Abstract
With increasing application of targeted therapies, and the development of acquired resistance, much attention is being focused on developing in vitro and in vivo patient-specific tumor models for individualized therapeutic evaluation of cancers. Circulating tumor cells (CTCs), provide a source of non-invasively and sequentially sampled invasive cancer cells suitable for propagation in vitro. We review the advantages and challenges associated with ex vivo culture of tumor cells circulating in the blood of cancer patients.
The blood-borne spread of cancer, from its primary site to distal metastases underlies most cancer related mortality. The mechanisms responsible for the seeding into the bloodstream of tumor cells, either as single invasive cells or as clusters or microemboli, followed by their survival in the vascular space, and ultimately invasion and proliferation in distant tissues, are all the subject of intensive investigation. In humans, such studies have been limited by the difficulty in accessing different stages of the metastatic cascade, particularly the transient intravascular phase. The analysis of circulating tumor cells (CTCs) thus provides a window into blood borne metastasis, albeit one that has been complicated by the fact that these cells are very rare, estimated at one tumor cell admixed with a billion normal blood cells, and hence their isolation poses a significant technological challenge (1-3). Recent technological advances, particularly in the field of microfluidics, are poised to revolutionize these studies by providing more efficient isolation of CTCs, which are in better condition for either molecular or functional studies. Beyond their role in enabling studies of the process of metastasis, CTCs also provide a potential source for sampling tumor cells during the course of treatment. This so-called “blood biopsy” provides a way for non-invasive monitoring of cancer, a challenge that has become particularly important given the advent of new powerful targeted cancer therapies. These new therapies may induce dramatic initial tumor responses, but they also select for the emergence of resistant clones whose altered genetic or epigenetic features must be understood before “second line” therapies may be administered. In this more interventionist world of “real time” cancer monitoring, CTCs may soon play an important role in guiding clinical therapies. This review will focus on this application of CTCs, in light of the newly reported ability to culture CTCs ex vivo. Additional CTC monitoring platforms, including circulating tumor DNA (ctDNA) have been discussed elsewhere (2-4)
The FDA-approved CTC isolation platform, CellSearch, has laid much of the groundwork for early clinical trials using CTCs, although next generation technologies are poised to take the field further. CellSearch uses antibody against the epithelial cell surface molecule EpCAM to magnetically enrich for CTCs that have been fixed within a tube of whole blood. While reliable and robust, the technology has been challenged both by its relatively low sensitivity, and by the fact that CTCs are fixed and hence unavailable for functional analyses. In addition, epithelial to mesenchymal transition (EMT), which has been implicated in cancer cell invasion, raises the possibility that only a fraction of CTCs of interest are interrogated using this EpCAM-dependent isolation strategy. The addition of other cell surface antigens, representing lineage or oncogenic drivers, such as HER2 and EGFR and other tumor cell specific markers have been used to partially overcome this limitation (5-9). Replacing antibody-based selection with physical separation of cells based on size has also been tested. Such approaches have involved simple filters as well as more complex microfluidic approaches (2-4), but they are based on the assumption that CTCs are much larger than normal blood cells. While immortalized cancer cell lines commonly studied in the laboratory may be much larger than normal blood cells, the size distribution of primary patient-derived CTCs is considerable. In fact, across many cancers, the diameter of primary CTCs overlaps significantly with that of normal leukocytes (10, 11). For all these reasons, the most appealing strategy may be to avoid positive selection of tumor cells based on their often variable characteristics, and instead remove normal blood cells from the clinical specimens. The very large excess of normal hematopoietic cells poses a significant technological challenge to their depletion, but their size and cell surface epitopes are well established and invariant, thereby overcoming a major biological hurdle resulting from tumor heterogeneity.
Traditional CTC isolation technologies that have involved removal of normal blood components have relied on chemical lysis of red blood cell (RBCs) followed by depletion of CD45 positive leukocytes, using either fluorescence activated cell sorting (FACS) or commercially available kits for cell aggregation (12-16). These approaches are limited by extensive cell loss under relatively harsh sample processing conditions that decrease the number and viability of tumor cells circulating in the blood. Technologies requiring minimal sample handling and fewer steps to deplete RBCs and WBCs have been developed to circumvent these issues (17). Among these, the recently developed CTC-iChip is a microfluidic platform that includes an initial size-based lateral displacement of nucleated cells from platelets and RBCs as they flow through precisely spaced microscopic pillars. The nucleated cells, including a mixture of leukocytes tagged with magnetic bead-conjugated antibodies against CD45, CD66b and CD14 together with rare unlabeled CTCs, are then focused into a single file as they flow through a series of specially curved channels. The curvatures take advantage of the biophysical principle of inertial focusing to achieve the precise alignment of each cell in a single row within the microfluidic stream. In turn, this enables the highly effective deflection of magnetic bead-bearing leukocytes as they pass through an external magnetic field. Together, this sophisticated microfluidic technology minimizes cell loss and preserves the viability of CTCs, which are unfixed, free in solution, and have not been manipulated with antibodies or magnetic beads. Among the downstream analyses that are made possible by this gentle isolation platform is the ability to culture viable CTCs (11, 18, 19).
With the advent of targeted cancer therapies and the inevitable development of acquired drug resistance, there is an emergent need to personalize treatment through preclinical modeling. For example, initial treatment selection in BRAF-mutant melanoma or EGFR-mutant non-small cell lung cancer is readily made on the basis of tumor genotyping alone, since the vast majority of mutation-positive cancers respond as expected to suppression of these driving oncogenes. The acquisition of drug resistance, however, is less well understood and not as predictable, with an array of genetic and epigenetic alterations that may affect sensitivity to second line therapies. Major breakthroughs have resulted from serial biopsy studies in cases with acquired drug resistance (20) and the increasing use of CTC and ctDNA-based genotyping may provide a noninvasive strategy for “real time” monitoring of evolving tumor genotypes. In this regard, in addition to being noninvasive and hence readily repeated during a patient's course of treatment, blood-based genotyping has the theoretical advantage of representing multiple sites of disease and hence being more representative of tumor heterogeneity than a single tumor biopsy sampling a single accessible site of disease. Culturing viable tumor cells, either from tumor biopsies (21) or from CTCs (19) complements these genetic studies by correlating tumor cell drug sensitivity with genotypes.
While CTCs presumably include metastatic precursors capable of initiating distant lesions, most of the tumor cells isolated from the bloodstream even using gentle microfluidic conditions are not viable. The successful ex vivo culture of CTCs has thus presented a Holy Grail in the field, which would provide exceptional reagents to study cancer metastasis as well as perform individualized preclinical testing for drug susceptibility. In this context, immortalization of CTCs using viral oncogenes such as SV40 large T antigen, the E6/E7 papilloma proteins, or the human Telomerase reverse transcriptase (hTERT) to bypass cellular senescence (22), would compromise critical signaling pathways and can confound downstream analyses. Conditions that have recently been optimized for culture of epithelial cancer cells, including the use of Rho Kinase inhibitors and feeder layers (23) or organoid cultures (24) are promising, although these techniques have been optimized using large numbers of cells available from tumor biopsies, rather than very rare CTCs in a blood specimen.
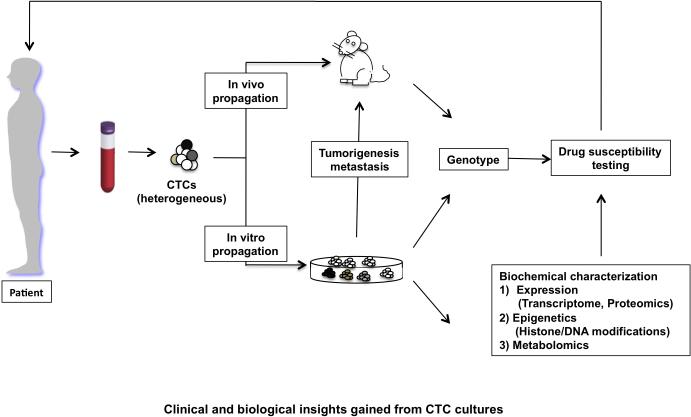
Initial studies in mouse models have shown that CTCs derived from xenografts generated from already immortalized cancer cell lines (mouse mammary cancer cells 4T1 and human lung cancer H460) can be cultured using standard RPMI1640 supplemented with fetal bovine serum (FBS) (25, 26), documenting the preserved viability of tumor cells recovered from the bloodstream. These culture conditions also appear to support short term proliferation of some CTCs from patients with mesothelioma, esophageal and bladder cancer, following isolation using the size-based MetaCell assay, as assessed by simple cytomorphology (27-29). Short-term cultures were established from CTCs isolated from 14 out of 19 lung cancer patients using coculture with cancer associated fibroblasts and extracellular matrix consisting of collagen and matrigel. These CTCs survived for 14 days in culture, expanding to a total of about 10-500 tumor cells, enabling some genotyping, RNA analysis, and cellular invasion assays (30). Direct inoculation of CTCs into immune compromised mice has also been explored as a possible method to propagate isolated CTCs. Buffy coat preparations from some prostate and colon cancer patients have been reported to form tumors in mice, although the resulting tumors were not subjected to molecular characterization (31). In a recent study, human breast CTCs isolated (using RosetteSep kit depletion of hematopoietic cells) from three out of 110 patients with very high CTC counts (>1,000 EpCAM-positive cells per 7.5 mls of blood), formed metastases in bone, liver and lung following direct injection into the mouse femur. The tumor cells were enriched for expression of EpCAM, CD44, CD47 and MET, but were not capable of proliferation in vitro (32). In another study, cultures of EpCAM-negative human breast CTCs isolated by HER2+/EGFR+/HPSE+/ Notch1+ multi-parameteric FACS analysis produced brain metastases following intravenous or intracardiac injection (33). Finally, in the highly metastatic small cell lung cancer (SCLC), CTCs isolated from patients with a high CTC burden (>400 CTCs per 7.5 mls of blood) formed tumors in mice, whose response to platinum and etoposide chemotherapy mirrored clinical observations (34). All together, these initial studies point to the promise of using immunosuppressed mice as CTC ‘incubators’, potentially generating preclinical models matched to individual patient tumors (Figure 1).
Figure 1.
outlines the approaches that can be used to propagate CTCs ex vivo. CTCs could be inoculated directly into mice or grown in vitro. The CTC cultures grown in vitro offer a more versatile system for genotyping and comprehensive biochemical and functional analysis of these tumor cells providing insight into putative therapeutic targets, which can be further investigated using drug sensitivity assays. Identification of drug sensitivity profiles can be informative in personalized clinical management of the patient.
Stable long-term in vitro CTC cultures provide an opportunity for high throughput preclinical testing of therapeutic regimens but until recently this has had limited success. One prostate CTC line was established from more than a hundred CTCs, following depletion of blood components using cell aggregation, followed by culture in vitro for > 9months using organoid conditions modified to support the growth of prostate tumor cells. (35). Organoid cultures involve use of a three dimensional matrix supplemented with growth factors that can sustain adult stem cells allowing self-organization into epithelia that appear to mimic the tissue of origin (24). The prostate-specific 3D-culture condition was able to sustain tumor cell growth from ~20% of prostate cancer biopsies, but it had lower efficiency at supporting the growth of prostate CTCs, which involve a much smaller tumor cell inoculum (35). The CTC-derived prostate organoid was tumorigenic in mice, with a high proportion of mutations (67%) corresponding to those present in lymph node metastases from the patient from which they were derived, and showing further enrichment for some mutant alleles.
A long term (>1 year) culture from a colon cancer patient with a high CTC burden (>300 CTCs per 7.5 mls of blood) was also recently established using depletion of normal blood components by cell aggregation. The cells were cultured under non-adherent conditions and were tumorigenic in mice, showing shared chromosomal aberrations as the primary tumor and enrichment for stem-like properties and an osteomimetic signature. (36)
In our own studies, we established non-adherent in vitro cultures from six of 35 (16.5%) women with metastatic luminal breast cancer using the CTC-iChip platform which achieves highly efficient microfluidic depletion of normal blood cells (19). These CTC cell lines have been maintained in vitro for over one year in serum free media supplemented with epidermal growth factor (EGF) and basic fibroblast growth factor (FGF) under non-adherent, hypoxic conditions. Using these conditions, we were also able to generate serial CTC cultures from individual patients monitored over multiple months of therapy. CTC cultures were most successfully achieved when patients exhibited clinical resistance to treatment regimens and were less successful in responsive patients. Three of five breast CTC cell lines tested formed tumors in mice with relatively short latency (within 2-3 months), following inoculation of only a few thousand cells, suggesting enrichment for tumor initiating cells. Most significantly, the in vitro cultures of CTC lines enabled the correlation of detailed genotyping with drug response testing.
Mutational analysis of 1,000 cancer genes revealed a host of non-synonymous sequence variants in the breast CTC cell lines (19). Many of these were missense variants of unknown significance in known “cancer genes”, hence comparison with matched normal DNA was essential to exclude germline polymorphisms. In addition, to exclude the possibility of in vitro derived mutations, wherever possible, we analyzed serial CTC lines from the same patient, confirming the mutation in two independently isolated specimens. The study was remarkable for the identification of clinically unsuspected mutations in genes such as PIK3CA, FGFR2 and the estrogen receptor ESR1, which were presumably acquired during the prolonged course of treatment with anti-hormonal and chemotherapy agents (37, 38). Of note, each of these “driving and drugable” mutations was present within a mutational context of 10-20 other somatic mutations; hence, the ability to test a drug within the precise mutational background of a patient's tumor is a powerful strategy to address inter-individual variability in known drug/gene sensitivity patterns. Indeed, in a CTC line harboring both PIK3CA and FGFR2 mutations (50% allele frequency for both mutations), treatment with PI3K or FGFR inhibitors resulted in cell killing, with the two drugs together demonstrating a synergistic effect. In a mouse xenograft model derived from this CTC line, treatment with either individual drug suppressed tumor growth, while simultaneous treatment with the two agents shrank the mouse tumor (19). Similarly, we identified mutations in ESR1 conferring ligand independent activation of the estrogen receptor and potential cooperative drug effects between estrogen modulators and HSP90 inhibitors. The heterogeneity of the CTC lines was apparent in the variable allele frequency of ESR1 mutations among different patients, consistent with the emergence of hormone-resistant clones and their representation within the oligoclonal CTC cultures. While none of the few patients whose CTCs were studied in this way were able to benefit from such preclinical drug sensitivity testing, this proof-of-concept study illustrates potential future applications in personalized clinical management of patients with cancer.
The next critical step to achieve clinical applicability of CTC cultures involves further optimization of conditions to robustly support in vitro outgrowth of limited numbers of CTCs from patients with breast cancer as well as other solid tumors, including improving blood collection/stabilization protocols, as well as testing various culture conditions. Recently, culture conditions, including the use of a fibroblast feeder layer and treatment with the Rho kinase inhibitor Y-27632, have been shown to effectively support the growth of normal and tumor cells of epithelial origin (23). These growth conditions were recently used to establish cell cultures from biopsies obtained from lung cancer patients being treated with EGFR or ALK tyrosine kinase inhibitors, and genotyping cultured tumor cells enabled the identification of various drug combinations that suppressed their growth (21). As noted above, three dimensional matrix supplemented with growth factors can sustain adult stem cells allowing self-organization into epithelia representing the tissue of origin, and these have enabled the culture of at least one prostate cancer CTC line (24) (35). However, both by virtue of their small numbers, as well as potential biological differences, robust and reliable culture conditions for CTCs remain to be established. Once these are in place, they could ultimately set the stage for clinical trials based on individual preclinical predictions. Past attempts at predicting sensitivity to chemotherapy regimens, the so-called “Salmon Assays” (39) were unsuccessful in the clinical setting for a number of reasons: the very small subset of cancer cells that could be cultured in the 1980's were often not representative of the larger heterogeneous tumor mass, and before the advent of genotype-directed targeted therapies, the diversity of potentially useful chemotherapeutic agents was limited. Future studies will therefore be needed to document whether high efficiency culture of CTCs, combined with genotype-directed targeted therapeutics, will prove predictive of clinical response in this new age of cancer treatment. Critical to testing this model will be novel clinical research designs that enable controlled trials of investigational agents based on individualized preclinical testing (ie. “N of one” studies). If successful, this strategy may open the door to routine preclinical personalized drug sensitivity testing in cancer.
Acknowledgements
The authors apologize that space constraints prevent them from citing all relevant publications. This work was supported by grants from the Breast Cancer Research Foundation (D.A.H), Stand Up to Cancer (D.A.H., M.T., S.M), the Wellcome Trust (D.A.H), National Foundation for Cancer Research (D.A.H), NIH CA129933 (D.A.H), NIBIB EB008047 (M.T., D.A.H), Susan G. Komen for the Cure KG09042 (S.M), NCI-MGH Proton Federal Share Program (S.M), and the Howard Hughes Medical Institute (D.A.H).
References
- 1.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–22. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med. 2014 doi: 10.15252/emmm.201303698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–82. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4:650–61. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Albuquerque A, Kubisch I, Breier G, Stamminger G, Fersis N, Eichler A, et al. Multimarker gene analysis of circulating tumor cells in pancreatic cancer patients: a feasibility study. Oncology. 2012;82:3–10. doi: 10.1159/000335479. [DOI] [PubMed] [Google Scholar]
- 6.Fierer JO, Veggiani G, Howarth M. SpyLigase peptide-peptide ligation polymerizes affibodies to enhance magnetic cancer cell capture. Proc Natl Acad Sci U S A. 2014;111:E1176–81. doi: 10.1073/pnas.1315776111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pecot CV, Bischoff FZ, Mayer JA, Wong KL, Pham T, Bottsford-Miller J, et al. A novel platform for detection of CK+ and CK- CTCs. Cancer Discov. 2011;1:580–6. doi: 10.1158/2159-8290.CD-11-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan Y, Tan J, Asghar W, Kim YT, Liu Y, Iqbal SM. Velocity effect on aptamer- based circulating tumor cell isolation in microfluidic devices. J Phys Chem B. 2011;115:13891–6. doi: 10.1021/jp205511m. [DOI] [PubMed] [Google Scholar]
- 9.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coumans FA, van Dalum G, Beck M, Terstappen LW. Filter characteristics influencing circulating tumor cell enrichment from whole blood. PLoS One. 2013;8:e61770. doi: 10.1371/journal.pone.0061770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balasubramanian P, Lang JC, Jatana KR, Miller B, Ozer E, Old M, et al. Multiparameter analysis, including EMT markers, on negatively enriched blood samples from patients with squamous cell carcinoma of the head and neck. PLoS One. 2012;7:e42048. doi: 10.1371/journal.pone.0042048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Fusi A, Schmittel A, Tinhofer I, Schneider A, Keilholz U. Eradication of EGFR-positive circulating tumor cells and objective tumor response with lapatinib and capecitabine. Cancer Biol Ther. 2010;10:860–4. doi: 10.4161/cbt.10.9.13323. [DOI] [PubMed] [Google Scholar]
- 14.Lustberg M, Jatana KR, Zborowski M, Chalmers JJ. Emerging technologies for CTC detection based on depletion of normal cells. Recent Results Cancer Res. 2012;195:97–110. doi: 10.1007/978-3-642-28160-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sergeant G, van Eijsden R, Roskams T, Van Duppen V, Topal B. Pancreatic cancer circulating tumour cells express a cell motility gene signature that predicts survival after surgery. BMC Cancer. 2012;12:527. doi: 10.1186/1471-2407-12-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A, et al. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol Bioeng. 2009;102:521–34. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sajay BN, Chang CP, Ahmad H, Chung WC, Puiu PD, Rahman AR. Towards an optimal and unbiased approach for tumor cell isolation. Biomed Microdevices. 2013;15:699–709. doi: 10.1007/s10544-013-9757-9. [DOI] [PubMed] [Google Scholar]
- 18.Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–18. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–20. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014 doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipps C, May T, Hauser H, Wirth D. Eternity and functionality - rational access to physiologically relevant cell lines. Biol Chem. 2013;394:1637–48. doi: 10.1515/hsz-2013-0158. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Kang JH, Krause S, Tobin H, Mammoto A, Kanapathipillai M, Ingber DE. A combined micromagnetic-microfluidic device for rapid capture and culture of rare circulating tumor cells. Lab Chip. 2012;12:2175–81. doi: 10.1039/c2lc40072c. [DOI] [PubMed] [Google Scholar]
- 26.Kolostova K, Zhang Y, Hoffman RM, Bobek V. In vitro culture and characterization of human lung cancer circulating tumor cells isolated by size exclusion from an orthotopic nude-mouse model expressing fluorescent protein. J Fluoresc. 2014;24:1531–6. doi: 10.1007/s10895-014-1439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bobek V, Kacprzak G, Rzechonek A, Kolostova K. Detection and cultivation of circulating tumor cells in malignant pleural mesothelioma. Anticancer Res. 2014;34:2565–9. [PubMed] [Google Scholar]
- 28.Bobek V, Matkowski R, Gurlich R, Grabowski K, Szelachowska J, Lischke R, et al. Cultivation of circulating tumor cells in esophageal cancer. Folia Histochem Cytobiol. 2014;52:171–7. doi: 10.5603/FHC.2014.0020. [DOI] [PubMed] [Google Scholar]
- 29.Cegan M, Kolostova K, Matkowski R, Broul M, Schraml J, Fiutowski M, et al. In vitro culturing of viable circulating tumor cells of urinary bladder cancer. Int J Clin Exp Pathol. 2014;7:7164–71. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Shiratsuchi H, Lin J, Chen G, Reddy RM, Azizi E, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget. 2014;5:12383–97. doi: 10.18632/oncotarget.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pretlow TG, Schwartz S, Giaconia JM, Wright AL, Grimm HA, Edgehouse NL, et al. Prostate cancer and other xenografts from cells in peripheral blood of patients. Cancer Res. 2000;60:4033–6. [PubMed] [Google Scholar]
- 32.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539–44. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 35.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–87. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cayrefourcq L, Mazard T, Joosse S, Solassol J, Ramos J, Assenat E, et al. Establishment and Characterization of a Cell Line from Human Circulating Colon Cancer Cells. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-2613. [DOI] [PubMed] [Google Scholar]
- 37.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–45. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alberts DS, Chen HS, Salmon SE. In vitro drug assay: pharmacologic considerations. Prog Clin Biol Res. 1980;48:197–207. [PubMed] [Google Scholar]