and in vitro (Budde et al. 2010).
Interestingly, genetic ablation of Dicer in oligodendrocyte progenitors did not affect proliferation (Dugas, 2010; Zhao, 2010) but impaired differentiation and caused myelin disruption in mutant mice (Dugas et al. 2010; Zhao et al. 2010). The inhibitory effect on differentiation was mostly attributed to decreased levels of two critical microRNAs: miR219 and miR338, which were also identified as the most up-regulated microRNAs in differentiating oligodendrocytes (Dugas et al. 2010; Lau et al. 2008; Zhao et al. 2010). MicroRNAs miR219 and miR338 were necessary and sufficient to induce differentiation by targeting genes related to the progenitor stage, myelin inhibitors or other cell lineages. For instance, it was shown that Pdgfra, Hes5, Sox6, Zfp238, and FoxJ3 were main targets for miR219, while Sox6, Hes5, NeuroD1, Isl1, Otx2, and Fgfr2 were targets of miR388 (Dugas et al. 2010; Wienholds et al. 2005), thereby revealing a role for microRNAs in repressing progenitor-essential genes or differentiation inhibitors. In contrast, miR138 was defined as playing a role only during a specific window of differentiation, by promoting early stages of differentiation but suppressing myelin formation (Dugas et al. 2010). The molecular mechanism are unknown, but bioinformatics analysis revealed that potential target genes for this micro RNA are Sox4 and Uhrf1bp1, both associated with inhibition of differentiation(Dugas et al. 2006; Stolt et al. 2006).
5. TRANSCRIPTON FACTORS AND CHROMATIN CHANGES DURING THE PROGRESSION FROM PROGENITOR TO MYELINATING OLIGODENDROCYTE
Figure 1 depicts a schematic summary of the transcriptional events and post-translational modifications of the histones occurring in the nuclei of proliferating oligodendrocyte progenitors, which are mainly characterized by relaxed euchromatin, abundant distribution of acetylated histone marks and high accessibility of transcription factors to the DNA. Direct binding of transcription factors to their cis-elements is favored by the access of the recognition sequences in the DNA (Fig. 1). Progenitors are highly proliferative cells and they can be actively maintained in the cell cycle by the action of mitogens (e.g. FGF2 and PDGF-AA), which stimulate E2F-dependent transcription of genes involved in proliferation (Magri et al. 2014b). This important role mediated by E2F1 is also associated with the transcriptional activation of genes involved in chromatin architecture including the histone variant H2A.Z and the DNA binding molecule UHRF1 that preferentially binds to hemi-methylated DNA (Magri et al. 2014a).
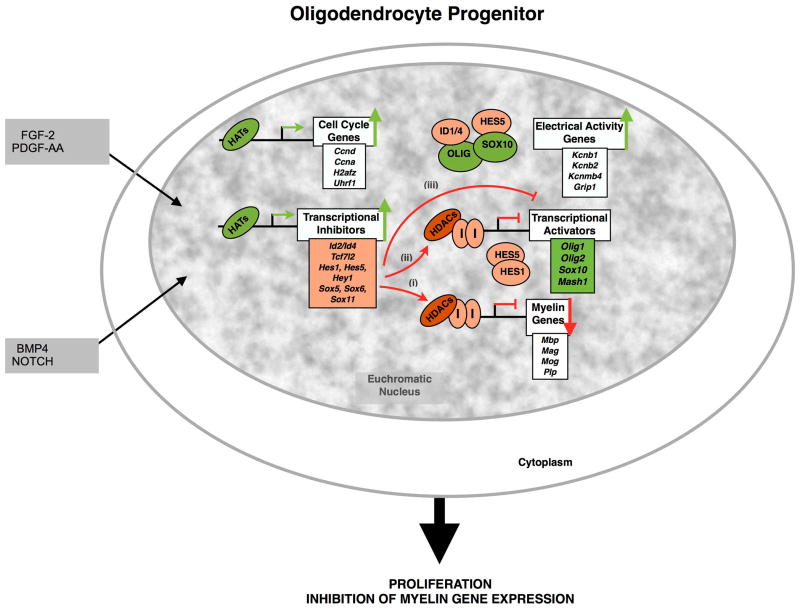
Figure 1. The euchromatic nucleus of oligodendrocyte progenitors is characterized by the expression of cell cycle genes and high levels of transcriptional inhibitors.
Oligodendrocyte progenitors are highly proliferative bipolar cells, actively responding to mitogenic signals (PDGF-AA, FGF-2). At the molecular level, these progenitors are characterized by a transcriptionally active chromatin state characterized by the abundant deposition of acetyl marks by histone acetyl transferases (HATs) at genes regulating proliferation and transcriptional inhibitors (orange box) of myelin genes (white box) and their activators (green box). As consequence, the expression levels of cell cycle genes and transcriptional inhibitors of genes associated with the differentiated state are very high. Three mechanisms of actions have been previously described for the action of the inhibitors (HES5, ID2/4 and SOX5/6/11) and are represented in the figure: (i) direct recruitment of HDAC-containing repressive complexes to the promoter of myelin genes; or (ii) to the promoter of transcriptional activators. (iii) A third mechanism consists on the sequestration of transcriptional activators by the inhibitors through protein-protein interactions. In green are represented the events resulting in transcriptional activation and in red those resulting in transcriptional repression. Green arrows indicate high levels of transcripts while red arrows indicate low levels.
These proliferative cells are also undifferentiated and characteristically defined by high levels of transcriptional inhibitors, including the HLH family members HES1, HES5, ID2, ID4 and the SOX family members of subgroup C (e.g.SOX11), subgroup D (e.g. SOX5 and 6) and possibly also members of the HMG family of transcription factors (e.g.TCF7L2/TCF4). This vast repertoire of transcriptional inhibitors prevents differentiation in multiple ways: (i) by direct binding to negative regulatory elements on myelin genes or (ii) oligodendrocyte specific transcription factors or (iii) by sequestration of positive regulators of myelin genes (Fig. 1).
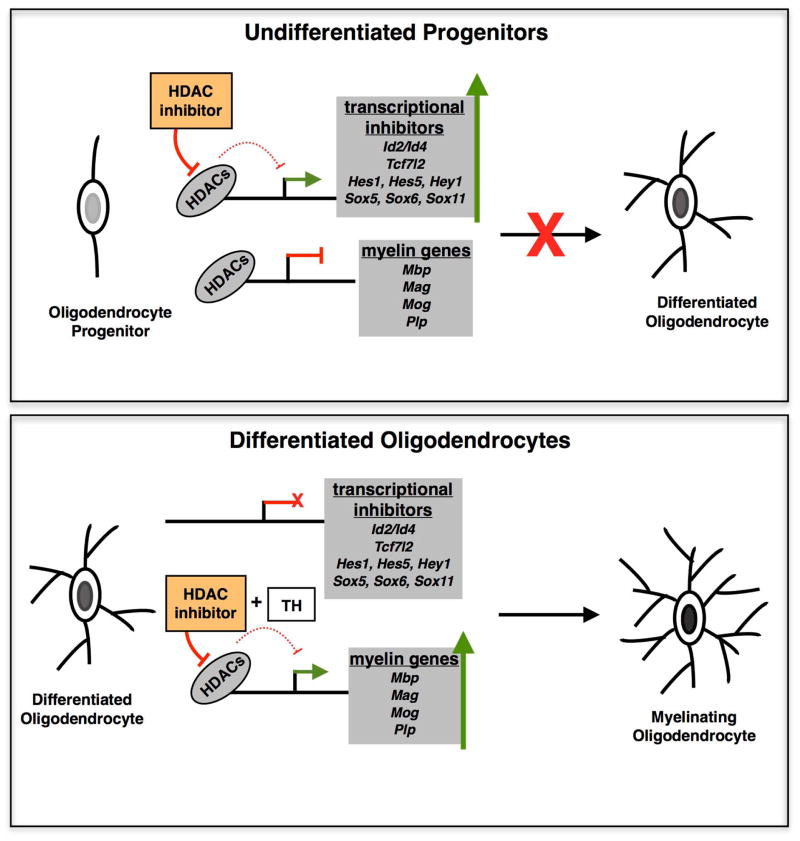
The fact that transcriptional inhibitors of differentiation were shown to recruit HDAC-containing repressive complexes to myelin genes (Liu et al. 2006) suggested the possibility that inhibition of HDAC activity, using specific pharmacological inhibitors could increase myelin gene expression. However, treatment of cultured oligodendrocyte lineage cells or neonatal pups with HDAC inhibitors prevented myelin gene expression and impaired development of myelinating tracts, respectively, due to up-regulation of transcriptional inhibitors (Marin-Husstege et al. 2002),(Shen et al. 2005) (Swiss et al. 2011). A large body of evidence supported that HDAC activity is necessary for earlier stages of oligodendrocyte differentiation during developmental myelination (Fig. 2) and remyelination in order to remove the inhibitory “brakes” to myelin gene expression, defined by the transcriptional inhibitors of differentiation (Popko 2008; Shen and Casaccia-Bonnefil 2008). In support of this model, high levels of the transcriptional inhibitors Hes5 and Id4 were detected in the brains of old mice, characterized by defective remyelination and impaired recruitment of HDACs to chromatin loci containing genes encoding for transcriptional inhibitors (Shen and Casaccia-Bonnefil 2008). High levels of these inhibitors and impaired histone acetylation were also detected in the brain of multiple sclerosis patients (Pedre et al. 2011), thereby further reinforcing the importance of HDAC-dependent removal of inhibitory constrains to myelin gene expression during repair. Interestingly, HDAC inhibition in cultured neural progenitor cells has been shown to promote myelin gene expression and later markers of differentiation, if combined with treatment with thyroid hormone (Castelo-Branco et al. 2014a). These results were consistent with the previously reported evidence that the last stages of differentiation of oligodendrocytes are characterized by the presence of increased histone acetylation marks on myelin genes (Liu et al. 2010). The translational implications of this finding will need further investigation in disease models of demyelination, given the positive results of acute co-treatment of T3 and HDAC inhibitors in a rat model of EAE (Castelo-Branco et al. 2014b). Together these results support the concept that inhibition of histone deacetylation is detrimental during a selective temporal window of progenitor differentiation (Shen et al. 2005), when it serves the main purpose of “removing the brakes” of myelination, while it may be beneficial at later time points when its inhibition serves the purpose of enhancing myelin gene expression.
Figure 2. The functional role of HDACs in oligodendrocyte lineage cells depends on the differentiation stage.
The functional role of HDACs in oligodendrocyte differentiation is stage-dependent. The pharmacological inhibition of HDACs at early stages of progenitor differentiation (TOP panel) results in increased levels of transcriptional inhibitors of oligodendrocyte differentiation and impaired differentiation. Pharmacological inhibition of HDACs at later stages of oligodendrocyte differentiation (BOTTOM panel) and in the presence of thyroid hormone (TH) enhances the transcriptional activity of myelin gene promoters and favors the attainment of a pro-myelinating phenotype.
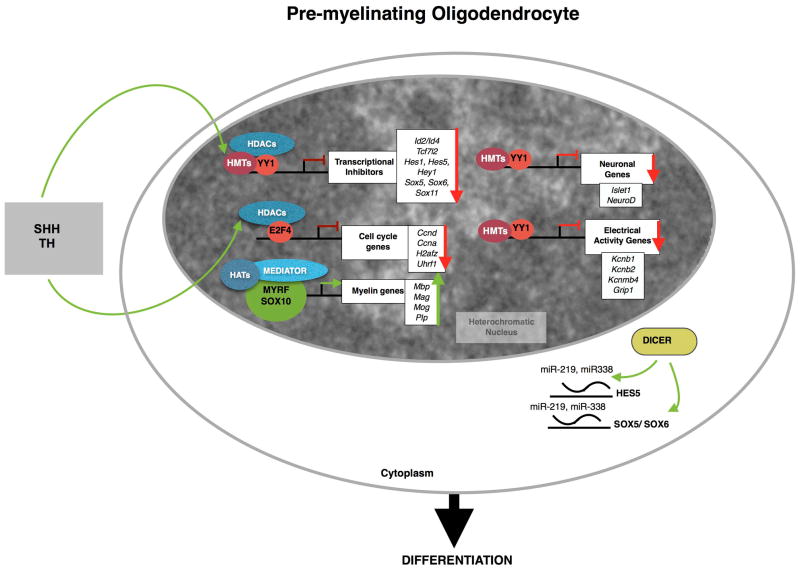
The process of differentiation of OP into oligodendrocytes is characterized by the progressive formation of heterochromatin starting at the nuclear periphery and radially converging towards the center of the nuclei (Fig. 3). This is associated with high levels of histone deacetylase activity during the early stages of differentiation and high levels of K9 histone methyltransferases, leading to transcriptional repression of several gene categories, including: transcriptional inhibitors of myelin genes (e.g. Hes5, Sox5), transcription factors regulating neuronal lineage genes (e.g. NeuroD and Islet1). The transcript levels of the inhibitors are further reduced by the up-regulation of microRNAs targeting the inhibitory bHLH molecules (e.g. Hes5) and Sox members (e.g Sox5, Sox6) (Fig. 3). Therefore, the process of myelination is characterized by de-repression of myelin genes and their activators including SOX10 and MYRF. Additional molecules, including ZFP191 and members of the nuclear hormone receptor family members, such as PPAR, RAR and RXR all contribute to these late stages of oligodendrocyte maturation with ZFP191 and MYRF also modulating the stage of axonal wrapping.
Figure 3. The process of differentiation into oligodendrocytes is characterized by repressive events mediated by post-translational histone modifications and the action of microRNAs.
The differentiation of progenitors towards oligodendrocytes includes branching of cellular processes and the progressive formation of heterochromatin in the nucleus. The underlying molecular mechanism is triggered by the action of extracellular signals (e.g. SHH, TH) that stimulate (green arrows) the activity of chromatin modifying enzymes including K9 histone methyltransferases (HMT) and histone deacetylases (HDACs). These enzymes are recruited into repressive complexes with transcription factors (i.e. E2F4, YY1) which target them to loci encoding for genes expressing transcriptional inhibitors of myelin genes as well as genes encoding for modulators of cell proliferation, neuronal lineage and electrical properties. Transcriptional repression at these loci is assocated with differentiation. The concerted action of micro-RNAs, generated by Dicer contributes to the decreased levels of differentiation inhibitors (e.g.Hes5, Sox5, Sox6).
5. CONCLUDING REMARKS
The most important message of this review is the description of distinctive nuclear landscapes defining progenitors and differentiated cells.
From a developmental perspective, the acknowledgement of the existence of distinctive nuclear landscapes in proliferating and differentiating progenitors is of great interest because it allows considering the critical importance of stage-specific cellular context, especially when interpreting the phenotype of mice with specific gene ablation at distinct time points.
From a translational perspective, it allows to take in consideration the modalities of responsiveness of targeted cell populations. At least two types of approaches can be envisioned. Targeting of the progenitor pool must take into account that these cells are amenable to transcription factor manipulations based on the relaxed chromatin structure and relatively easy access to the DNA. However these cells are also characterized by an active transcription program regulating cell proliferation and therefore need to be signaled to exit from the cell cycle. Targeting later stages of differentiation may also include the stimulation of specific transcription factors and nuclear receptors (PPARs, RXRS, TH) that have been associated with myelination (Billon et al. 2002; Feinstein et al. 2002; Huang et al. ; Niino et al. 2001). However, therapeutic approaches targeting later stages of differentiation may require the use of epigenome modulatory compounds that, in contrast to early stages of differentation, may enhance the access of transcription factors to DNA (Castelo-Branco et al. 2014a; Gacias et al. 2014).
Acknowledgments
PC is the recipient of grants R01-NS052738 and R37-NS42925 from NIH-NINDS. MH is the recipient of the fellowship F31NS083344. The authors are grateful to Dr. B. Barres and WD Richardson for providing helpful feedback and valuable suggestions and to Drs. M. Wegner, J. Goldman and B. Popko for sharing opinions and unpublished results.
GLOSSARY
- OP
Oligodendrocyte Progenitors
- bHLH (Basic helix-loop-helix)
Protein structural domain in transcription factors, composed by two alpha helices connected by a loop, that allows protein-protein interactions and the formation of homo or heterodimers. This domain is distinct from the DNA binding-motif which is rich in basic aminoacids and localized to the N-terminus
- E-proteins
Class I of basic helix-loop-helix transcription factors that are ubiquitously expressed. They are able to heterodimerize with other transcription factors with the bHLH domain
- E-boxes
DNA consensus sequence (CANNTG) recognized by E-proteins
- SOX
SOX proteins are SRY-Related HMG-Box transcription factors characterized by having a HMG (High Mobility Group) domain
- High Mobility Group (HMG-box)
Protein structural motif characterized by the presence of three alpha helices separated by loops found in transcription factors and subunits of chromatin-remodeling complexes. Two types of DNA binding proteins containing this domain have been identified: those that bind non-specifically to DNA (i.e. HMGB) which is in a non-B conformation (i.e. bent, kinked and unwound) and transcription factors (i.e. SOX family members) with sequence-specific DNA binding
- N-boxes
DNA consensus sequence (CACNAG) recognized by HES transcription factors
- Zinc finger
Protein structural motif characterized by a tertiary structure stabilized by a Zn ion. The best-characterized mammalian Zn finger transcription factors are characterized by the presence of spaced cysteine and histidine aminoacid residues which typically define repeats, with each repeat forming a “finger” with the ability to bind in the major groove of DNA
- Peroxisome proliferator-activated receptors (PPAR)
Peroxisome proliferator-activated receptor defines a nuclear receptor family with the ability to form dimers with RXR and bind to the DNA consensus sequence: AGGTCANAGGTCA to promote transcriptional activity, after binding to the respective ligand (fatty acids for PPARα, prostaglandins for PPARγ)
- Retinoic acid receptor (RAR)
It includes a family of nuclear receptors (RAR α, β and γ) with the ability to bind the retinoic acid response elements (RARE) corresponding to a tandem sequence AGGTCA. In the absence of the ligand, the RAR form RXR-RAR heterodimers with repressive activity, while in the presence of the ligands (all-trans retinoic acid or 9-cis retinoic acid) they modulate transcriptional activity
- Retinoid X receptor (RXR)
The retinoid X receptor superfamily of transcription factors includes RXR α, β and γ and forms repressive complexes with RAR in the absence of the ligand (9-cis retinoic acid). In the presence of the ligand, they have the ability to bind to other classes of nuclear receptors, including TR, VDR and PPARs
- Chromatin remodeling
The process of modifying chromatin structure by nucleosome repositioning (or sliding) along the DNA
- Euchromatin
It is a relaxed chromatin structure that favors transcription factor access to DNA. It includes the most active regions of the genome, which are actively transcribed
- Heterochromatin
It is a compact form of chromatin that can be constitutive or facultative and is associated with transcription repression
- Histone acetyltransferases (HATs)
Group of enzymes responsible for the acetylation of lysine residues on nucleosomal histones and additional substrates, including enzymes and transcription factors
- Histone deacetylases (HDACs)
Group of enzymes responsible for the removal of acetyl groups from lysine residues on nucleosomal histones to promote transcription repression. Besides nucleosomal histones, HDACs can also modulate the activity of other proteins, including transcription factors, other enzymes and even cytoskeletal molecules, depending on their nuclear or cytosolic distrubution
- Histone lysine methyltransferases (HMTs)
Enzymes with the ability to specifically methylate lysine residues at precise genomic locations (e.g. K4, K9, K27 on histone H3) by transferring one, two or three methyl groups
- Histone lysine demethylases (KDM)
Enzymes catalyzing the removal of methyl groups from lysine residues on nucleosomal histones
- Protein arginine methyltransferases (PRMTs)
Family of enzymes that methylate arginine residues in histones or non-histone proteins. Three types of methylations have been identified: mono-methylation, asymmetric dimethylation and symmetric dimethylation
- Polycomb group of proteins (PcG)
Family of proteins with distinctive repressive functions including the PRC1 and PRC2 protein complexes, which catalyze the formation of the H3K27me3 repressive histone mark
Footnotes
The authors declare no conflict of interest.
References
- Akazawa C, Sasai Y, Nakanishi S, Kageyama R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992;267(30):21879–85. [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–94. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306(5704):2111–5. doi: 10.1126/science.1103709. [DOI] [PubMed] [Google Scholar]
- Avery OT, Macleod CM, McCarty M. Studies on the Chemical Nature of the Substance Inducing Transformation of Pneumococcal Types : Induction of Transformation by a Desoxyribonucleic Acid Fraction Isolated from Pneumococcus Type Iii. J Exp Med. 1944;79(2):137–58. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120(5):1097–108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lassar A, Tapscott S, Thayer M, Lockshon D, Weintraub H. Id: a negative regulator of helix-loop-helix DNA binding proteins. Control of terminal myogenic differentiation. Ann N Y Acad Sci. 1990a;599:1–11. doi: 10.1111/j.1749-6632.1990.tb42359.x. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990b;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Berndt JA, Kim JG, Tosic M, Kim C, Hudson LD. The transcriptional regulator Yin Yang 1 activates the myelin PLP gene. J Neurochem. 2001;77(3):935–42. doi: 10.1046/j.1471-4159.2001.00307.x. [DOI] [PubMed] [Google Scholar]
- Billon N, Jolicoeur C, Tokumoto Y, Vennstrom B, Raff M. Normal timing of oligodendrocyte development depends on thyroid hormone receptor alpha 1 (TRalpha1) EMBO J. 2002;21(23):6452–60. doi: 10.1093/emboj/cdf662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof M, Weider M, Kuspert M, Nave KA, Wegner M. Brg1-Dependent Chromatin Remodelling Is Not Essentially Required during Oligodendroglial Differentiation. J Neurosci. 2015;35(1):21–35. doi: 10.1523/JNEUROSCI.1468-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30(1):73–6. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9(23):2888–902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7(4):592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16(8):4349–56. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15(1):66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M. Control of oligodendroglial cell number by the miR-17–92 cluster. Development. 2010;137(13):2127–32. doi: 10.1242/dev.050633. [DOI] [PubMed] [Google Scholar]
- Bujalka H, Koenning M, Jackson S, Perreau VM, Pope B, Hay CM, Mitew S, Hill AF, Lu QR, Wegner M, et al. MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol. 2013;11(8):e1001625. doi: 10.1371/journal.pbio.1001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat Neurosci. 2003;6(11):1162–8. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45(1):41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, Pietrosanti L, De Smaele E, Ferretti E, Miele E, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12(2):132–42. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco G, Lilja T, Wallenborg K, Falcao AM, Marques SC, Gracias A, Solum D, Paap R, Walfridsson J, Teixeira AI, et al. Neural stem cell differentiation is dictated by distinct actions of nuclear receptor corepressors and histone deacetylases. Stem Cell Reports. 2014a;3(3):502–15. doi: 10.1016/j.stemcr.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco G, Stridh P, Guerreiro-Cacais AO, Adzemovic MZ, Falcao AM, Marta M, Berglund R, Gillett A, Hamza KH, Lassmann H, et al. Acute treatment with valproic acid and l-thyroxine ameliorates clinical signs of experimental autoimmune encephalomyelitis and prevents brain pathology in DA rats. Neurobiol Dis. 2014b;71:220–33. doi: 10.1016/j.nbd.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Chew LJ, Shen W, Ming X, Senatorov VV, Jr, Chen HL, Cheng Y, Hong E, Knoblach S, Gallo V. SRY-box containing gene 17 regulates the Wnt/beta-catenin signaling pathway in oligodendrocyte progenitor cells. J Neurosci. 2011;31(39):13921–35. doi: 10.1523/JNEUROSCI.3343-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118(5):545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- De Nuccio C, Bernardo A, De Simone R, Mancuso E, Magnaghi V, Visentin S, Minghetti L. Peroxisome proliferator-activated receptor gamma agonists accelerate oligodendrocyte maturation and influence mitochondrial functions and oscillatory Ca(2+) waves. J Neuropathol Exp Neurol. 2011;70(10):900–12. doi: 10.1097/NEN.0b013e3182309ab1. [DOI] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52(6):953–68. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585(13):2024–31. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65(5):597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Ibrahim A, Barres BA. The T3-induced gene KLF9 regulates oligodendrocyte differentiation and myelin regeneration. Mol Cell Neurosci. 2012;50(1):45–57. doi: 10.1016/j.mcn.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26(43):10967–83. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26(2–4):148–65. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138(1):172–85. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, Kaing S, Sanai N, Franklin RJ, Rowitch DH. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23(13):1571–85. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–40. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Feinstein DL, Galea E, Gavrilyuk V, Brosnan CF, Whitacre CC, Dumitrescu-Ozimek L, Landreth GE, Pershadsingh HA, Weinberg G, Heneka MT. Peroxisome proliferator-activated receptor-gamma agonists prevent experimental autoimmune encephalomyelitis. Ann Neurol. 2002;51(6):694–702. doi: 10.1002/ana.10206. [DOI] [PubMed] [Google Scholar]
- Feller C, Forne I, Imhof A, Becker PB. Global and Specific Responses of the Histone Acetylome to Systematic Perturbation. Mol Cell. 2015 doi: 10.1016/j.molcel.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12(12):1052–8. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Fields RD. Oligodendrocytes changing the rules: action potentials in glia and oligodendrocytes controlling action potentials. Neuroscientist. 2008;14(6):540–3. doi: 10.1177/1073858408320294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzsch M, Stolt CC, Lommes P, Wegner M. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression. Development. 2008;135(4):637–46. doi: 10.1242/dev.010454. [DOI] [PubMed] [Google Scholar]
- Funfschilling U, Supplie LM, Mahad D, Boretius S, Saab AS, Edgar J, Brinkmann BG, Kassmann CM, Tzvetanova ID, Mobius W, et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature. 2012;485(7399):517–21. doi: 10.1038/nature11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacias M, Gerona-Navarro G, Plotnikov AN, Zhang G, Zeng L, Kaur J, Moy G, Rusinova E, Rodriguez Y, Matikainen B, et al. Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem Biol. 2014;21(7):841–54. doi: 10.1016/j.chembiol.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J Neurosci. 2005;25(36):8311–21. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–8. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39(5):749–65. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Grbavec D, Stifani S. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem Biophys Res Commun. 1996;223(3):701–5. doi: 10.1006/bbrc.1996.0959. [DOI] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75(3):463–76. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55(2):217–30. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Kim JY, Dupree J, Tewari A, Melendez-Vasquez C, Svaren J, Casaccia P. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat Neurosci. 2010;13(12):1472–80. doi: 10.1038/nn.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20(14):1848–67. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80(4):583–92. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hoffmann SA, Hos D, Kuspert M, Lang RA, Lovell-Badge R, Wegner M, Reiprich S. Stem cell factor Sox2 and its close relative Sox3 have differentiation functions in oligodendrocytes. Development. 2014;141(1):39–50. doi: 10.1242/dev.098418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Mechanisms for the control of gene activity during development. Biol Rev Camb Philos Soc. 1990;65(4):431–71. doi: 10.1111/j.1469-185x.1990.tb01233.x. [DOI] [PubMed] [Google Scholar]
- Howng SY, Avila RL, Emery B, Traka M, Lin W, Watkins T, Cook S, Bronson R, Davisson M, Barres BA, et al. ZFP191 is required by oligodendrocytes for CNS myelination. Genes Dev. 2010;24(3):301–11. doi: 10.1101/gad.1864510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JK, Jarjour AA, Nait Oumesmar B, Kerninon C, Williams A, Krezel W, Kagechika H, Bauer J, Zhao C, Evercooren AB, et al. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat Neurosci. 2011;14(1):45–53. doi: 10.1038/nn.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton SR, Pevny LH. SOX2 expression levels distinguish between neural progenitor populations of the developing dorsal telencephalon. Dev Biol. 2011;352(1):40–7. doi: 10.1016/j.ydbio.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5(6):226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238(11):2800–12. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim H. Recruitment and biological consequences of histone modification of H3K27me3 and H3K9me3. ILAR J. 2012;53(3–4):232–9. doi: 10.1093/ilar.53.3-4.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Armstrong RC, v Agoston D, Robinsky A, Wiese C, Nagle J, Hudson LD. Myelin transcription factor 1 (Myt1) of the oligodendrocyte lineage, along with a closely related CCHC zinc finger, is expressed in developing neurons in the mammalian central nervous system. J Neurosci Res. 1997;50(2):272–90. doi: 10.1002/(SICI)1097-4547(19971015)50:2<272::AID-JNR16>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;9(8):597–604. [PubMed] [Google Scholar]
- Koenning M, Jackson S, Hay CM, Faux C, Kilpatrick TJ, Willingham M, Emery B. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci. 2012;32(36):12528–42. doi: 10.1523/JNEUROSCI.1069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000;127(14):2989–98. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004;18(23):2963–72. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13(5):297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184(4139):868–71. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28(45):11720–30. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11(5):754–70. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268(5212):836–44. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Leslie AG, Arnott S, Chandrasekaran R, Ratliff RL. Polymorphism of DNA double helices. J Mol Biol. 1980;143(1):49–72. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- Li H, de Faria JP, Andrew P, Nitarska J, Richardson WD. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron. 2011;69(5):918–29. doi: 10.1016/j.neuron.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27(52):14375–82. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25(20):4833–42. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33(4):193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Dietz K, Deloyht JM, Pedre X, Kelkar D, Kaur J, Vialou V, Lobo MK, Dietz DM, Nestler EJ, et al. Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat Neurosci. 2012;15(12):1621–3. doi: 10.1038/nn.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Magri L, Zhang F, Marsh NO, Albrecht S, Huynh JL, Kaur J, Kuhlmann T, Zhang W, Slesinger PA, et al. Chromatin Landscape Defined by Repressive Histone Methylation during Oligodendrocyte Differentiation. J Neurosci. 2015;35(1):352–65. doi: 10.1523/JNEUROSCI.2606-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sandoval J, Doh ST, Cai L, Lopez-Rodas G, Casaccia P. Epigenetic modifiers are necessary but not sufficient for reprogramming non-myelinating cells into myelin gene-expressing cells. PLoS One. 2010;5(9):e13023. doi: 10.1371/journal.pone.0013023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JML, Zhang F, Marsh NO, Albrecht S, Huynh JL, Kaur J, Kuhlmann T, Zhang W, Slesinger PA, Casaccia P. Chromatin landscape defined by repressive histone methylation during oligodendrocyte differentiation. J Neurosci. 2014 doi: 10.1523/JNEUROSCI.2606-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4(10):973–4. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109(1):75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog--regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25(2):317–29. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87(1):43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23(1):1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- Magri L, Gacias M, Wu M, Swiss VA, Janssen WG, Casaccia P. c-Myc-dependent transcriptional regulation of cell cycle and nucleosomal histones during oligodendrocyte differentiation. Neuroscience. 2014a;276:72–86. doi: 10.1016/j.neuroscience.2014.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri L, Swiss VA, Jablonska B, Lei L, Pedre X, Walsh M, Zhang W, Gallo V, Canoll P, Casaccia P. E2F1 coregulates cell cycle genes and chromatin components during the transition of oligodendrocyte progenitors from proliferation to differentiation. J Neurosci. 2014b;34(4):1481–93. doi: 10.1523/JNEUROSCI.2840-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinodan M, Rosen KM, Ito S, Corfas G. A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science. 2012;337(6100):1357–60. doi: 10.1126/science.1220845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, He Y, Li J, Kondo T, Sablitzky F, Casaccia-Bonnefil P. Multiple roles of Id4 in developmental myelination: predicted outcomes and unexpected findings. Glia. 2006;54(4):285–96. doi: 10.1002/glia.20385. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22(23):10333–45. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19(4):662–71. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Wang H, Liu S, Niu J, Wang L, He Y, Etxeberria A, Chan JR, Xiao L. Stage-specific deletion of Olig2 conveys opposing functions on differentiation and maturation of oligodendrocytes. J Neurosci. 2013;33(19):8454–62. doi: 10.1523/JNEUROSCI.2453-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliori V, Phalke S, Bezzi M, Guccione E. Arginine/lysine-methyl/methyl switches: biochemical role of histone arginine methylation in transcriptional regulation. Epigenomics. 2010;2(1):119–37. doi: 10.2217/epi.09.39. [DOI] [PubMed] [Google Scholar]
- Ming X, Chew LJ, Gallo V. Transgenic overexpression of Sox17 promotes oligodendrocyte development and attenuates demyelination. J Neurosci. 2013;33(30):12528–42. doi: 10.1523/JNEUROSCI.0536-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll NM, Hong E, Fauveau M, Naruse M, Kerninon C, Tepavcevic V, Klopstein A, Seilhean D, Chew LJ, Gallo V, et al. SOX17 is expressed in regenerating oligodendrocytes in experimental models of demyelination and in multiple sclerosis. Glia. 2013;61(10):1659–72. doi: 10.1002/glia.22547. [DOI] [PubMed] [Google Scholar]
- Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J Biol Chem. 2007;282(10):7632–40. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickol J, Behe M, Felsenfeld G. Effect of the B--Z transition in poly(dG-m5dC). poly(dG-m5dC) on nucleosome formation. Proc Natl Acad Sci U S A. 1982;79(6):1771–5. doi: 10.1073/pnas.79.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JA, Berndt JA, Hudson LD, Armstrong RC. Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells. Mol Cell Neurosci. 2004;25(1):111–23. doi: 10.1016/j.mcn.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Niino M, Iwabuchi K, Kikuchi S, Ato M, Morohashi T, Ogata A, Tashiro K, Onoe K. Amelioration of experimental autoimmune encephalomyelitis in C57BL/6 mice by an agonist of peroxisome proliferator-activated receptor-gamma. J Neuroimmunol. 2001;116(1):40–8. doi: 10.1016/s0165-5728(01)00285-5. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31(5):773–89. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Ogata T, Ueno T, Hoshikawa S, Ito J, Okazaki R, Hayakawa K, Morioka K, Yamamoto S, Nakamura K, Tanaka S, et al. Hes1 functions downstream of growth factors to maintain oligodendrocyte lineage cells in the early progenitor stage. Neuroscience. 2011;176:132–41. doi: 10.1016/j.neuroscience.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18(8):2196–207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, Temple S. Cell types to order: temporal specification of CNS stem cells. Curr Opin Neurobiol. 2009;19(2):112–9. doi: 10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Paes de Faria J, Kessaris N, Andrew P, Richardson WD, Li H. New Olig1 null mice confirm a non-essential role for Olig1 in oligodendrocyte development. BMC Neurosci. 2014;15:12. doi: 10.1186/1471-2202-15-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79(5):805–15. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pedre X, Mastronardi F, Bruck W, Lopez-Rodas G, Kuhlmann T, Casaccia P. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J Neurosci. 2011;31(9):3435–45. doi: 10.1523/JNEUROSCI.4507-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122(4):517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Popko B. Epigenetic control of myelin repair. Nat Neurosci. 2008;11(9):987–8. doi: 10.1038/nn0908-987. [DOI] [PubMed] [Google Scholar]
- Pozniak CD, Langseth AJ, Dijkgraaf GJ, Choe Y, Werb Z, Pleasure SJ. Sox10 directs neural stem cells toward the oligodendrocyte lineage by decreasing Suppressor of Fused expression. Proc Natl Acad Sci U S A. 2010;107(50):21795–800. doi: 10.1073/pnas.1016485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128(14):2723–33. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Riccio A. Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat Neurosci. 2010;13(11):1330–7. doi: 10.1038/nn.2671. [DOI] [PubMed] [Google Scholar]
- Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. J Neurochem. 2005;93(6):1444–53. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluja I, Granneman JG, Skoff RP. PPAR delta agonists stimulate oligodendrocyte differentiation in tissue culture. Glia. 2001;33(3):191–204. [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131(17):4131–42. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Schlierf B, Werner T, Glaser G, Wegner M. Expression of connexin47 in oligodendrocytes is regulated by the Sox10 transcription factor. J Mol Biol. 2006;361(1):11–21. doi: 10.1016/j.jmb.2006.05.072. [DOI] [PubMed] [Google Scholar]
- Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6(4):a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw NN, Arya DP. Recognition of the unique structure of DNA:RNA hybrids. Biochimie. 2008;90(7):1026–39. doi: 10.1016/j.biochi.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Shen S, Casaccia-Bonnefil P. Post-translational modifications of nucleosomal histones in oligodendrocyte lineage cells in development and disease. J Mol Neurosci. 2008;35(1):13–22. doi: 10.1007/s12031-007-9014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169(4):577–89. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJ, Casaccia-Bonnefil P. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008;11(9):1024–34. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Amor S, Gerritsen W, Baker D, Jackson SL, Boddeke E, Copray S. Intraventricularly injected Olig2-NSCs attenuate established relapsing-remitting EAE in mice. Cell Transplant. 2012 doi: 10.3727/096368911X637443. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15(4):272–86. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311(5762):844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Sock E, Schmidt K, Hermanns-Borgmeyer I, Bosl MR, Wegner M. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol Cell Biol. 2001;21(20):6951–9. doi: 10.1128/MCB.21.20.6951-6959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Natale J, Chew LJ, Belachew S, Cheng Y, Aguirre A, Lytle J, Nait-Oumesmar B, Kerninon C, Kanai-Azuma M, et al. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci. 2006;26(38):9722–35. doi: 10.1523/JNEUROSCI.1716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundarapandian MM, Selvaraj V, Lo UG, Golub MS, Feldman DH, Pleasure DE, Deng W. Zfp488 promotes oligodendrocyte differentiation of neural progenitor cells in adult mice after demyelination. Sci Rep. 2011;1:2. doi: 10.1038/srep00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternglanz R. Histone acetylation: a gateway to transcriptional activation. Trends Biochem Sci. 1996;21(10):357–8. [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131(10):2349–58. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17(13):1677–89. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16(2):165–70. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgartner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11(5):697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Schmitt S, Lommes P, Sock E, Wegner M. Impact of transcription factor Sox8 on oligodendrocyte specification in the mouse embryonic spinal cord. Dev Biol. 2005;281(2):309–17. doi: 10.1016/j.ydbio.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11(12):996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Parras CM, Nakatani H, Lebel M, Guillemot F, Nakafuku M. Ascl1 is required for oligodendrocyte development in the spinal cord. Development. 2008;135(7):1271–81. doi: 10.1242/dev.015370. [DOI] [PubMed] [Google Scholar]
- Sun T, Dong H, Wu L, Kane M, Rowitch DH, Stiles CD. Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J Neurosci. 2003;23(29):9547–56. doi: 10.1523/JNEUROSCI.23-29-09547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Echelard Y, Lu R, Yuk DI, Kaing S, Stiles CD, Rowitch DH. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr Biol. 2001;11(18):1413–20. doi: 10.1016/s0960-9822(01)00441-9. [DOI] [PubMed] [Google Scholar]
- Swiss VA, Nguyen T, Dugas J, Ibrahim A, Barres B, Androulakis IP, Casaccia P. Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PLoS One. 2011;6(4):e18088. doi: 10.1371/journal.pone.0018088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thier M, Worsdorfer P, Lakes YB, Gorris R, Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nothen MM, et al. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10(4):473–9. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- van Grunsven LA, Taelman V, Michiels C, Verstappen G, Souopgui J, Nichane M, Moens E, Opdecamp K, Vanhomwegen J, Kricha S, et al. XSip1 neuralizing activity involves the co-repressor CtBP and occurs through BMP dependent and independent mechanisms. Dev Biol. 2007;306(1):34–49. doi: 10.1016/j.ydbio.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Vana AC, Lucchinetti CF, Le TQ, Armstrong RC. Myelin transcription factor 1 (Myt1) expression in demyelinated lesions of rodent and human CNS. Glia. 2007;55(7):687–97. doi: 10.1002/glia.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Timmers HT. Grasping trimethylation of histone H3 at lysine 4. Epigenomics. 2010;2(3):395–406. doi: 10.2217/epi.10.11. [DOI] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem. 1999;274(29):20489–98. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- Vogl MR, Reiprich S, Kuspert M, Kosian T, Schrewe H, Nave KA, Wegner M. Sox10 cooperates with the mediator subunit 12 during terminal differentiation of myelinating glia. J Neurosci. 2013;33(15):6679–90. doi: 10.1523/JNEUROSCI.5178-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183(4676):1654–5. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41(1):10–3. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29(3):603–14. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Dulin J, Wu H, Hurlock E, Lee SE, Jansson K, Lu QR. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133(17):3389–98. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–8. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. Stage-specific expression of myelin basic protein in oligodendrocytes involves Nkx2.2-mediated repression that is relieved by the Sp1 transcription factor. J Biol Chem. 2005;280(16):16284–94. doi: 10.1074/jbc.M500491200. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251(4995):761–6. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Weng Q, Chen Y, Wang H, Xu X, Yang B, He Q, Shou W, Higashi Y, van den Berghe V, Seuntjens E, et al. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron. 2012;73(4):713–28. doi: 10.1016/j.neuron.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Pruss D. Targeting chromatin disruption: Transcription regulators that acetylate histones. Cell. 1996;84(6):817–9. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- Wu M, Hernandez M, Shen S, Sabo JK, Kelkar D, Wang J, O’Leary R, Phillips GR, Cate HS, Casaccia P. Differential modulation of the oligodendrocyte transcriptome by sonic hedgehog and bone morphogenetic protein 4 via opposing effects on histone acetylation. J Neurosci. 2012;32(19):6651–64. doi: 10.1523/JNEUROSCI.4876-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Front Biosci. 2006;11:344–55. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25(6):1354–65. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21(17):5979–91. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12(7):829–38. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115(3):281–92. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–57. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152(1–2):248–61. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21(8):1273–83. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol. 2011;13(11):1295–304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65(5):612–26. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109(1):61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31(5):791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25(2):331–43. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zuo H, Maher BJ, Serwanski DR, LoTurco JJ, Lu QR, Nishiyama A. Olig2-dependent developmental fate switch of NG2 cells. Development. 2012;139(13):2299–307. doi: 10.1242/dev.078873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolova OE, Wight PA. YY1 negatively regulates mouse myelin proteolipid protein (Plp1) gene expression in oligodendroglial cells. ASN Neuro. 2011;3(4) doi: 10.1042/AN20110021. [DOI] [PMC free article] [PubMed] [Google Scholar]