Abstract
RIG-I and MDA5 are well-conserved cytoplasmic pattern recognition receptors that detect viral RNAs during infection and activate the type I interferon (IFN)-mediated antiviral immune response. While much is known about how these receptors recognize viral RNAs, how they interact with their common signaling adaptor molecule MAVS and activate the downstream signaling pathway had been less clear. Previous studies have shown that the signaling domains (tandem CARDs or 2CARDs) of RIG-I and MDA5 must form homo-oligomers in order to interact with MAVS, and that their interactions lead to filament formation of MAVS, a prerequisite for downstream signal activation. More recent data suggest that multiple mechanisms synergistically promote tetramer formation of RIG-I 2CARD, and that this tetramer resembles a lock-washer, which serves as a helical template to nucleate the MAVS filament. We here summarize these recent findings and discuss the current understanding of the signal activation mechanisms of RIG-I and MDA5.
Introduction
RIG-I and MDA5 are well-conserved cytoplasmic pattern recognition receptors that detect viral RNAs during infection and activate a series of antiviral signaling pathways, leading to the production of type I interferons (IFNs) and other pro-inflammatory cytokines [1]. RIG-I and MDA5 play non-redundant roles by detecting largely distinct groups of viruses and by recognizing distinct features of viral RNAs [2-4]. They are paralogous receptors, sharing the same domain architecture and the downstream adaptor molecule, MAVS (also known as IPS-1, VISA and Cardif) [5-8]. Both RIG-I and MDA5 have the N-terminal tandem caspase activation recruitment domain (tandem CARD or 2CARD), which interacts with MAVS and activates the antiviral signaling pathway (Fig. 1A). Next to 2CARD are the central helicase domain and CTD, which together function as an RNA recognition unit (Fig. 1A). Studies have shown that in the absence of viral RNA, 2CARD of RIG-I (and likely MDA5) is masked by the intramolecular interaction with the helicase domain, but upon viral RNA binding, it is exposed to interact with the single CARD domain of MAVS (Fig. 1A) [9]. Note that while MAVS is anchored to the outermembrane of mitochondria and peroxisomes through the C-terminal transmembrane (TM) region [5-8, 10], the N-terminal CARD is in the cytoplasm and accessible to RIG-I/MDA5. Upon its interaction with RIG-I/MDA5 2CARD, MAVS recruits TRAF2, 5 and 6 to its linker between CARD and TM (Fig. 1A), which in turn activates the further downstream antiviral signaling pathway [11-13].
Fig. 1.

(A) Schematic of viral RNA recognition and antiviral signal activation by RIG-I and MDA5. Both receptors utilize MAVS on mitochondria (or peroxisome) for activation of the downstream signaling pathway. This review focuses on the interactions between the CARD domains of RIG-IMDA5 and MAVS. See (B) for details. (B) Antiviral signal activation requires monomer-to-filament transition of MAVS CARD, which in turn requires homo-oligomerization of RIG-I/MDA5 2CARDs. There are at least two mutually non-exclusive mechanisms that induce 2CARD oligomerization; ubiquitin-mediated and filament-induced mechanisms.
While much attention has been paid to the mechanisms by which RIG-I and MDA5 recognize their cognate viral RNAs, relatively little had been known until recently as to how they interact with MAVS, whether this interaction is direct and how this interaction leads to “activation” of MAVS. In this review, we will summarize our current understanding of the signal activation mechanism of RIG-I/MDA5, with the focus on the oligomeric/multimeric assemblies of RIG-IMDA5 2CARDs and MAVS CARD, the key molecular event during their signal activation processes.
Filament formation of MAVS CARD
CARD belongs to the Death Domain superfamily, which is composed of the subfamilies of Death Domains (DDs), Death Effector Domains (DEDs) and Pyrin domains (PYD) along with CARDs [14]. They form a compact (∼10 kDa) globular domain with the common protein fold of six antiparallel α-helices. These domains are often involved in protein-protein interactions, in particular homotypic, sub-family restricted interactions, and play important roles in many cell death and inflammatory signaling pathways [14]. Biochemical and structural analysis of CARDs along with other members of the DD superfamily is often challenging due to their propensity to aggregate or to form large oligomeric assemblies.
Aggregation or oligomerization of CARD was also shown to be central to the signaling pathway of RIG-I/MDA5 and MAVS. Since its discovery, MAVS was reported to form detergent-insoluble aggregates on the surface of mitochondria upon activation of RIG-IMDA5 [6, 15]. In 2011, Zhijian J. Chen group showed that this aggregate is in fact a highly ordered filament formed by MAVS CARD and that this filament formation is necessary for recruitment of TRAF molecules and activation of the downstream signaling pathway (Fig. 1B) [16]. Filament formation of MAVS CARD is also sufficient for signal activation; induction of MAVS filament formation by seeding with pre-formed filaments is sufficient to induce dimerization of IRF3 [16]. Mutational studies showed close correlation between filament formation in vitro and the signaling activity in cells [17-19], providing additional support for the notion that the filament architecture underlies the assembly of the “active” conformation of MAVS.
How can MAVS CARD form filaments while anchored to the outer membrane of mitochondria? Between the N-terminal CARD and the C-terminal TM is the ∼400 amino acid-long linker (Fig. 1A), which would provide sufficient flexibility for CARD filament assembly on mitochondrial surface. Cellular imaging analysis revealed formation of ∼400 nm long rod-shaped aggregates of MAVS on mitochondria, which likely represent short filaments [19]. Reasons for the limited elongation of the MAVS filament is unclear, but this could be due to limited number of the MAVS molecules per mitochondrion (or fused mitochondria) or as-yet-unknown cellular mechanisms to regulate filament propagation. Mitochondrial elongation/fusion was also shown to accompany MAVS activation [20, 21], but causal relationship and its impact on MAVS filament assembly remains to be further investigated.
Together, these findings have defined the filamentous form as the “active” state of MAVS and have laid a foundation for a series of investigations by us and others as to how RIG-I/MDA5 induces MAVS filament formation. The crucial breakthrough was in vitro reconstitution of the signaling events by the Z. J. Chen group (using purified mitochondria or cell extract) [22-24] and our group (using purified components) [17, 18, 25, 26]. In particular, our reconstitution from the purified components demonstrated that the interaction between RIG-I/MDA5 2CARD and MAVS CARD is direct, and that this interaction is sufficient to induce filament formation of MAVS [17, 25]. Below, we first describe the mechanism of how RIG-I 2CARD induces MAVS filament formation at the level of biochemistry, structure and function, followed by our current understanding of MDA5-mediated signal activation.
Ub-dependent oligomerization of RIG-I 2CARD
A series of studies (to be described below) have shown that oligomerization of 2CARD is required to induce filament formation of MAVS CARD. There appear to be at least two mutually non-exclusive mechanisms to induce 2CARD oligomerization, one that utilizes K63-linked polyubiquitin chains (K63-Ubn) and the other mediated by filament formation of the RNA-binding domain (helicase domain and CTD). We first describe the Ub-dependent mechanism in this section.
Ubiquitin (Ub) plays important roles in diverse cellular functions. In particular, K63-Ubn has been shown to function as an activation switch in many immune signaling pathways [27, 28]. In 2007, Jae U. Jung and his colleagues showed that RIG-I is covalently modified with K63-Ubn by an E3 ligase, Trim25 [29]. They identified six Lys residues in 2CARD that are modified with K63-Ubn, and showed that these modifications, in particular that of Lys172, are important for efficient antiviral signaling activity of RIG-I. Subsequently, Z. J. Chen and his colleagues showed that RIG-I 2CARD can non-covalently bind unanchored K63-Ubn, and this non-covalent interaction alone can lead to oligomerization of 2CARD and subsequent activation (i.e. filament formation) of MAVS [22]. The Chen group demonstrated that the addition of both unanchored chain of K63-Ubn and recombinant RIG-I 2CARD to the cell extract, but neither alone, can induce efficient dimerization of IRF3 [22]. Since then, one of the central questions in the field had been how covalent and non-covalent interactions with K63-Ubn lead to activation of RIG-I signaling and what their relative contributions are. The challenge in dissecting their roles came, in part, from the lack of a structural model and identification of mutations that selectively impair Ub-binding without affecting its Ub-conjugation or interaction with another RIG-I 2CARD or MAVS.
In 2014, we reported the crystal structure of the 2CARD tetramer non-covalently bound by K63-Ub2 [18]. The structure explained how the non-covalent interaction with K63-Ubn leads to tetramerization of 2CARD and identified residues involved in Ub binding, 2CARD homo-oligomerization and MAVS activation, which allowed testing the importance of individual factors. In the structure, 2CARD forms the core tetramer and three chains of K63-Ub2 bind the periphery of the 2CARD tetramer, bridging between the adjacent 2CARD subunits (Fig. 2A). In each 2CARD, there are two distinct Ub binding sites (proximal and distal sites, based on its proximity to the Ub-conjugation sites), the relative orientation of which is compatible only with the K63-linkage and no other Lys linkages. While the structure was determined with diubiquitin chains, which is inefficient in inducing tetramerization of 2CARD in solution, biochemical analysis showed that longer Ub chains bind 2CARD using the same interface as K63-Ub2, and can better stabilize the 2CARD tetramer through high avidity binding (i.e. wrapping around the tetramer, as shown in the right panel of Fig. 2B) [18]. Mutation of Ub-binding sites (e.g. R71, D75, K95 and E98) showed the importance of Ub-binding in RIG-I signaling, especially with isolated 2CARD or with full-length RIG-I stimulated by short dsRNA [18].
Fig. 2.
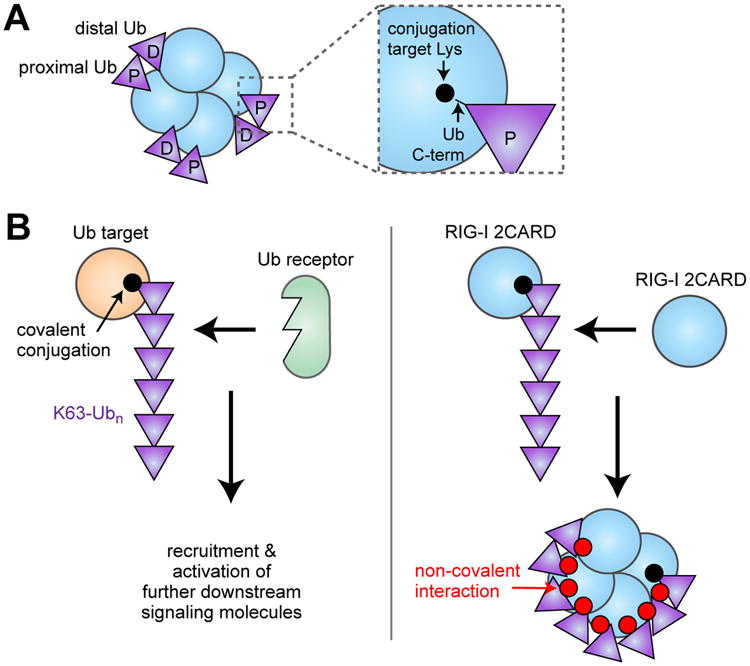
Mechanism by which K63-Ubn mediates oligomerization of RIG-I 2CARD. (A) Schematic of the structure of the RIG-I 2CARD tetramer non-covalently bound by K63-Ub2. The proximal and distal Ub's are bound to 2CARD in such a way that the proximal Ub can be covalently conjugated to the known Lys targets of 2CARD. (B) (left) It was previously thought that K63-Ubn conjugation activates a signaling pathway by recruiting specific Ub binding receptors to the target protein. In the case of RIG-I (right), K63-Ubn conjugation triggers self-assembly of 2CARDs due to its dual property as both the Ub conjugation target and the Ub receptor.
What does the structure tell us about the role of covalent K63-linked ubiquitination of RIG-I 2CARD? While the structure was determined with non-covalently bound Ub, the C-terminus of the proximal Ub is within the covalent conjugation distance from the six Lys residues known to be modified with K63-Ubn (Fig. 2B) [18]. This observation led to two predictions, both of which turned out to be true. First, the covalently conjugated K63-Ubn would bind the 2CARD tetramer in the same manner as the unanchored K63-Ubn. Second, having Ub covalently anchored immediately adjacent to its binding site would significantly enhance its ability to induce 2CARD tetramerization. Consistent with this prediction, covalently conjugated K63-Ubn also induces 2CARD tetramerization using the same 2CARD:Ub interface [18]. Furthermore, K63-Ub2, which is inefficient in inducing RIG-I 2CARD tetramerization and MAVS filament formation, becomes a robust stimulant upon covalent conjugation [18]. These results explain the observed importance of both covalent and non-covalent interactions between RIG-I and Ub in cells. In addition, they suggest that covalent and non-covalent interactions in fact cooperate in formation of a stable 2CARD tetramer and in robust activation of the antiviral signaling pathway (Fig. 2B).
Ub-independent, filament-mediated oligomerization of RIG-I 2CARD
Several lines of evidence suggest that K63-Ubn is not the sole mechanism to induce oligomerization of RIG-I 2CARD (Fig. 1B). The RNA-binding domain (helicase-CTD) of RIG-I forms short filamentous oligomers near dsRNA ends during ATP hydrolysis [17, 30], and the consequent proximity of 2CARDs alongside the filament can induce oligomerization of 2CARD in the absence of K63-Ubn (Fig. 3A) [17]. The linker between 2CARD and the core filament (∼50 amino acids) is in fact long enough to allow tetramerization of RIG-I 2CARD within the filamentous architecture (Fig. 3A). We found that the 2CARD oligomer formed in the filament also induces MAVS filament formation and utilizes the same inter-CARD interface as that formed with K63-Ubn, suggesting their structural similarity [17]. The importance of proximity-induced oligomerization was further supported by the observation that bridging of multiple 2CARDs through an artificial fusion protein was sufficient to enhance its antiviral signaling activity [17]. In further support of the importance of the proximity-induced oligomerization mechanism, isolated RIG-I 2CARD without the dimeric fusion protein, GST, is less efficient in signal activation than with GST (unpublished data).
Fig. 3.

The role of RIG-IMDA5 filament formation in signaling. (A) Model of filament-induced (i.e. proximity-induced) oligomerization of RIG-IMDA5 2CARDs. For both RIG-I and MDA5, the linker between 2CARD and the helicase domain is long enough to allow oligomerization of 2CARD alongside the filament. (B) The IFN-β promoter activity of wild-type RIG-I or mutants defective for Ub-binding or -conjugation. While Ub-binding and -conjugation are important for RIG-I-mediated signaling in response to 21 bp dsRNA, their importance diminishes with increasing dsRNA length. This suggests that filament formation (on long dsRNA) compensates for the loss of Ub-mediated oligomerization of 2CARD. (C) Synergism between the filament-induced and Ub-mediated (conjugation and binding) mechanisms for stable oligomerization of 2CARD and robust signaling by RIG-I. The relative importance of each of these mechanisms appears dependent on the dsRNA length. Images in (A) and (B) were adopted from Refs. [18, 25].
How can we reconcile the above two mechanisms of RIG-I activation, i.e. filament-mediated vs. Ub-dependent mechanisms? Several lines of evidence suggest that neither is a sole universal mechanism. For example, activation of RIG-I does not absolutely require filament formation as short duplex dsRNA (>20 bp) can still activate RIG-I-mediated signaling [31, 32], albeit less efficiently than longer dsRNA (e.g. ∼100-500 bp, when compared at the same molar concentration) [30, 33]. Similarly, mutations that impair Ub binding or conjugation do not abrogate signaling (in particular on long dsRNA)[17, 18], although future analysis is required to have mutations that simultaneously abrogate Ub binding and conjugation. We instead propose that filament-dependent and Ub-dependent mechanisms in fact cooperate for the robust signaling activity of RIG-I. While K63-Ubn plays an important role in signal activation in response to short dsRNAs, its importance diminishes with increasing length of dsRNA as filament formation can gradually compensate for the loss of Ub conjugation or binding (Fig. 3B) [18]. These results suggest that it is this combination of Ub-conjugation, Ub-binding and filament formation that allows RIG-I to efficiently recognize a broad repertoire of viral RNAs (Fig. 3C).
Mechanism for MAVS filament nucleation by the RIG-I 2CARD tetramer
How does the 2CARD tetramer induce filament formation of MAVS CARD? The first clue came from the assembly architecture of the RIG-I 2CARD tetramer, which displays a helical symmetry rather than the simple 4-fold (C4) symmetry typically seen with other tetrameric proteins (Fig. 4) [18]. The helical geometry can be described using the lock-washer model, in which a ring of the tetramer is split at one point and the two ends are displaced precisely by the half the thickness of the ring (Fig. 4). In this geometry, the first CARD of 2CARD forms the first helical turn, which is extended by the second CARD of 2CARD. The helical assembly of 2CARD is limited to the tetramer because the first and the second CARDs form a tight intramolecular interaction (Fig. 4), which makes the 2CARD as a rigid unit of the helical assembly. However, one could imagine that a single CARD, such as that of MAVS CARD, could be recruited and may extend the helical trajectory of 2CARD to form a longer filament (Fig. 4). This helical extension, in fact, turned out to be exactly how RIG-I 2CARD induces filament formation of MAVS CARD. Our 3.6Å electron microscopy reconstruction of the MAVS CARD filament showed that the helical geometry of the MAVS CARD filament is identical to that of the RIG-I 2CARD tetramer [26]. In addition, our recent crystal structure of the 2CARD tetramer in complex with the first helical turn of the MAVS CARD also showed that MAVS CARD is recruited to the RIG-I 2CARD tetramer by extending the helical trajectory pre-defined by the 2CARD tetramer (Fig. 4) [26].
Fig. 4.

Mechanism by which RIG-I 2CARD nucleates the MAVS CARD filament. In the absence of viral RNA, RIG-I 2CARD exists as a monomer, in which the first and second CARDs form a tight intramolecular interaction. Upon viral RNA binding, 2CARD assembles into a helical tetramer (either mediated by K63-Ubn or filament) that resembles a lock-washer. The 2CARD tetramer nucleates the MAVS CARD filament by recruiting MAVS CARD along the helical trajectory pre-defined by the 2CARD tetramer. Images were adopted from Ref [18].
Thus, the three independent structural analyses (the RIG-I 2CARD tetramer, the MAVS CARD filament and the RIG-I 2CARD:MAVS CARD complex) consistently demonstrated that RIG-I activates the signaling pathway by assembling 2CARDs into a helical template that nucleates the MAVS filament. This “lock-washer” mechanism also indicates that the physical nature of the signal that is passed on from RIG-I to MAVS is the assembly architecture of CARD, a previously unrecognized form of the molecular signal.
Independent EM studies by Qiu-Xing Jiang and his colleagues reported the ∼9.6 Å MAVS filament architecture vastly different from our near atomic resolution structure [19]. Unlike our filament model in which inter-CARD interactions match those observed in the RIG-I 2CARD tetramer and other assembly structures of the Death Domain superfamily [26], Jiang's filament involves previously unseen inter-CARD interactions. Could both structures be right? It is highly unlikely. Independent analysis of Jiang's data by Edward H. Egelman suggests that the filaments used by these two studies most likely share the same structure and Jiang's model involved incorrect assignment of the helical symmetry [34], a common mistake in helical reconstruction at low resolution [35].
Mechanism for 2CARD oligomerization and MAVS filament induction by MDA5
Mechanisms by which MDA5 oligomerizes its 2CARD and stimulates MAVS filament formation is less clear in comparison to RIG-I. While it is probable that MDA5 utilizes a similar lock-washer mechanism for inducing MAVS filament formation, evidence suggests subtle differences between RIG-I and MDA5 in the 2CARD oligomerization mechanism and perhaps the size of the 2CARD oligomers.
As with RIG-I 2CARD, MDA5 2CARD was proposed to utilize K63-Ubn for oligomerization and for subsequent MAVS activation [23]. In our hands, MDA5 2CARD has significantly lower affinity for K63-Ubn than RIG-I [25]. Since MDA5 2CARD is not covalently modified with K63-Ubn [29], it raises a question as to whether the low affinity interaction with K63-Ubn alone can provide sufficient stability to the 2CARD oligomer. We have found that purified MDA5 2CARD can spontaneously oligomerize in a concentration-dependent, Ub-independent manner and this oligomer can induce MAVS filament formation [25]. Since MDA5 forms filaments during dsRNA recognition [36, 37], we proposed that proximity of 2CARDs within the filamentous architecture enables oligomerization of 2CARD (Fig. 3A) [25]. The difference is that, unlike RIG-I, filament formation is absolutely required for dsRNA recognition by MDA5 [25, 36] and thus is expected to play more important roles in its 2CARD oligomerization. Dissection of relative importance of Ub-mediated and filament-mediated oligomerization of MDA5 2CARD requires future investigation.
Does MDA5 2CARD also form a tetramer as with RIG-I 2CARD? In MDA5, the linker between 2CARD and the helicase domain (∼100 amino acids) is twice as long as in RIG-I (Fig. 3A), and thus would allow formation of oligomers composed of 8-10 2CARDs. Interestingly, the length (but not sequence) of this linker is well conserved in MDA5 across species, suggesting that the size of the MDA5 2CARD oligomers may be also conserved and may require to be greater than a tetramer. In support of this notion, mutagenesis analysis suggests that MDA5 2CARD utilizes the tail surface (namely the surface opposite from the putative MAVS recruitment surface) for signal activation, whereas RIG-I 2CARD does not [18]. It is tempting to speculate that MDA5 2CARD forms a larger oligomer, perhaps an octamer formed by two lock-washer tetramers joined tail-to-tail. Future research is required to understand the oligomerization mechanism and structure of the MDA5 2CARD oligomers.
Outstanding questions and implications for other biological systems
There are still several important questions as yet to be answered. For example, how does the filament formation of MAVS CARD activate the downstream signaling pathway? The Z. J. Chen group showed that TRAF2, 5 and 6 were recruited to MAVS only when MAVS is in the filamentous state [16]. Based on the known preference of TRAFs for oligomerized target proteins [38], one may speculate that the MAVS filament recruits TRAF molecules by juxtaposing multiple TRAF binding sites in proximity. Consistent with this view, Takashi Fujita and his colleagues showed that the mutant MAVS where CARD is replaced by tandem repeats of FKBP can robustly activate antiviral signaling when its oligomerization is induced by a FKBP-crosslinker [39]. If simple proximity of a few TRAF binding sites can mediate the downstream signal activation, what is the significance of the filamentous architecture of MAVS as opposed to other oligomeric structures? We do not know the full answer to this question, but it is tempting to speculate that the filament structure, in comparison to a finite oligomeric structure, is particularly useful in signal amplification as the small number of RIG-I oligomers can induce oligomerization of a large number of MAVS.
Could the lock-washer mechanism illustrated in this review (Fig. 4) be unique to RIG-I/MDA5 or more generally applicable to other biological systems? Intriguingly, analogous mechanisms have been observed for nucleation of microtubules [40] and assembly of the Myddosome in the Toll-like receptor signaling pathway [41], another branch of innate immune pathway functioning parallel to RIG-I and MDA5. In both cases, the nucleator, the γ-tublin ring complex or MyD88, forms a helical template to recruit α/β tubulins or IRAK4/2 by extending the helical trajectory pre-defined by the respective nucleator. Although IRAK4/2 do not form extended filaments as with α/β tubulins or MAVS, the similarity in the underlying mechanism is remarkable. It is tempting to speculate that a similar nucleation mechanism may apply to a broader range of molecules, in particular to an emerging class of innate immune receptors, such as the inflammasomes [42, 43], that utilize filament formation of the respective downstream molecules as a switch to turn on the downstream signaling pathway. Future research is required to define the ubiquity of the lock-washer mechanism in macromolecular assembly and signal propagation.
Highlights.
RIG-I and MDA5 oligomerize the respective 2CARDs upon viral RNA recognition.
There are ubiquitin-dependent and -independent mechanisms to oligomerize 2CARDs.
RIG-I 2CARD forms a lock-washer-like tetramer.
The 2CARD tetramer serves as a helical template for nucleating the MAVS filament.
Acknowledgments
BW was supported by Charles A. King Trust Fellowship. SH was a Pew Scholar and was funded by NIH R01 grants (AI106912 and AI111784).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immuno Rev. 2011;243:91–8. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 2.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 3.Loo MY, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–45. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103(22):8459–64. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19(6):727–40. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 6*.Seth RB, et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–82. doi: 10.1016/j.cell.2005.08.012. This paper reports for the first time that MAVS aggregation accompanies its signal activation. [DOI] [PubMed] [Google Scholar]
- 7.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437(7062):1167–72. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 8.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–8. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 9*.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–35. doi: 10.1016/j.cell.2011.09.039. This study presents the crystal structure of 2CARD in the auto-repressed state, i.e. in the ligand-free, full-length RIG-I. [DOI] [PubMed] [Google Scholar]
- 10.Dixit E, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141(4):668–81. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha SK, et al. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 2006;25:3257–63. doi: 10.1038/sj.emboj.7601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paz S, et al. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res. 2011;21:895–910. doi: 10.1038/cr.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S, et al. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife. 2013;2:e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22(2):241–7. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang ED, Wang CY. MAVS Self-Association Mediates Antiviral Innate Immune Signaling. J Virol. 2009;83(8):3420–8. doi: 10.1128/JVI.02623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Hou F, et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–61. doi: 10.1016/j.cell.2011.06.041. This paper demonstrates that MAVS forms filaments upon activation of the RIG-I/MDA5 signaling pathway and that this filament formation is important for the downstream signal activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Peisley A, et al. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell. 2013;51(5):573–83. doi: 10.1016/j.molcel.2013.07.024. This work provides the evidence supporting filament-induced (i.e. proximity-induced) oligomerization of RIG-I 2CARD. [DOI] [PubMed] [Google Scholar]
- 18**.Peisley A, et al. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509(7498):110–114. doi: 10.1038/nature13140. This paper presents the crystal structure of the RIG-I 2CARD tetramer non-covalently bound by K63-Ubn. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. Elife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castanier C, et al. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010;11:133–8. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onoguchi K, et al. Virus-Infection or 5′ppp-RNA Activates Antiviral Signal through Redistribution of IPS-1 Mediated by MFN1. PLoS Pathog. 2010;6:e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141(2):315–30. doi: 10.1016/j.cell.2010.03.029. This paper reports the first reconstitution of the signaling event using cell extract and purified mitochondria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang X, et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36(6):959–73. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng W, et al. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–25. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Wu B, et al. Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5. Cell. 2013;152(1-2):276–89. doi: 10.1016/j.cell.2012.11.048. This study reports the first reconstitution of the signaling event entirely from the purified components of RIG-I/MDA5 2CARD and MAVS CARD. They also report the evidence supporting filament-induced (i.e. proximity-induced) oligomerization of MDA5 2CARD. [DOI] [PubMed] [Google Scholar]
- 26**.Wu B, et al. Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.06.010. This work reports the near atomic resolution structure of the MAVS CARD filament and the complex of RIG-I 2CARD and MAVS CARD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harhaj EM, Dixit VM. Regulation of NF-κB by deubiquitinases. Immunol Rev. 2012;246:107–24. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunol Rev. 2012;246:95–106. doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. doi: 10.1038/nature05732. This paper provided the first demonstration of the involvement of ubiquitin in RIG-I signaling pathway. They showed that RIG-I 2CARD is covalently modified with K63-Ubn by Trim25 and that this modification is important for its signaling activity. [DOI] [PubMed] [Google Scholar]
- 30*.Patel JR, et al. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14(9):780–7. doi: 10.1038/embor.2013.102. This paper showed the importance of RIG-I oligomerization through RNA binding and ATP hydrolsyis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlee M, et al. Recognition of 5′triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohlway A, et al. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013;14:772–9. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binder M, et al. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J Biol Chem. 2011;286(31):27278–87. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egelman EH. Ambiguities in Helical Reconstruction. eLIFE. 2014;3:e04969. doi: 10.7554/eLife.04969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egelman EH. Reconstruction of helical filaments and tubes. Methods Enzymol. 2010;482:167–83. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peisley A, et al. Cooperative Assembly and Dynamic Disassembly of MDA5 Filaments for Viral dsRNA Recognition. Proc Natl Acad Sci U S A. 2011;108(52):21010–5. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;7:1714–26. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H. Assembly of post-receptor signaling complexes for the tumor necrosis factor receptor superfamily. Adv Protein Chem. 2004;68:225–79. doi: 10.1016/S0065-3233(04)68007-7. [DOI] [PubMed] [Google Scholar]
- 39.Takamatsu S, et al. Functional Characterization of Domains of IPS-1 Using an Inducible Oligomerization System. PLos One. 2013;8:e53578. doi: 10.1371/journal.pone.0053578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kollman JM, et al. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–82. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–90. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu A, et al. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell. 2014;156:1194–206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai X, et al. Prion-like Polymerization Underlies Signal Transduction in Antiviral Immune Defense and Inflammasome Activation. Cell. 2014;156:1207–22. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]


