Abstract
Targeted cancer therapies offer great clinical promise, but treatment resistance is common, and basic research aimed at overcoming this challenge is limited by reduced genomic and biological complexity in artificially induced rodent tumors compared to their human counterparts. Animal models that more faithfully recapitulate genotype-specific human pathology could improve the predictive value of these investigations. Here, a newly identified animal model for oncogenic BRAF-driven cancers is described. With 20,000 new cases in the United States each year, canine invasive transitional cell carcinoma of the bladder (InvTCC) is a common, naturally occurring malignancy that shares significant histological, biological, and clinical phenotypes with human muscle invasive bladder cancer. In order to identify somatic drivers of canine InvTCC, the complete transcriptome for multiple tumors was determined by RNAseq. All tumors harbored a somatic mutation that is homologous to the human BRAF(V600E) mutation, and an identical mutation was present in 87% of 62 additional canine InvTCC tumors. The mutation was also detectable in the urine sediments of all dogs tested with mutation-positive tumors. Functional experiments suggest that, like human tumors, canine activating BRAF mutations potently stimulate the mitogen activated protein kinase (MAPK) pathway. Cell lines with the mutation have elevated levels of phosphorylated MEK, compared to a line with wild type BRAF. This effect can be diminished through application of the BRAF(V600E) inhibitor vemurafenib. These findings set the stage for canine InvTCC as a powerful system to evaluate BRAF-targeted therapies, as well as therapies designed to overcome resistance, which could enhance treatment of both human and canine cancers
Keywords: Bladder Cancer, BRAF Mutation, Comparative Oncology, Canine Cancer
Introduction
Cancer continues to be a major cause of death worldwide, even in the setting of the most advanced care (1). This is true not only in human populations but also in the domestic dog, where approximately 25% of the population will develop cancer in their lifetime and, like humans, the incidence increases rapidly with age (2,3). Many naturally occurring cancers in domestic dogs closely mimic their human counterparts in both histopathological features and biological behavior, including distant metastasis, response to therapy, and extensive intratumor heterogeneity (4–6). Breed-specific risk paired with the simplified canine genetic architecture has enabled identification of susceptibility loci for several malignancies including renal cell carcinoma, histiocytic sarcoma, osteosarcoma, and squamous cell carcinoma of the digit (7–10). Furthermore, the existing veterinary medical infrastructure and the availability of a canine genome reference sequence offer tremendous opportunities to integrate naturally occurring canine cancer into mainstream oncology research (11).
Naturally-occurring, high-grade, invasive transitional cell carcinoma (InvTCC), also referred to as invasive urothelial carcinoma, has received considerable attention in comparative oncology research because of its similarity to human invasive urothelial carcinoma in both appearance and behavior (6,12). Each year, this cancer kills tens of thousands of people and dogs worldwide (6,13). Although organ-confined bladder cancer can be successfully treated, distant metastases occur in 50% of cases, and the metastatic disease is almost uniformly fatal in both species (6,13).
Recent advances in genomics, technology, and expertise are leading to a much more comprehensive understanding of cancer at the molecular level (14). However, molecularly-targeted therapies have not yet approached their full potential as anticancer agents. The development of resistance to targeted drugs often occurs quickly, due in large part to the tremendous complexity and heterogeneity within a given patient’s tumor (15,16). The presence of multiple signaling pathways capable of producing similar effects within cancer cells, and cross talk between pathways generate numerous opportunities for the dysregulation of signaling routes that bypass the drug target (15,16).
Clearly, maximizing the success of molecular-based treatments will require relevant in vivo systems in which common molecular perturbations are found in natural, heterogeneous tumors that mimic what is observed in human cancers. The domestic dog can provide such a system. Due to the similarities in histology, biological responses and presentation, canine cancers have already provided important clinical information to improve detection and treatment of human cancers(17). Canine lymphomas and bladder carcinomas have been used to determine dosage levels and test new chemotherapeutic agents while osteosarcoma in dogs proved invaluable for improving treatment strategies that preserve patient limbs(18). Studies of tumor karyotypes show that cancers in the dog undergo many of the same rearrangements that typify human tumors, suggesting that both diseases develop in the same manner (19). Indeed, the first cancer to be mapped in the dog, RCND, was later found to be the exact same gene that causes the human cancer syndrome Birt-Hogg-Dube(20). The system has also proven useful for understanding how non-coding mutations can cause cancers, as in the case of squamous cell carcinoma of the digit (21) making it particularly tantalizing for understanding tissue specificity of mutations.
Through complete transcriptome sequencing of InvTCC tumors, we have identified a mutation in the canine BRAF gene that is identical to the BRAF(V600E) mutation reported with high frequency in several human cancers. Further genotyping confirmed the mutation was present in nearly 85% of canine InvTCC tumors. The ubiquitous nature of the BRAF(V600E) mutation has made it a common target of molecular cancer therapies.
Although BRAF mutations are rare in human bladder cancers, they are very common in other tumor types. The finding of homologous BRAF(V600E) mutations in the vast majority of canine InvTCC tumors has tremendous implications for comparative cancer research and sets the stage for the domestic dog to provide a highly relevant system in which to further develop BRAF-targeted therapy strategies, as well as advance our understanding of molecular events leading to BRAF mutation-associated cancer in both humans and dogs.
Materials and Methods
Sample Collection
Sample collection was performed following approval of the Animal Care and Use Committees of the collecting institutions, and owners of all participating dogs signed an informed consent document. Tumor samples were obtained through surgical biopsy or cystoscopy as directed by evaluation and treatment protocols decided upon through owner/oncologist consultations, and diagnosis of InvTCC made by histopathology. Tissues were snap frozen and stored at −80°C until extraction. Whole blood samples were collected in 3–6 ml EDTA or ACD tubes and stored at 4°C prior to extraction. Genomic DNA was isolated from blood using a standard phenol-chloroform protocol. DNA and RNA were extracted from tumor tissue samples using the AllPrep kit (Qiagen Corp., Alameda, CA). RNA for RNAseq was extracted from flash frozen tumor tissue using the AllPrep kit (Qiagen Corp. Alameda, CA). A small number of samples were extracted from FFPE blocks or slides using the RecoverAll kit (Ambion, Grand Island, NY) with pretreatment in 100% Xylene. Free catch urine samples were collected from 16 dogs that were diagnosed with InvTCC of the bladder and from three control dogs. Sediment was obtained by centrifugation in 15 mL conical tubes at 3300 rpm for 30 min. Sediment was washed with cold PBS twice before extracting DNA using the Gentra Puregene kit following the bodily fluids protocol or the Qiamp DNA Micro kit (Qiagen Alameda, CA). All samples were stripped of identifiers, numerically coded, and aliquoted for long-term storage at −80°C.
RNAseq
RNA samples were chosen from four InvTCC confirmed bladder tumors and two normal bladder tissue samples. RNA was quantified using a Qubit spectrophotometer (Qiagen, Alameda, CA), and quality was assessed as RNA integrity number seven or greater by Bioanalyzer (Agilent, Santa Clara, CA). Sequencing was performed on the Illumina HiSeq 2000 (San Diego, CA) using paired-end library preparation. Libraries were barcoded, pooled, and run on two sequencing lanes producing 106.4–131.9 million reads per sample. Sequence reads were aligned to the canfam3.1 reference using Bowtie and the Ensembl v72 canine gene model (22). Single nucleotide genotypes were called using Samtools mpileup and Unified Genotyper from the Genome Analysis Tool Kit (23,24). Only variants called in both programs were included in further analyses. Common germline polymorphisms and platform-specific errors were filtered out by comparison to whole genome sequences from 19 pure-bred dogs, three mixed breed dogs, and four wolves, in addition to the transcriptome sequences from the two normal bladder tissues.
Sanger sequencing and RFLP genotyping
The BRAF mutation was initially confirmed using Sanger sequencing following PCR amplification from genomic DNA on 59 tumors, 7 normal bladder tissues, and 96 germline DNA samples. Because the mutant allele is not found in all cells of the tumor and is often difficult to see in a peak-trace chromatogram, genotyping of all samples that were initially called wild-type were repeated using an RFLP method. An 830 bp region was first amplified using standard PCR protocols and primers F- AATAAATGGGTTTGCATGAGAG and R- TGGCCTCAATTCTTACCATCCAC using 1.8% GC Melt reagent (Clontech Laboratories, Inc., Mountain View, CA). The segment was designed to contain two cut sites for the BtsIMutI restriction enzyme (New England Biolabs, Ipswich, MA), one that cuts all canine DNA samples, and one that cuts only wild-type BRAF sequences at the mutation site. Wild-type sequences produce three bands of sizes 564/190/76 whereas the mutant sequences produce two bands of sizes 640/190. DNA extracted from 16 urine samples was also genotyped using the RFLP method.
Amplicon enrichment and next generation sequence genotyping
A 160bp segment surrounding the mutation was amplified via PCR in six urine samples, 12 tumor samples, one positive tumor and one negative germline control. In order to test sensitivity, four samples were designed with ratios of mutant allele to wild-type allele of 1:1, 1:10, 1:100, and 1:1000. Sample specific barcodes and stagger sequences were added to the sequence specific primers F- CATGAAGACCTCACAGTAAA and R- GCACCTCAGGGTCCAA along with the Illumina (San Diego, CA) sequencing adapter. All amplicons were combined into one library, products were cleaned using the AMPure XP bead kit (Beckman-Coulter, Indianapolis, IN) and sequenced on an Illumina MiSeq instrument according to the manufacturer’s protocol. Reads were aligned to the CanFam 3.1 reference sequence with BWA (23), and allelic depth was assessed using the Integrative Genome Viewer (http://www.broadinstitute.org/software/igv/). At the position of interest, the reference allele is a T and the mutant base is an A. Since the systematic sequencing error rate is substitution-specific (for example, the T>A error rate differs from the T>C error rate), we determined the background T>A substitution rate per 10,000 reads at each of 33 T positions in the amplicon, and then normalized these values against that sample’s mean T>A substitution rate. Samples were determined to be mutation positive when the normalized substitution rate at the position of interest was a statistical outlier compared to the substitution rate at all other positions via Grubb’s test (critical Z-score of 3.29 for 33 positions and P >0.01).
Both tumor and urine samples were obtained from ten dogs. To verify the source of the samples before comparing mutation status, a set of four microsatellite markers were genotyped on each tissue and urine sample and matching germline DNA when available. Microsatellites markers were chosen based on the frequency of heterozygotes in a previously published multi-breed set. The markers, C09.474, FH3072, REN112G10, and REN293N22 were amplified in the presence of a dye-labeled third primer and fragment sizes separated by capillary electrophoresis as described previously (25). Genotypes were called using Genemapper 4.0 (Life Technologies, Pleasanton, CA). Sample sets were retained if alleles matched at all markers. The genotypes from one triplet were inconclusive, and it was therefore removed from further analysis.
Western blots
Four canine InvTCC cell lines (26) were treated with vemurafenib (1 μM) or vehicle control for 2 and 24 hours. Drug concentration and treatment times were selected from unpublished pilot data and from published studies of vemurafenib. (27–29) Total protein (50 μg) from lysates of treated canine InvTCC cells was separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane overnight. Membranes were blocked with 5% BSA in Tris-buffered saline with tween-20 (TBS-T) for 1 hour and incubated with antibodies for MEK (Cell Signaling, Boston, MA) and phosphorylated MEK (Cell Signaling, Boston, MA) proteins diluted in 5% BSA in TBS-T overnight with shaking at 4°C. Membranes were subsequently incubated with secondary antibody (goat anti-mouse conjugated with horseradish peroxidase) at 1:10,000 dilution for 1 hour. Protein was detected on UVP ChemiDoc-It®TS2 Imager (UVP LLC, Upland, CA) using chemiluminescence (Super Signal, Pierce Biotechnology, Rockford, IL). All cell lines were developed and authenticated as described in (26).
Cell proliferation assays
The effect of the BRAF(V600E) inhibitor vemurafenib (30) was determined in the four canine InvTCC cell lines described above using a sulforhodamine B assay, as described by Skehan et al. (31). Briefly, canine InvTCC cells (1,000–1,500 cells/well in 96 well plates) were cultured in DMEM/F12 media with 10% FBS at 37°C for 24 hours (27–29). Vemurafenib (0.001–10μM) or vehicle control was added to the wells, and the plates incubated for 72 hours. Cells were fixed and stained with sulforhodamine B solution followed by washing. The bound stain was solubilized with Tris buffer, and the optical density measured at 490 nm. The optical densities of wells for each set of conditions (different drug concentrations or vehicle control) were averaged, and the percent growth of control was calculated as described (31).
Results
We sequenced the complete transcriptomes of four histologically confirmed canine InvTCC tumors with an average of 109.7–137.2 X coverage of the Ensembl annotated exons. The four cases represented two Scottish terriers, one West Highland white terrier, and one Shetland sheepdog. A total of 45,061 variant positions were identified, with 23,625–26,190 raw variants per tumor. Since matched normal genetic material was not available, germline polymorphisms and systematic sequencing errors were removed by filtering against 24.3 million variants uncovered in whole genome sequencing of 26 canids or concurrently sequenced normal bladder samples. We then filtered genotypes with quality <13 or positions with more than two tumors uncalled, leaving 538–912 variants per sample (Table 1). Somatic variants were annotated with the Variant Effect Predictor (32) and protein-changing mutations were analyzed (Supplemental Table 1). Mutations were found in 15 genes that have been associated with tumor development in humans (Table 2).
Table 1.
Variant filtration and annotation in the four InvTCC tumors. Filtering polymorphic positions and low-quality variants narrowed candidate somatic mutations, and functional annotation highlighted variants for further analysis.
| a SCOT1 | SCOT2 | WHWT | SHLT | |
|---|---|---|---|---|
| Raw Variants | 26,190 | 25,009 | 25,968 | 23,625 |
| Polymorphism Filter | 1,105 | 750 | 989 | 874 |
| Quality Filter | 912 | 538 | 759 | 704 |
| Synonymous | 233 | 136 | 211 | 204 |
| Missense | 338 | 144 | 244 | 205 |
| Stop Gained | 5 | 5 | 2 | 3 |
SCOT1 and SCOT2 = Scottish Terrier, WHWT = West Highland White Terrier, SHLT = Shetland Sheepdog.
Table 2.
Predicted deleterious somatic mutations identified in cancer associated genes.
| Gene Symbol | Cancer Associationa | Consequence | SIFT score | Tumors Affected |
|---|---|---|---|---|
| BRAF | KCM | missense | 0 | 4 |
| MLLT4 | C | missense | 0.02 | 1 |
| KIF5B | C | missense | 0.04 | 1 |
| ATM | C | missense | 0.01 | 1 |
| CREBB | KC | missense | 0 | 1 |
| NUP214 | C | missense | 0 | 1 |
| CDKN2B | K | missense | 0.01 | 1 |
| EGFR | KCM | missense | 0 | 1 |
| P2RY14 | M | missense | 0.01 | 1 |
| BRCA2 | KC | missense | 0 | 1 |
| NFKB2 | KC | missense | 0.04 | 1 |
| PIK3CA | KCM | stop | NA | 1 |
| BCOR | C | missense | 0.01 | 1 |
| MED12 | C | missense | 0 | 1 |
| ELF4 | C | stop | NA | 1 |
K=KEGG cancer pathway, C=Cosmic cancer gene census, M=MAPK pathway
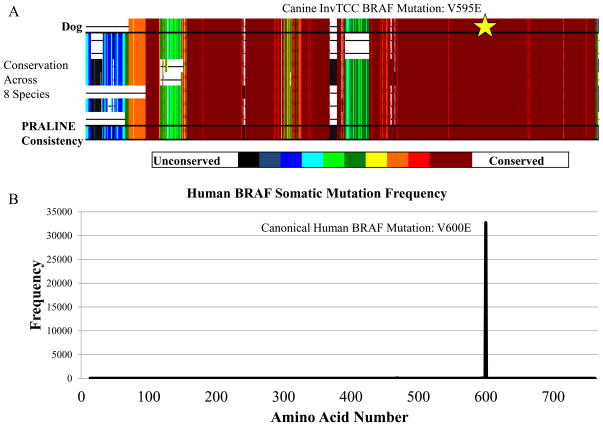
All four tumors were found to harbor a non-synonymous, single nucleotide variant at genomic position 8296284 on Canis familiaris chromosome 16 (CFA16), that results in a valine to glutamic acid substitution at codon 595 of canine BRAF (Figure 1a). This mutation, BRAF(V595E), is homologous by multiple sequence alignment to the oncogenic human BRAF(V600E) mutation (33) (Figure 1b). Sanger sequencing of each tumor confirmed the mutation. We genotyped 62 additional histologically-confirmed InvTCC tumors using Sanger sequencing and restriction fragment length polymorphism (RFLP) assays. The mutation was found in 49 of these additional samples, for a total of 80.3% (53 of 66) (Supplemental Table 2).
Figure 1.
Position of BRAF mutation and conservation across species. A) Multiple sequence alignment of the BRAF protein sequences from eight species including dog, human, chimpanzee, mouse, rat, cat, zebrafish, and chicken. Colors indicate level of conservation with black being no conservation and maroon being a perfect match. Alignment and conservation scoring were performed with the PRALINE multiple sequence alignment tool (http://www.ibi.vu.nl/programs/pralinewww/). The star highlights the canonical mutation. B) Frequency of somatic BRAF mutations by protein position among human tumors in the COSMIC database (34).
To confirm that the mutation was somatic rather than inherited, we genotyped DNA isolated from peripheral blood samples taken from 96 dogs with InvTCC, including 42 with tumors positive for the BRAF(V595E) mutation. Breeds included in the analysis are shown in Table 3. We did not observe the mutation in any germline sample, nor did we find the mutation in a panel of nine healthy bladder tissues. In addition, we genotyped two intronic SNPs within 300 BPs of the mutation (CFA16: 8296009 and 8296465), which revealed almost complete concordance between the tumor and normal DNA. The lone discrepancy between the tumor and normal DNA was in a case that was heterozygous for the intronic polymorphisms in the germline, but homozygous for those alleles as well as BRAF(V595E) in the tumor. It is likely that this tumor has a deletion of the wild-type locus.
Table 3.
Breed distribution of InvTCC and the BRAF(V595E) mutation in tumors and peripheral blood cell DNA.
| Tumor DNA | Germline DNA | |||
|---|---|---|---|---|
|
|
||||
| Breed | BRAF(V595E) | BRAF(wt) | BRAF(V595E) | BRAF(wt) |
| Mixed breed | 13 | 4 | 0 | 23 |
| Scottish terrier | 11 | 1 | 0 | 18 |
| Shetland sheepdog | 7 | 0 | 0 | 13 |
| Beagle | 10 | 0 | 0 | 10 |
| West Highland white terrier | 2 | 1 | 0 | 5 |
| German shepherd dog | 2 | 0 | 0 | 3 |
| Jack Russell terrier | 3 | 0 | 0 | 1 |
| Miniature pinscher | 2 | 0 | 0 | 2 |
| Other breedsa | 8 | 2 | 0 | 21 |
Breeds with 3 or fewer representatives: Dachshund, Fox terrier, Border collie, Australian shepherd, German Shorthaired pointer, Shih Tzu, Standard Schnauzer, Miniature Schnauzer, Newfoundland, Brittany, Cairn terrier, Chesapeake Bay retriever, Greyhound, Petit Basset Griffon Vendeen, Staffordshire Bull terrier, Yorkshire terrier, Labrador retriever
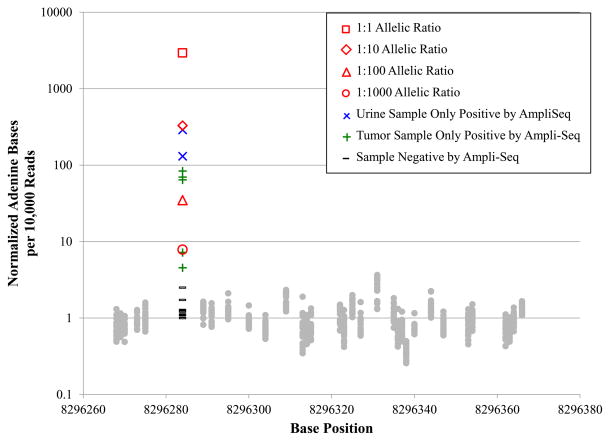
To determine whether the mutant allele was detectable in DNA shed through urine, we used the RFLP method to genotype DNA urine sediment of three control dogs and 16 dogs affected with InvTCC. The BRAF(V595E) mutation was conclusively detected in eight of 16 affected dogs (50%), seven were negative and one was inconclusive. None of the three healthy control dogs were positive for the mutation. Matching tumor tissue was genotyped from nine of the affected dogs and the urine genotypes mirrored the tumor genotypes in eight of nine cases. To improve the sensitivity of the urine mutation detection, we used targeted amplicon enrichment and ultra-high depth sequencing (mean coverage = 1,268,644X) to probe for the mutation. Using this method we were able to detect 100% (9 of 9) of the tumor genotypes in the matched urine samples. We created serial dilutions of BRAF(V595E) negative and positive DNA samples, we found that a sample with a predicted 1:1000 mutant to wild-type ratio displayed a statistically significant 7.8-fold enrichment (Grubb’s test, Z-score=5.39, P<0.01) for the variant allele compared to background T>A mutation rates (Figure 2). Two additional samples with 4.5 and 7.2-fold enrichment of the mutant allele were also significant (Grubb’s test Z-score=4.86 and 4.99, respectively, P<.01) (Table 4), thus, the limit of detection by this method is somewhat less than one mutant read per 1,000. Furthermore, using the targeted method to repeat the genotyping of all tumors that were negative for the BRAF mutation using Sanger sequencing and RFLP assays, we detected five additional tumors that were positive for the mutation (Figure 2) bringing the total to 58 out of 66 or 87.9% of all tumors. Comparing these reads to the dilutions suggests a sensitivity limit for the RFLP assay in tumors of nearly 100 mutant alleles per 1000 wild-type alleles, whereas the sequencing-based approach identified the mutation at fewer than one mutant base per 1000 wild-type alleles.
Figure 2.
Detection of variant alleles in ultra-high depth amplicon sequencing (mean coverage = 1,268,644X). Values are reported as mutant alleles per 10,000 reads normalized to the sample-specific background T>A substitution rate (mean background rate = 2.11 Adenine bases/10,000 reads). The normalized substitution rates for serially diluted samples are shown in red. The mutation was detected in the most dilute sample, so the lower limit of detection for this assay is below 1 mutant base to 1000 reference alleles. Five tumor samples and two urine samples, depicted in blue and green, respectively, were negative for the mutation by other genotyping approaches, but positive in the amplicon sequencing experiment. Samples shown in gray were negative for the mutation by all genotyping methods.
Table 4.
Genotyping results from BRAF targeted amplicon sequencing.
| Samplea | Total Reads | #Alt Bases | %Alt Bases | Alt Basesb per 10,000 reads | Z-scorec |
|---|---|---|---|---|---|
| 1:1 Ratio | 1200288 | 551584 | 47.94% | 2936.4 | 5.57 |
| 1:10 Ratio | 1274184 | 105318 | 8.63% | 329.7 | 5.57 |
| 1:100 Ratio | 1204844 | 10472 | 0.91% | 34.9 | 5.56 |
| 1:1000 Ratio | 1266088 | 1793 | 0.15% | 7.8 | 5.39 |
| AUSS_877 - T | 1620812 | 336 | 0.02% | 1.7 | 1.31 |
| AUSS_877 - U | 1266688 | 286 | 0.02% | 1.1 | 0.22 |
| BEAG_314 - U | 1353652 | 31119 | 2.37% | 130.8 | 5.57 |
| BEAG_927 - T | 1159434 | 1293 | 0.13% | 7.2 | 4.99 |
| MINP_700 - T | 1206636 | 20495 | 1.83% | 83.6 | 5.57 |
| MIX_022 - T | 1206484 | 252 | 0.02% | 1.0 | 0.02 |
| MIX_022 - U | 1444658 | 336 | 0.02% | 1.0 | 0.01 |
| MIX_038 - T | 1302402 | 402 | 0.03% | 1.3 | 0.51 |
| MIX_366 - T | 1207812 | 318 | 0.03% | 1.2 | 0.32 |
| MIX_366 - U | 1373150 | 294 | 0.02% | 1.1 | 0.14 |
| MIX_707 - U | 1163154 | 37984 | 7.83% | 289.9 | 5.57 |
| MSNZ_415 - T | 1205018 | 14554 | 1.25% | 69.8 | 5.57 |
| SCOT_055 - T | 1255306 | 13400 | 1.10% | 64.3 | 5.57 |
| SCOT_353 - T | 1180946 | 826 | 0.08% | 4.5 | 4.86 |
| SCOT_408 - T | 1372440 | 654 | 0.05% | 2.5 | 2.58 |
| WEST_309 - T | 1200474 | 338 | 0.03% | 1.7 | 1.31 |
| WEST_309 - U | 1177048 | 278 | 0.02% | 1.3 | 0.41 |
|
| |||||
| Mean | 1268644 | ||||
Bolded samples are statistically significant outliers and thus positive for the mutation.
T = Tumor DNA, U= Urine Sediment DNA.
Normalized to the background T>A substitution rate at 32 other sequenced positions.
Statistical outliers via Grubb’s Test have Z-score >3.29 of 33 datapoints, P<0.01)
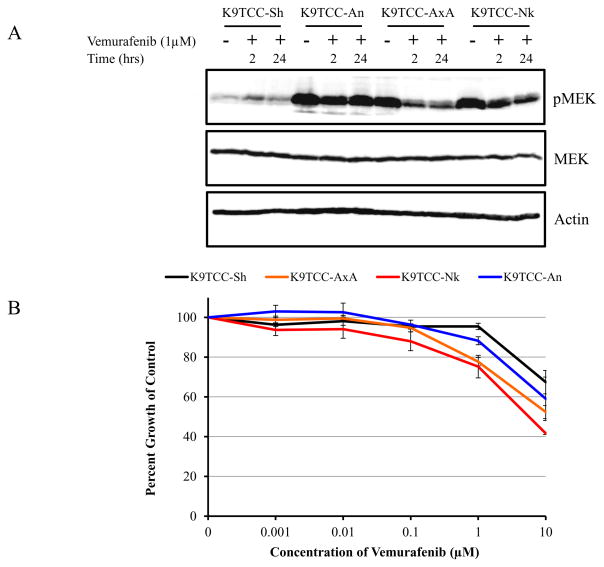
To assess whether the canine BRAF(V595E) mutation recapitulates the downstream MAPK pathway dysregulation mechanism of its human homolog, we assayed the levels of MAPK/ERK kinase (MEK) and phosphorylated MEK (pMEK) with Western blots of lysates from cells treated with either the BRAF(V600E) specific inhibitor, vemurafenib (1 μMol) or a vehicle control. At baseline, three canine InvTCC cell lines that express BRAF(V595E) showed high levels of pMEK. Two of the three lines displayed a clear reduction in pMEK at two and 24 hours of vemurafenib exposure, while levels of MEK remained constant. One BRAF mutation positive cell line, K9TCC-An, showed negligible response to vemurafenib treatment and continued to show very high levels of pMEK. In contrast, an InvTCC cell line that does not carry the mutation (K9TCC-Sh), displayed a low baseline level of pMEK and also showed no change in response to the inhibitor (Figure 3).
Figure 3.
MEK activation and vemurafenib response in cell lines with and without the BRAF(V595E) mutation. A) Western blotting was used to detect pMEK and total MEK protein in canine InvTCC cells lines treated with vehicle control (lanes 1, 4, 7, and 10) or vemurafenib (1μMol) for 2 hours (lanes 2, 5, 8, and 11) and 24 hours (lanes 3, 6, 9, and 12). Four cell lines were tested, three with the BRAF(V595E) mutation (K9TCC-An, AxA, and Nk) and one without (K9TCC-Sh). Actin was used as a loading control. Data shown are representative of three independent experiments. B) Four proliferating canine InvTCC cell lines were treated for 72 hours with vemurafenib at concentrations of 0–10 μMol. Data are expressed as the percent growth of vehicle control treated cells. The K9TCC-PU-Sh cells express only the wild-type BRAF, and the other three cell lines express BRAF(V595E) mutation. Proliferation of cell lines with the BRAF(V595E) mutation was significantly inhibited by vemurafenib compared to the wild-type cell-line at 1μM (P=0.00043, 0.0089, and 0.019 for K9TCC-AxA, K9TCC-An and K9TCC-Nk, respectively).
Since vemurafenib depressed MEK activation, we tested whether the drug inhibited proliferation of canine InvTCC cells. Although the response was modest with one μMol vemurafenib, the cell lines with the BRAF(V595E) mutation were significantly more sensitive to the vemurafenib than the cell line with wild-type BRAF (K9TCC-Sh) (P<0.02, Student’s t-Test, 2-tailed). As expected, K9TCC-An, the cell line that did not display a reduction in pMEK after vemurafenib administration, also demonstrated a less dramatic inhibition of proliferation than the other two mutation-positive cell lines, averaging 88% of control over triplicate experiments compared to 75% and 78% of control in the more sensitive cell lines.
Discussion
Advances in sequencing technology and analysis techniques have enabled discoveries of genes and pathways that are commonly mutated in tumors. Some human cancer types have been found to harbor frequently recurrent somatic mutations in a small number of genes, which can accelerate basic and translational research in these malignancies (34); however, similar progress in canine cancers has been limited. Thus far, mutations found in human tumors have rarely been found in canine cancers. For example, non-synonymous changes in KRAS were reported in canine non-small cell lung cancers (NSCLC) a decade ago (35,36). The cases were spontaneous neoplasms identified in pet dogs, and of the 20 tumors examined, four had point mutations in the 12th codon of KRAS while the fifth had an activating mutation in codon 61. This finding was replicated in a larger study of 117 tumors (37), but not in 1996 study of 28 spontaneous and induced canine lung cancers in a closed colony of beagles, suggesting possible environmental or genetic contributions to mutation development (38). Since then, canine lymphomas, mammary tumors, fibrosarcomas, and melanomas have been screened, and human mutation hot-spots are generally not enriched for somatic mutations in canines (reviewed in: (39)).
The BRAF(V600E) mutation creates a valine-to-glutamate substitution in the activation segment of the kinase domain, which leads to constitutive cell signaling, growth factor-independent proliferation, and anti-apoptotic signaling in the tumor (40,41). Prior to this study, canine melanomas, but no other malignancies, have been assessed for activating BRAF mutations. In two studies covering a total of 29 canine melanoma cases, none harbored an activating BRAF(V600E) mutation (42,43). By comparison, we found that the homologous canine BRAF(V595E) mutation is present in approximately 85% of canine InvTCC tumors tested, making the mutation more common in canine InvTCC than in any single type of human cancer except hairy cell leukemia (reviewed in (44)). At the same time, BRAF(V600E) is an extremely important mutation across multiple cancer types and is present, collectively, in approximately 8% of all human cancers (33,40,41,45–48). Our findings indicate that naturally-occurring canine InvTCC offers an unparalleled opportunity in comparative oncology research to test and develop more effective therapies addressing BRAF and MAPK signaling alterations; define strategies to circumvent drug resistance in a tumor environment that rivals human cancer in heterogeneity; to study the molecular mechanisms of carcinogenesis involving BRAF that could apply to dogs and humans; and to develop better approaches for cancer detection and treatment to benefit dogs.
Multiple drugs have been developed to selectively target the BRAF(V600E) mutation (41), including vemurafenib, which prolongs survival in patients with BRAF(V600E) positive melanomas (49). In a phase III trial of 675 melanoma patients, the vemurafenib response rate was 57%, compared to 9% with standard dacarbazine treatment, and progression-free survival was significantly longer in patients receiving vemurafenib (6.9 months vs 1.6 months in the control arm) (50). However, some BRAF(V600E) positive cancers, including colorectal cancer, are inherently resistant to BRAF inhibitors (51), and even initially sensitive tumors adeptly develop vemurafenib resistance. Ongoing trials seek to delay resistance via co-administration with other drugs, including other MAPK pathway inhibitors(29,52). Our data suggest that canine bladder tumors may provide insight into additional strategies to address inherent resistance.
The complexity, heterogeneity, and cross talk between signaling pathways in cancer offers multiple opportunities for BRAF inhibitor resistance to develop, and models that recapitulate human intra- and inter-tumor heterogeneity are crucial. Heterogeneity in canine InvTCC has been well documented (5). In this study, heterogeneity was observed in the canine tumors not only in the presence or absence of the BRAF(V595E) mutation but in response to the inhibitor as well. All three cell lines with activating BRAF mutations demonstrated greatly increased baseline pMEK levels, but only two of the lines showed dramatic declines in pMEK and reduced proliferation in response to treatment. The concentration of vemurafenib that was applied to the canine InvTCC cells was lower than steady state concentrations achieved in the plasma of humans (53), however, this conservative approach was taken to limit off-target effects of the drug (reviewed in (54)). In the less responsive K9TCC-An line, it is likely that other somatic or inherited mutations modify the effect of the BRAF mutation, and further investigation will elucidate the role of these variants. Because multiple pathways are likely involved in InvTCC proliferation, it was not surprising to find that there was no correlation between the BRAF(V595E) mutation status and response to current canine InvTCC treatments (Supplemental Table 3).
As a model for BRAF targeted therapies, canine InvTCC could inform treatment strategies for several types of human cancer. However, we also expect our work to contribute to the invasive bladder cancer field. Although BRAF(V600E) mutations are rare in human urothelial cancers, occurring in <1% of tumors (34), more than one third of human bladder tumors have activating mutations in the RTK/RAS/RAF signaling pathway, including 11% that show amplification of EGFR and 6% bearing activating mutations in the RAS genes that activate BRAF (48). Combination therapies involving RTK inhibitors are some of the more promising lines of targeted therapies for urothelial carcinoma and other cancers (55,56). Identifying contributors to innate or acquired resistance to BRAF(V600E) inhibitors in canine InvTCC will reveal additional genes and proteins important in the development of bladder cancer and will directly inform treatment strategies. In addition, BRAF negative bladder cancer in dogs may replicate the genetics and biology of a significant subset of human urothelial carcinomas.
The high frequency of the BRAF mutation in canine InvTCC also opens the door for clinical and environmental modeling of all BRAF(V600E) positive tumors. Canine InvTCC risk has a strong inherited component, with breed-specific relative risks up to 20-fold (6,57). However, the somatic BRAF mutation shows no breed specificity, suggesting that environmental exposures may contribute to this mutation. It has been hypothesized that environmental factors promote the high frequency of BRAF mutations in melanoma, though the obvious candidate, UV exposure, has been ruled out (58). Canine InvTCC in high-risk breeds has been associated with exposure to lawn chemicals (59), although the specific chemicals and mechanisms involved have not been elucidated. The canine BRAF(V595E) positive tumors may prove to be an excellent system in which to evaluate the role of environmental exposures in oncogenesis.
Mutation testing using a restriction fragment length assay yielded 89% concordance between urine sediment DNA and matched tumor samples from the same dogs. Concordance was improved to 100% when discordant samples were tested with a much more sensitive targeted next-generation sequencing method. This approach also enabled detection of the mutation in tumors that initially tested negative by RFLP and Sanger sequencing, bringing final mutation frequency to 87.9% across 66 tumors. We speculate that detection by the other methods may have been hampered by either low sample purity or subclonality of the BRAF(V595E) mutation in a small subset of tumors. Additional tumor sequencing will likely provide the answer.
Because of the sensitivity of the assay, targeted next-generation amplicon sequencing could be used to develop a non-invasive, urine-screening test to detect InvTCC before it becomes clinically apparent. The same strategy could also be used as a post-diagnostic stratification tool, as dogs with BRAF-positive tumors would be expected to benefit from targeted approaches, which are likely to become part of the standard of care, while the minority of dogs lacking the mutation (12% of dogs) would be better served by different treatment. Such a test could also be evaluated in tracking treatment response or detecting disease relapse. Together, these applications exemplify the anticipated direction of modern personalized medicine.
In broader terms, this work represents an approach to meet two key challenges of pre-clinical models of any cancer type: replicating tumor heterogeneity and the microenvironment of the host (56,60). Heterogeneity is lost in individual cell-lines, and can only be accounted for by examining up to 100s of cell-lines per cancer type (61). Patient-derived tumor xenografts in mice can replicate some of the inherent heterogeneity of the tumor, but cannot reproduce the microenvironment of the original tumor or the complex immune system of the human patient. As in human cancers, naturally occurring tumors in the dog grow in their original microenvironment, and can be very similar to a human tumor developed from the same tissue source (60,62). In addition, the canine immune system rivals that of humans, which was an irreplaceable asset in the development of bone marrow transplant protocols that have preserved the lives of countless leukemia, lymphoma, and multiple myeloma patients (63).
The data summarized here identify a common and targetable mutation in canine InvTCC that will not only open up new treatment options but may also allow for non-invasive diagnosis of the cancer for a majority of dogs. However the study has some weaknesses. We do not yet know the time-line for early detection. The collection of urine samples from healthy dogs as well as pre-sympotomatic patients will be needed to determine the stage at which InvTCC can be detected through shedding of tumor cells in the urine. In addition, we have yet to identify a driver or marker for non-BRAF TCC, which makes up 12% of canine InvTCC. Sequencing of BRAF negative tumors will be needed to determine genetic profiles for this more rare type of tumor. It will be interesting to observe whether these later experiments provide additional insights into human TCC.
Our findings suggest that the dog is once again on the cusp of a cancer therapy breakthrough. The identification of a canine mutation that also commonly drives human tumors reminds us that cancers take no heed of tissue source or species line. Rather, they perturb growth pathways and take advantage of pliable and potent pathways that lend themselves to modification. Our initial success with targeted therapies demonstrates that the molecular defect, rather than the tissue or even the species, can serve as a starting point for investigating tumor biology and therapeutics. Cancers dysregulate some of the same pathways irrespective of species, and mutations shared across species boundaries set the stage for mutually informative efforts to manipulate oncogenic pathways, forcing them to work for us, rather than for the cancer.
Supplementary Material
Acknowledgments
We thank the many dog breeders, dog owners, and clinicians who submitted samples and data in support of this study. We would like to acknowledge the work of the NIH Intramural Sequencing Center for producing the next-generation sequencing data and Dr. Shelly Hoogstrander-Miller for assistance with sample collection. This work was supported by the Intramural Program of the National Human Genome Research Institute at NIH. Additional support was received from the AKC-Canine Health Foundation grants 754, 1336 and 1577 (E.A.O., H.G.P., and D.W.K.); and private donations made for bladder cancer research at Purdue University (D.W.K.).
Footnotes
Conflict of Interest Disclosure Statement: The authors report no conflicts of interest.
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronson RT. Variation in age at death of dogs of different sexes and breeds. Am J Vet Res. 1982;43(11):2057–9. [PubMed] [Google Scholar]
- 3.Dobson JM, Samuel S, Milstein H, Rogers K, Wood JL. Canine neoplasia in the UK: estimates of incidence rates from a population of insured dogs. J Small Anim Pract. 2002;43(6):240–6. doi: 10.1111/j.1748-5827.2002.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 4.Cadieu E, Ostrander EA. Canine genetics offers new mechanisms for the study of human cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2181–3. doi: 10.1158/1055-9965.EPI-07-2667. [DOI] [PubMed] [Google Scholar]
- 5.Ranieri G, Gadaleta CD, Patruno R, Zizzo N, Daidone MG, Hansson MG, et al. A model of study for human cancer: Spontaneous occurring tumors in dogs. Biological features and translation for new anticancer therapies. Crit Rev Oncol Hematol. 2013;88(1):187–97. doi: 10.1016/j.critrevonc.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Knapp DW. Urinary bladder cancer in dogs, a naturally occurring model for cancer biology and drug development. ILAR J. 2014;55(1):100–18. doi: 10.1093/ilar/ilu018. [DOI] [PubMed] [Google Scholar]
- 7.Jonasdottir TJ, Mellersh CS, Moe L, Heggebo R, Gamlem H, Ostrander EA, et al. Genetic mapping of a naturally occurring hereditary renal cancer syndrome in dogs. Proc Natl Acad Sci U S A. 2000;97(8):4132–7. doi: 10.1073/pnas.070053397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shearin AL, Hedan B, Cadieu E, Erich SA, Schmidt EV, Faden DL, et al. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol Biomarkers Prev. 2012;21(7):1019–27. doi: 10.1158/1055-9965.EPI-12-0190-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson EK, Sigurdsson S, Ivansson E, Thomas R, Elvers I, Wright J, et al. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 2013;14(12):R132. doi: 10.1186/gb-2013-14-12-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karyadi DM, Karlins E, Decker B, vonHoldt BM, Carpintero-Ramirez G, Parker HG, et al. A copy number variant at the KITLG locus likely confers risk for canine squamous cell carcinoma of the digit. PLoS genetics. 2013;9(3):e1003409. doi: 10.1371/journal.pgen.1003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker HG, Shearin AL, Ostrander EA. Man’s best friend becomes biology’s best in show: genome analyses in the domestic dog. Annu Rev Genet. 2010;44:309–36. doi: 10.1146/annurev-genet-102808-115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knapp DW. Animal models: naturally occurring canine urinary bladder cancer. In: Lerner SP, Schoenberg MP, Sternberg CN, editors. Textbook of Bladder Cancer. Oxon, United Kingdom: Taylor and Francis; 2006. pp. 171–75. [Google Scholar]
- 13.Griffiths TR. Current perspectives in bladder cancer management. Int J Clin Pract. 2013;67(5):435–48. doi: 10.1111/ijcp.12075. [DOI] [PubMed] [Google Scholar]
- 14.Collisson EA, Cho RJ, Gray JW. What are we learning from the cancer genome? Nat Rev Clin Oncol. 2012;9(11):621–30. doi: 10.1038/nrclinonc.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orphanos G, Kountourakis P. Targeting the HER2 receptor in metastatic breast cancer. Hematol Oncol Stem Cell Ther. 2012;5(3):127–37. doi: 10.5144/1658-3876.2012.127. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Fu P, Fan W. Novel targeted therapies to overcome trastuzumab resistance in HER2-overexpressing metastatic breast cancer. Curr Drug Targets. 2013;14(8):889–98. doi: 10.2174/13894501113149990161. [DOI] [PubMed] [Google Scholar]
- 17.Cekanova M, Rathore K. Animal models and therapeutic molecular targets of cancer: utility and limitations. Drug Des Devel Ther. 2014;8:1911–21. doi: 10.2147/DDDT.S49584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Withrow SJ, Thrall DE, Straw RC, Powers BE, Wrigley RH, Larue SM, et al. Intra-arterial cisplatin with or without radiation in limb-sparing for canine osteosarcoma. Cancer. 1993;71(8):2484–90. doi: 10.1002/1097-0142(19930415)71:8<2484::aid-cncr2820710810>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 19.Breen M, Modiano JF. Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans--man and his best friend share more than companionship. Chromosome Res. 2008;16(1):145–54. doi: 10.1007/s10577-007-1212-4. [DOI] [PubMed] [Google Scholar]
- 20.Lingaas F, Comstock KE, Kirkness EF, Sorensen A, Aarskaug T, Hitte C, et al. A mutation in the canine BHD gene is associated with hereditary multifocal renal cystadenocarcinoma and nodular dermatofibrosis in the German Shepherd dog. Hum Mol Genet. 2003;12(23):3043–53. doi: 10.1093/hmg/ddg336. [DOI] [PubMed] [Google Scholar]
- 21.Karyadi DM, Karlins E, Decker B, vonHoldt BM, Carpintero-Ramirez G, Parker HG, et al. A copy number variant at the KITLG locus likely confers risk for canine squamous cell carcinoma of the digit. PLoS Genet. 2013;9(3):e1003409. doi: 10.1371/journal.pgen.1003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, et al. Genetic structure of the purebred domestic dog. Science. 2004;304(5674):1160–4. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 26.Dhawan D, Ramos-Vara JA, Stewart JC, Zheng R, Knapp DW. Canine invasive transitional cell carcinoma cell lines: In vitro tools to complement a relevant animal model of invasive urinary bladder cancer. Urol Oncol. 2009;27(3):284–92. doi: 10.1016/j.urolonc.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 28.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2(3):227–35. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 30.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105(8):3041–6. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82(13):1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 32.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 34.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;Chapter 10(Unit 10):11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraegel SA, Gumerlock PH, Dungworth DL, Oreffo VI, Madewell BR. K-ras activation in non-small cell lung cancer in the dog. Cancer Res. 1992;52(17):4724–27. [PubMed] [Google Scholar]
- 36.Gumerlock PH, Meyers FJ, Foster BA, Kawakami TG, deVere White RW. Activated c-N-ras in radiation-induced acute nonlymphocytic leukemia: twelfth codon aspartic acid. Radiat Res. 1989;117(2):198–206. [PubMed] [Google Scholar]
- 37.Griffey SM, Kraegel SA, Madewell BR. Rapid detection of K-ras gene mutations in canine lung cancer using single-strand conformational polymorphism analysis. Carcinogenesis. 1998;19(6):959–63. doi: 10.1093/carcin/19.6.959. [DOI] [PubMed] [Google Scholar]
- 38.Tierney LA, Hahn FF, Lechner JF. p53, erbB-2 and K-ras gene alterations are rare in spontaneous and plutonium-239-induced canine lung neoplasia. Radiat Res. 1996;145(2):181–7. [PubMed] [Google Scholar]
- 39.Richter A, Murua Escobar H, Gunther K, Soller JT, Winkler S, Nolte I, et al. RAS gene hot-spot mutations in canine neoplasias. J Hered. 2005;96(7):764–5. doi: 10.1093/jhered/esi121. [DOI] [PubMed] [Google Scholar]
- 40.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6(4):313–9. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 41.Mandala M, Voit C. Targeting BRAF in melanoma: biological and clinical challenges. Crit Rev Oncol Hematol. 2013;87(3):239–55. doi: 10.1016/j.critrevonc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Shelly S, Chien MB, Yip B, Kent MS, Theon AP, McCallan JL, et al. Exon 15 BRAF mutations are uncommon in canine oral malignant melanomas. Mamm Genome. 2005;16(3):211–7. doi: 10.1007/s00335-004-2441-x. [DOI] [PubMed] [Google Scholar]
- 43.Fowles JS, Denton CL, Gustafson DL. Comparative analysis of MAPK and PI3K/AKT pathway activation and inhibition in human and canine melanoma. Vet Comp Oncol. 2013 doi: 10.1111/vco.12044. [DOI] [PubMed] [Google Scholar]
- 44.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19(11):1401–9. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 45.Jung CK, Little MP, Lubin JH, Brenner AV, Wells SA, Jr, Sigurdson AJ, et al. The increase in thyroid cancer incidence during the last four decades is accompanied by a high frequency of BRAF mutations and a sharp increase in RAS mutations. J Clin Endocrinol Metab. 2014;99(2):E276–85. doi: 10.1210/jc.2013-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iyer G, Al-Ahmadie H, Schultz N, Hanrahan AJ, Ostrovnaya I, Balar AV, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol. 2013;31(25):3133–40. doi: 10.1200/JCO.2012.46.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45(12):1459–63. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Network TCGAR. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Connolly K, Brungs D, Szeto E, Epstein RJ. Anticancer activity of combination targeted therapy using cetuximab plus vemurafenib for refractory BRAF (V600E)-mutant metastatic colorectal carcinoma. Curr Oncol. 2014;21(1):e151–4. doi: 10.3747/co.21.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson DB, Flaherty KT, Weber JS, Infante JR, Kim KB, Kefford RF, et al. Combined BRAF (Dabrafenib) and MEK Inhibition (Trametinib) in Patients With BRAFV600-Mutant Melanoma Experiencing Progression With Single-Agent BRAF Inhibitor. J Clin Oncol. 2014;32(33):3697–704. doi: 10.1200/JCO.2014.57.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ribas A, Zhang W, Chang I, Shirai K, Ernstoff MS, Daud A, et al. The effects of a high-fat meal on single-dose vemurafenib pharmacokinetics. J Clin Pharmacol. 2014;54(4):368–74. doi: 10.1002/jcph.255. [DOI] [PubMed] [Google Scholar]
- 54.Holderfield M, Nagel TE, Stuart DD. Mechanism and consequences of RAF kinase activation by small-molecule inhibitors. Br J Cancer. 2014;111(4):640–5. doi: 10.1038/bjc.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guancial EA, Chowdhury D, Rosenberg JE. Personalized therapy for urothelial cancer: review of the clinical evidence. Clin Investig (Lond) 2011;1(4):546–55. doi: 10.4155/cli.11.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang M, Shen A, Ding J, Geng M. Molecularly targeted cancer therapy: some lessons from the past decade. Trends Pharmacol Sci. 2014;35(1):41–50. doi: 10.1016/j.tips.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Knapp D, Glickman N, DeNicola D, Bonney P, Lin T, Glickman L. Naturally-occurring canine transitional cell carcinoma of the urinary bladder: A relevant model of human invasive bladder cancer. Urol Oncol. 2000;5:47–59. doi: 10.1016/s1078-1439(99)00006-x. [DOI] [PubMed] [Google Scholar]
- 58.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glickman LT, Raghavan M, Knapp DW, Bonney PL, Dawson MH. Herbicide exposure and the risk of transitional cell carcinoma of the urinary bladder in Scottish Terriers. J Am Vet Med Assoc. 2004;224(8):1290–7. doi: 10.2460/javma.2004.224.1290. [DOI] [PubMed] [Google Scholar]
- 60.Kumar S, Mokhtari RB, Yeger H, Baruchel S. Preclinical models for pediatric solid tumor drug discovery: current trends, challenges and the scopes for improvement. Expert Opin Drug Discov. 2012;7(11):1093–106. doi: 10.1517/17460441.2012.722077. [DOI] [PubMed] [Google Scholar]
- 61.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paoloni M, Webb C, Mazcko C, Cherba D, Hendricks W, Lana S, et al. Prospective molecular profiling of canine cancers provides a clinically relevant comparative model for evaluating personalized medicine (PMed) trials. PLoS One. 2014;9(3):e90028. doi: 10.1371/journal.pone.0090028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ram R, Storb R. Pharmacologic prophylaxis regimens for acute graft-versus-host disease: past, present and future. Leuk Lymphoma. 2013;54(8):1591–601. doi: 10.3109/10428194.2012.762978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.