Abstract
Kidney cancer (or renal cell carcinoma [RCC]) is known as “the internist’s tumor” because it has protean systemic manifestations suggesting it utilizes complex, non-physiologic metabolic pathways. Given the increasing incidence of this cancer and its lack of effective therapeutic targets, we undertook an extensive analysis of human RCC tissue employing combined grade-dependent proteomics and metabolomics analysis to determine how metabolic reprogramming occurring in this disease allows it to escape available therapeutic approaches. After validation experiments in RCC cell lines that were wild-type or mutant for the VHL tumor suppressor, in characterizing higher grade tumors we found that the Warburg effect is relatively more prominent at the expense of the tricarboxylic acid cycle and oxidative metabolism in general. Further, we found that the glutamine metabolism pathway acts to inhibit reactive oxygen species, as evidenced by an upregulated glutathione pathway, while the β-oxidation pathway is inhibited leading to increased fatty acyl-carnitines. In support of findings from previous urine metabolomics analyses, we also documented tryptophan catabolism associated with immune suppression, which was highly represented in RCC compared to other metabolic pathways. Together, our results offer a rationale to evaluate novel anti-metabolic treatment strategies being developed in other disease settings as therapeutic strategies in RCC.
Keywords: Renal cell carcinoma, proteomics, metabolomics, energy metabolism, tryptophan metabolism
Introduction
Examination of clinical materials using several distinct and complementary techniques has the potential to yield new perspectives in physiology and pathophysiology which might not be evident using each technique in isolation. With the acceptance of mainstream omics technologies (1), it has become possible to probe the molecular biology of disease with a high degree of resolution. However, due to the highly specialized nature of each omics technique and its associated analysis, each investigator tends to have a relatively narrow range of expertise directed towards but a single omics strategy; for this reason, it is rare to find a merging of several of these powerful techniques in a single study. Indeed, such single omics studies can be problematic for interpretation, for example the quantity of proteins (e.g. enzymes) as the output of proteomics does not always correlate with their activities, and many gene transcripts undergo post-transcriptional processing. It is also common to see publications which focus on technical and analytical aspects of the omics process at the expense of the overarching biological interpretation. Because the interpretation of omics data can be easily misconstrued due to covariant and systemic effects which are not biologically relevant, it is critical to validate all findings obtained in this manner by in vitro experiments to confirm the hypotheses built upon the omics data. For these reasons, it is a logical extension in this field to combine data from several complementary omics techniques to attempt to eliminate some of the bias inherent in a single method, and it is critical to validate the proposed hypotheses which come out of the omics data both in vitro and in vivo (2, 3).
The vast majority of published basic research in oncology is based on the behavior of commercially available cell lines. In these situations, experiments have been performed on cells representing predominantly a single (usually high) grade of cancer. In a parallel situation, authors of studies using in vivo xenografts, derived from immortalized cell lines, frequently ignore original tumor grade information or that which could be inferred from cell line characteristics. It is becoming abundantly clear from work in our and other laboratories that the biology of tumors of different grades is markedly distinct, at least in renal cell carcinoma (RCC) (4, 5). Indeed, work in our laboratory is focused on determining whether personalized therapy based on such grade classification may have clinical utility, a concept which is often foreign to clinicians. The utility of integrating multiple omics approaches is especially important when evaluating grade dependence with respect to therapy, because of the subtleties that can be difficult to recognize in comparing morphologically similar tissues.
In our continuing pursuit of physiological alterations that occur in kidney cancer, which may enable us to discover and validate new biomarkers and targets for this highly morbid disease, we have previously utilized both metabolomics and proteomics separately in a variety of biofluids and tissues (4–8). In the present study, for the reasons discussed above, we have merged the proteomics and metabolomics datasets derived from human RCC tumors of distinct Fuhrman grade in order to expand the pathway data previously obtained (4, 5); for validation purposes, we utilize several commercial cell lines, both with and without Von Hippel–Lindau (VHL) mutations, since VHL status affects energy metabolism through hypoxia-inducible factor (HIF) regulation. Using this approach, we show here incontrovertible and validated evidence of grade-dependent metabolic reprogramming in human RCC, characterized by increasing utilization of the Warburg effect in higher grade, and we further show grade-dependent alterations in fatty acid, glutamine, and glutathione metabolism. In addition, we show that the tryptophan pathway feeds directly into known immunosuppressive metabolites, confirming a previous hypothesis formulated from our urinary metabolomics analysis (7).
From the data presented here, it appears that combining omics techniques leads to synergism in knowledge and yields concepts of grade-dependent tumor biology which would otherwise not be obvious using a single technique in isolation. In addition, our work addresses a major issue regarding therapy for RCC patients, namely that there is at present no grade or mutation-specific therapy, despite the fact that many approved targeted therapeutics have been recently developed. Thus, our work will lead to both a better understanding of RCC biology and important clinical advances especially related to potential grade-specific therapeutics.
Materials and Methods
Materials
Low glucose Dulbecco’s modified Eagle’s medium (DMEM), glucose free DMEM, glutamine free DMEM, and glucose were obtained from Life Technologies (Grand Island, NY). Dimethyl sulfoxide (DMSO), 2-deoxy-D-glucose (2-DG), etomoxir, and mouse monoclonal anti-β-actin antibody were obtained from Sigma (St. Louis, MO). Stabilized glutamine was obtained from Gemini Bio-Products (West Sacramento, CA). Goat anti-mouse and goat anti-rabbit HRP conjugated IgG were obtained from Bio-Rad (Hercules, CA). Human interferon γ (IFNγ) was purchased from PeProTech (Rocky Hill, NJ). Methylthiohydantoin-DL-tryptophan (MTH-trp) was purchased from Enzo Life Sciences (Farmingdale, NY). Anti-human IDO antibody was obtained from Cell Signaling Technology (Danvers, MA). After appropriate IRB approval at UC Davis or Memorial Sloan Kettering Cancer Center (MSKCC), and after pathological examination of partial or total nephrectomy samples, excess tissue was analyzed by shotgun proteomics and metabolomics, respectively (Supplemental Table 1).
Cell culture
The RCC cell lines Caki-1 (VHL-wt) and 786-O (VHL-mut) were obtained from the American Type Culture Collection (ATCC, Rockville, MD); the primary “normal human kidney” proximal tubular cells (NHK) were obtained from Lonza (Allendale, NJ). All ATCC and Lonza cell lines undergo extensive authentication tests during the accessioning process as described on their website; in addition all cells were frequently tested for mycoplasma in the author’s laboratory. The cells were maintained in DMEM supplemented with 10% fetal bovine serum, 100 units/mL streptomycin, and 100 mg/mL penicillin (complete medium). The cells were maintained at 37°C with 5% CO2.
Primary cell cultures
After appropriate IRB approval at the University of Milano-Bicocca, primary cell cultures were obtained from human normal cortex and RCC tissues, culture conditions and immunophenotypical characterization were performed as described (9).
Tryptophan and kynurenine Analysis
After the indicated treatments, cell media was collected and polar metabolites were extracted using a modification of a previous method (10). Briefly, media aliquots were extracted with ice-cold chloroform, methanol, and 3 mM PIPES-3 mM EDTA, pH 7.4, and vortexed. Extraction mixtures were then stored at −80°C. Immediately prior to analysis, samples were thawed and centrifuged for 10 minutes at 10,000 rpm. The upper, aqueous phase was subjected to High-performance liquid chromatography (HPLC) analysis using a modification of a previous method (11). HPLC separations were performed on an Agilent 1100 instrument (Agilent Technologies, Folsom, CA) using a Thermo 50 × 2.1 Hypersil GOLD C18 column. HPLC conditions were as follows: Mobile Phase: 15 mM Potassium Phosphate, pH 6.4, with 2.7% (v/v) acetonitrile; Isocratic flow rate: 0.8 mL/min; Injection volume: 20 μL; Run time: 10 minutes. Column temperature was maintained at 30° C. Detection was accomplished using an Agilent 1100 diode array detector, with detection of kynurenine at 360 nm and detection of tryptophan at 286 nm. Analyte concentrations were calculated using peak area under the curves (AUC) and standard curves generated using authentic standards for each metabolite of interest.
MTT assay
Cell viability assays were performed as described previously (12). Briefly, cells were plated in 96-well plates, and after appropriate treatments, the cells were incubated in MTT solution/media mixture. Then, the MTT solution was removed and the blue crystalline precipitate in each well was dissolved in DMSO. Visible absorbance of each well at 540 nm was quantified using a microplate reader.
Lactate assay
Cells were plated in 24 well plates, and after appropriate treatments, the media were processed to measure lactate levels using the Lactate Assay Kit from Biovision (Milpitas, CA) following the manufacturer’s instructions.
Oxidative metabolism measurements
Oxygen consumption rates (OCR) were measured using the XF24 Extracellular Flux Analyzer from (Seahorse Bioscience, Chicopee, MA). For experiments utilizing etomoxir or 2-deoxyglucose, cells were pretreated 30 minutes prior to running the plate on the XF24 instrument. Media was changed to glucose-free or glutamine-free media 30 minutes prior to running the assay.
Oil red O staining of RCC tissue
8–10 μm cryostat sections of RCC tissue samples of distinct Fuhrman grade were fixed in 10% neutral buffered formalin solution (Sigma-Aldrich) for 1.5 hours and rinsed with PBS. Staining was performed with freshly prepared Oil Red O (ORO) working solution, obtained by 3:2 dilution in H2O of ORO stock solution (0.35% in isopropanol), for 1.5 hours. The slides were then rinsed with H2O and counter-staining was performed with haematoxylin solution for 3 min. The stained slides were mounted with CC/Mount (Sigma, St. Louis, MO) and analyzed by Nikon Eclipse E800 with 20x objectives. Three to four pictures for each slide were randomly captured and analyzed by LuciaG Image Analysis System (Nikon, Melville, NY).
Oil red O staining and quantitative assay in vitro
Primary cell cultures and cell lines, plated in triplicate in 96 well plates, were treated with different etomoxir concentrations for 72 hours, fixed in 10% formalin for one hour and stained with ORO working solution for one hour. ORO stained lipids were quantified by extracting the dye from cells with 200 μl isopropanol and measuring the absorbance of each well at 510 nm using a microplate reader.
Immunoblotting
Immunoblotting was done as described previously (12). Briefly, after appropriate treatments, the cells were washed with PBS, lysed in lysis buffer, and cell lysates were immunoblotted. Membranes were blocked in 5% nonfat dry milk for one hour at room temperature and probed with appropriate antibodies. Membranes were then probed with HRP tagged anti-mouse or anti-rabbit IgG antibodies. Signal was detected using enhanced chemiluminescence solutions.
Oxidized glutathione (GSSG) and glutathione (GSH) measurement and quantitation
Cells were plated into 96 well plates (5000/well) and the next day treated with or without stabilized glutamine (Gemini Bio-Products, Sacramento, CA) for 24 hours. The GSH and GSSG concentrations were measured in each well of cells using the GSH/GSSG-Glo Assay kit (Promega, Madison, WI) following the manufacturer’s instructions..
Whole frozen tissues which were leftover from the metabolomics analysis were utilized to measure absolute GSH and GSSG levels using the GSH/GSSG-Glo Assay kit (Promega, Madison, WI) following the manufacturer’s instructions.
Acylcarnitine quantitation
Deuterium-labeled internal standards were added to the tissues and the mixture was solubilized in methanol followed by a crash extraction. The extracted mixture was injected onto an Atlantis HILIC Column connected to a Waters Xevo triple quadrupole mass spectrometer (Waters, MA). The analytes were ionized via positive electrospray and the mass spectrometer is operated in the tandem MS mode. The absolute concentration of each acylcarnitine was determined by comparing the peak to that of the relevant internal standard. The composition of 15 markers is included in this panel.
NADP/NADPH assay
After indicated treatments, the NADPH/NADP ratio was measured using the NADP/NADPH-Glo Assay kit (Promega) following the manufacturer’s instructions.
Non-targeted metabolomics analysis
For metabolomics analysis, after appropriate IRB approval at MSKCC, ccRCC tumors and normal kidney tissues from matched controls were obtained (Supplemental Table 1). Tumors used for the study were from archived frozen ccRCC specimens. The majority of the samples were snap frozen within one to two hours of resection (maximum up to four hours). Samples were inventoried and immediately stored at −80 °C. At the time of analysis samples were extracted and prepared for analysis using Metabolon’s standard solvent extraction method. The extracted samples were split into equal parts for analysis on the GC/MS and LC/MS/MS platforms. Samples were analyzed over nine consecutive run days. Also included were several technical replicate samples created from a homogeneous pool containing a small amount of all study samples (“Client Matrix”).
Metabolite extraction and measurement was conducted at Metabolon Inc. (Durham, NC) as previously described (6, 16). All samples were maintained at −80 °C until processed and raw data is shown in Supplemental Table 2 with statistics in Supplemental Table 3. The mass spectrometer platforms, sample extraction and preparation, instrument settings and conditions, and data handling have previously been described in detail (17).
The major components of the process can be summarized as follows: the sample preparation process was carried out using the automated MicroLab STAR® system (Hamilton, Reno, NV). Samples were prepared using a proprietary series of organic and aqueous extractions to remove the protein fraction while allowing maximum recovery of small molecules. A cocktail of recovery standards was added prior to the first step in the extraction process for QC purposes. The resulting extract was divided into three fractions for untargeted metabolic profiling and randomized for analysis. Each [sample was dried under vacuum to remove organic solvent. Samples were characterized using three independent platforms: ultrahigh-performance liquid chromatography/tandem mass spectrometry (UHPLC-MS/MS) in the negative ion mode, UHPLC-MS/MS in the positive ion mode and gas chromatography-mass spectrometry (GC-MS) after siaylation.. The reproducibility of the extraction protocol was assessed by the recovery of the xenobiotic compounds spiked in every tissue sample prior to extraction. The LC/MS portion of the platform was based on a Waters ACQUITY UPLC and a Thermo-Finnigan LTQ mass spectrometer, which consisted of an electrospray ionization (ESI) source and linear ion-trap (LIT) mass analyzer. One aliquot was analyzed using acidic positive ion optimized conditions and the other using basic negative ion optimized conditions in two independent injections using separate dedicated columns. Samples destined for GC/MS analysis were re-dried under vacuum desiccation for a minimum of 24 hours prior to being derivatized under dried nitrogen using bistrimethyl-silyl-triflouroacetamide (BSTFA). Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization.
Identification of known chemical entities was based on comparison to metabolomic library entries of purified standards based on chromatographic properties and mass spectra. As of this writing, more than 4000 commercially available purified standard compounds had been acquired and registered into the LIMS for distribution to both the LC and GC platforms for determination of their analytical characteristics. MS peaks were identified using Metabolon’s proprietary peak integration/identification software, by comparing MS peak data to that of a library of purified standard. The combination of chromatographic properties and mass spectra gave an indication of a match to the specific compound or an isobaric entity. The integrated ion peaks for each compound were normalized in MS run-day blocks by registering the medians to equal one (1.00) and normalizing each data point proportionately (termed the “block correction”). This integrated peak normalization generates a median scaled relative value for each metabolite measured in a given sample. Minimum median scaled metabolite values were imputed for specific samples in which the metabolite levels fell below the lower limit of quantitation. Following normalization procedures and imputation, statistical analysis was performed on log transformed data to identify significant differences in metabolite levels between comparator groups.
Treatment group samples were evenly distributed across all days of the study to avoid confounding the experimental group differences with any instrument interday sensitivity differences. For example, if there were three treatment groups with 16 subjects each, eight from each group were analyzed on one day and the remaining on the second day. As a result of the treatment groups being evenly distributed and balanced across the run-days, the expected values of the means for each day were likely equal. Hence, it was possible to divide each sample by the sample median to divide out any instrument sensitivity differences. This normalization was performed on a per-compound basis. Thus, the sample medians, per compound, were equal for each instrument run-day. Sample median was used as opposed to sample mean, because the sample mean can be greatly affected by outliers. The samples were randomized with regard to run-day and then randomized within the run-day. The raw data is in Supplemental Table 2 and statistics for tumors compared to matched controls are in Supplemental Table 3.
Proteomics analysis
Tissue protein levels were obtained from a previously published proteomics study in RCC tissues which was performed under appropriate UC Davis IRB approval as described (4). The raw data and statistics for protein levels in tumors compared to normal kidney tissues are in Supplemental Table 4.
Creation of pathway maps
Data from a prior non-targeted proteomics study on grade-dependent formalin fixed paraffin embedded ccRCC tissues (4) and metabolomics data described above (Supplemental Table 1) were used to compare proteins/enzymes with metabolic pathways. Quantitative changes of individual metabolites as compared with matched control tissues, and proteins in relation to adjacent “normal” tissue, were calculated and graphed on a KEGG-based pathway map as a function of grade. While it is true that levels of many enzymes are not altered when their catalytic activity is increased, by combining proteomics and metabolomics data, we were able to identify activity of enzymes that had no change in protein concentration since enzymatic activity is quantitatively proportional to the appearance of the end metabolite.
Statistics
Comparisons of mean values were performed using the independent samples t-test. For the human metabolomics data, a paired t-test analysis (tumor vs. non-tumor matched control) was performed on log transformed data. A p-value of < 0.05 was considered statistically significant unless stated.
Results and Discussion
The purpose of this study was to evaluate specific biochemical pathways associated with RCC, and moreover to do so at a high degree of resolution, in a continuing effort to attain new insights regarding metabolic reprogramming in these tumors, as well as to identify potentially new grade-dependent therapeutic targets for RCC. This work is unique from other metabolic studies in RCC due to its concurrent utilization of two distinct although complementary techniques to generate and validate reprogrammed energy metabolism and tryptophan pathways in RCC, thereby ameliorating some of the drawbacks and deficiencies of each omics technique as used in isolation.
Aerobic glycolysis is upregulated in high grade RCC
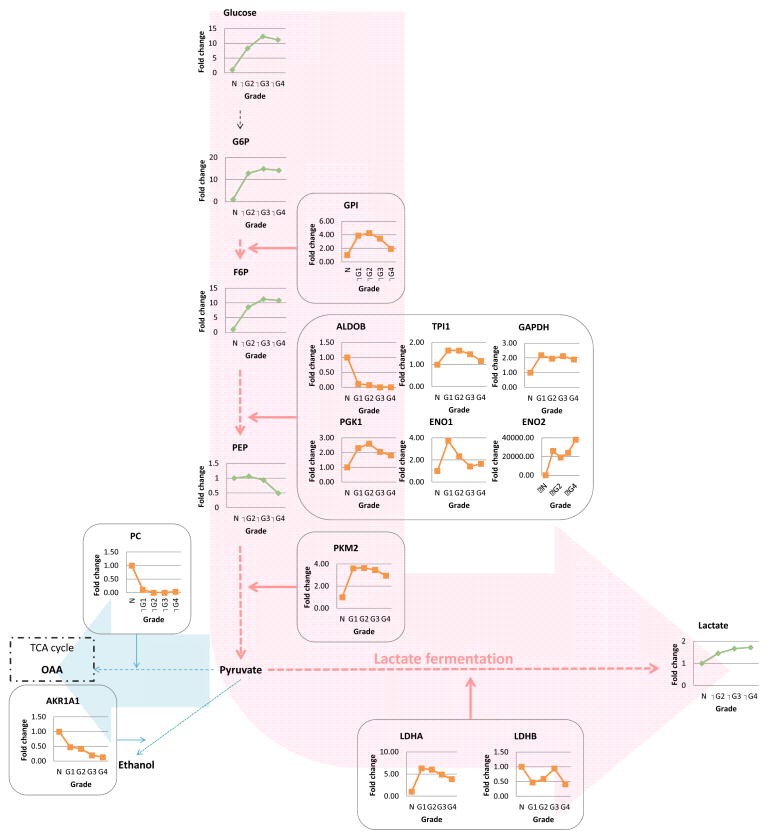
It has long been known that most solid tumors utilize aerobic glycolysis/lactic acid fermentation (the Warburg effect) in order to provide sufficient energy for cell metabolism, as well as to synthesize key membrane components and other cellular building materials (4, 18–20). Given the prominence of the Warburg effect observed in RCC in many studies (including our proteomics studies (4, 5)), we first asked whether there is evidence of aerobic glycolysis in RCC tissues using the combined metabolomics/proteomics approach. The glycolysis-relevant metabolites (glucose, glucose-6-phosphate, and fructose-6-phosphate) are increased significantly with higher grade, suggesting that glucose metabolism is more prominent with increasing tumor grade (Fig. 1, Supplemental Table 3 and 4). Consistent with these data, most of the enzymes involved in this process, especially glyceraldehyde 3-phosphate dehydrogenase (GAPDH), enolase 2, and pyruvate kinase-muscle-2 (PKM2), are increased in tumor as compared to normal tissues, although their levels do not show the same unambiguous grade dependence as most of the metabolites, likely due to the fact as previously noted that enzymatic activity can change without an alteration of enzyme (i.e. protein) concentration. L-lactate was similarly increased in a grade-dependent manner (up to two-fold in grade 4), suggesting that lactate fermentation was upregulated in RCC, particularly at higher grades. In support of these data, lactate dehydrogenase A (LDHA), involved in converting pyruvate to lactate, was increased in all grades of RCC as compared to control tissue possibly through increased HIF activity (21) due to VHL inactivation which is seen in more than 90% of RCC tumors (22). On the other hand, pyruvate carboxylase (PC), the enzyme which converts pyruvate to oxaloacetate in the tricarboxylic acid (TCA) cycle, was decreased in all grades, suggesting that glucose and sugars involved in glycolysis were utilized for lactate fermentation in RCC rather than for metabolism within the TCA cycle or other downstream oxidative metabolism pathways.
Figure 1. 1. Aerobic glycolysis is grade-dependently upregulated in RCC.
Combined proteomics and metabolomics data of human RCC tissue was overlayed onto a stylized KEGG-based pathway diagram. Green, metabolite; orange, enzyme; black dot arrow, metabolism; red arrow, upregulated pathway; blue arrow, downregulated pathway; G6P, glucose-6-phosphate; PEP, F6P, fructose 6-phosphate; phosphoenolpyruvate; OAA, oxaloacetate; GPI, glucose-6-phosphate isomerase; ALDOB, aldolase B; TPI1, triosephosphate isomerase 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PGK1, phosphoglycerate kinase 1; ENO, enolase; PKM, pyruvate kinase; PC, pyruvate carboxylase; AKR1A1, aldo-keto reductase family 1, member A1; LDH, lactate dehydrogenase.
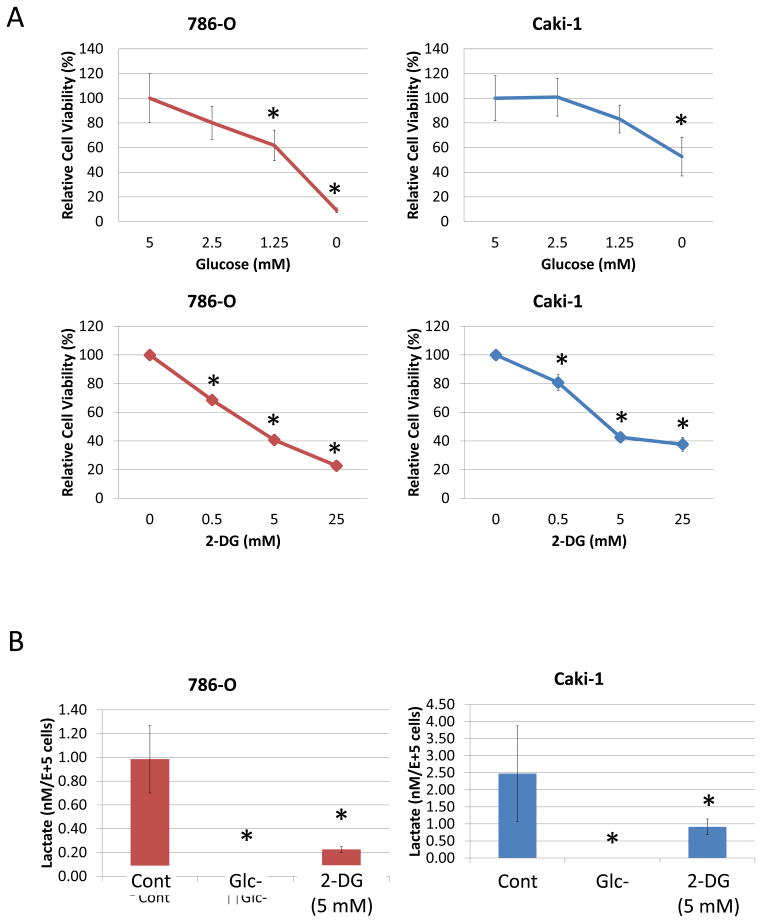
While the omics-derived pathway data are suggestive of biochemical and metabolic changes, we next asked whether such biology, which would occur as a result of such reprogramming, could be validated using available RCC cell lines. Both VHL-mut (786-O) and VHL-wt (Caki-1) human RCC cell lines showed decreased viability in decreased glucose concentrations or when incubated with 2-DG (a glycolysis inhibitor) at various concentrations (Fig. 2A). To confirm that, as a signature of aerobic glycolysis, lactate production in these cells was highly active under normoxia, both cell lines were incubated in 5 mM glucose media, glucose-depleted media, or with 2-DG and 5 mM glucose, and lactate concentrations were measured in the media. Both procedures (2-DG and glucose depletion) markedly decreased lactate production (Fig. 2B), demonstrating that both RCC cell lines studied rely primarily on aerobic glycolysis (Warburg effect), consistent with the data obtained from the human RCC tissues.
Figure 2. 2. Inhibition of glycolysis attenuates RCC cell viability and lactate production.
(A) 786-O (VHL-mut) and Caki-1 (VHL-wt) cells were treated with media at the indicated glucose concentration, or with the indicated concentration of 2-DG in 5 mM glucose media, for 72 hours and then subjected to MTT assays. *P < 0.05 compared to 5 mM glucose or 0 mM 2-DG. Error bars indicate standard deviation. Data are representative of three repeats.
(B) 786-O (VHL-mut) and Caki-1 (VHL-wt) cells were treated with glucose depleted media (Glc-) or 2-DG (5 mM) for 24 hours then lactate in the media was measured as described in Materials and Methods. *P < 0.05 compared to control (glucose 5 mM). Error bars indicate standard deviation. Data are representative of three repeats.
Acylcarnitines are increased, yet fatty acid β-oxidation mechanism is attenuated, with higher grade
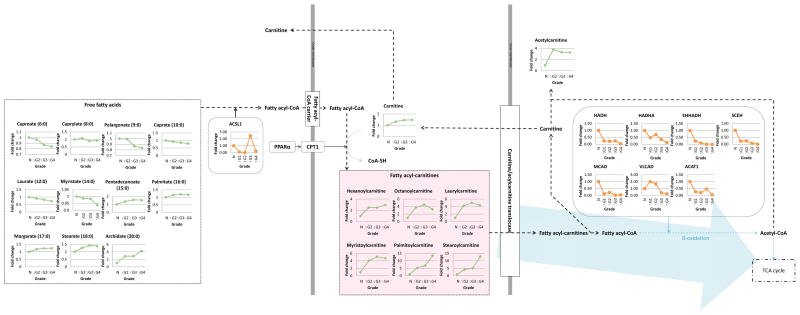
Many tumors and other tissues are able to oxidize fatty acids to satisfy their energy requirements (23). Free fatty acids (FFA) are esterified to fatty acyl-CoA and then transported into the mitochondria by carnitine palmitoyltransferase-1 (CPT1) and the carnitine system, at which point in normal tissue they are subjected to β-oxidation as fatty acyl-CoA to feed into the TCA cycle (24). To study this process in human RCC as a function of tumor grade, we evaluated the levels of the key components of this pathway as well as the enzymes that control their interconversions. While the levels of shorter chain FFAs (6:0, 8:0, 9:0, 10:0, and 12:0) were generally decreased in a grade-dependent manner, the levels of carnitine and the acylcarnitines were increased in all grades (Fig. 3; Supplemental Tables 3 and 5) consistent with previous human urinary metabolomics studies in our laboratory (25). Absolute quantitation of acylcarnitines in a selection of tumor and matched normal control tissues revealed that most of these metabolites were higher in RCC tissues and the increase was significant for the major metabolite L-carnitine (Supplemental Table 5). However, surprisingly, most of the enzymes of β-oxidation were decreased in high grade tissues suggesting a lack of oxidation of the acyl-CoAs in RCC tissue, which is a possible mechanism for the accumulation of acylcarnitines.
Figure 3. 3. Acylcarnitines are increased and β-oxidation is downregulated in RCC.
Combined proteomics and metabolomics data of human RCC tissue was overlayed onto a stylized KEGG-based pathway diagram. Green, metabolite; orange, enzyme; black dot arrow, metabolism; red, upregulated pathway; blue arrow, downregulated pathway. ACSL, acyl-CoA synthetase long-chain; PPAR, peroxisome proliferator-activated receptor; CPT, carnitine palmitoyltransferase; HADH, carnitine palmitoyltransferase alpha subunit; HADHA, hydroxyacyl-CoA dehydrogenase; EHHADH, enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase; SCEH, short-chain enoyl-CoA hydratase; MCAD, medium-chain specific acyl-CoA dehydrogenase; VLCAD, very long-chain specific acyl-CoA dehydrogenase; ACAT, acetyl-CoA acetyltransferase.
Adipogenesis with clear cytosol is a known signature of RCC (26), and increased activity of adipogenesis-related proteins, such as acyl-CoA:cholesterol acyltransferase, have been reported to be an indicator of aggressiveness (27). Thus we first hypothesized that the grade dependent decrease of FFAs is due to increased adipogenesis: accumulation of lipids (triglycerides and esterified fatty acids such as cholesterol and glycerol) in the cytosol. To test this hypothesis, we performed ORO staining in normal kidney tissues and grade 2 – 4 RCC tissues to measure triglycerides and esterified fatty acids. Surprisingly, the results showed a grade dependent decrease of ORO staining at higher grades, while grade 2 had significantly increased staining compared to normal kidney tissues (Supplemental Fig. 1A), suggesting that the grade dependent decrease of cellular FFAs is due to increased utilization of FFAs. To further support this finding, we examined whether inhibition of a FA transporter, CPT1, by etomoxir increases lipid storage in primary normal human kidney epithelial cells, primary RCC cells, and 786-O. In all cell lines tested, etomoxir increased ORO staining (Supplemental Fig. 1B), suggesting increased lipid storage in those cells upon CPT1 inhibition due to decreased FA utilization to fatty acyl-carnitines. Our finding that the majority of fatty acid β-oxidation enzymes were down-regulated in proportion to grade (Fig. 3) suggests that the acylcarnitines were utilized for other processes not directly related to energy metabolism, for example for their anti-inflammatory actions (25).
The glutamine pathway feeds the GSH/GSSG antioxidant system
There is increasing evidence that the glutamine pathway is essential in a variety of cancers, including RCC, to support efficient carbon metabolism of both anabolism and cell growth (28, 29). Indeed, VHL-mut RCC has been shown to be sensitive to glutaminase inhibition, as in some cases these tumors use glutamate to regulate the TCA cycle and subsequent lipogenesis (30). Moreover, the prominence of glutaminase may be a vestige of non-transformed renal epithelial cell biology in which it is utilized for systemic acid secretion through the renal tubules (31).
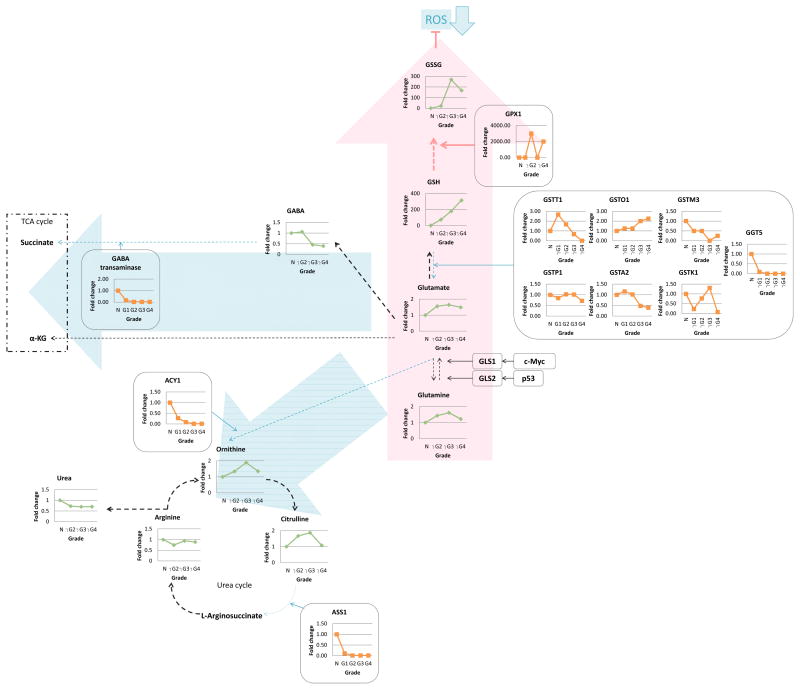
Glutamine feeds into various downstream metabolic pathways including the TCA and urea cycles as well as the GSH pathway. While all the enzymes detected by the metabolomics analysis, that catabolize glutamine or glutamate to feed into the TCA and urea cycles were decreased in our analysis, the metabolite levels in the pathway from glutamine to GSH and GSSG were grade-dependently increased and associated with generally decreased levels of the enzymes which catalyze GSH to glutamate, suggesting that glutamine is reprogrammed to the GSH pathway at higher grades (Fig. 4; Supplemental Fig. 2A). Indeed, the marked increase in GSH in the global metabolomics study (Fig. 4; Supplemental Table 2) was one of the highest magnitude changes observed in our entire metabolomics analysis. These data suggest that glutamine is being utilized by the cancer cell in large part to attenuate oxidative stress, rather than for energy and cell component production through the TCA cycle, thereby yielding a survival advantage to these cells. In fact this hypothesis is validated by the finding that the GSH/GSSG ratio in Grade 3 and 4 tumors, after direct quantitative measurement of these metabolites, was significantly higher than that of normal kidneys (Supplemental Fig. 2A and Supplemental Table 5), indicative of decreased oxidative stress in the high grade tumors.
Figure 4. 4. The glutamine pathway bolstered the glutathione system.
Combined proteomics and metabolomics data of human RCC tissue was overlayed onto a stylized KEGG-based pathway diagram. Levels of metabolites and enzymes were graphed grade dependently. Green, metabolite; orange, enzyme; black dot arrow, metabolism; red arrow, upregulated pathway; blue arrow, downregulated pathway; ROS, reactive oxygen species; GSSG, oxidized glutathione; GSH, glutathione; GPX1, glutathione peroxidase 1; GSTT1, glutathione S-transferase theta 1; GSTO1, glutathione S-transferase omega 1; GSTM3, glutathione S-transferase mu 3; GSTP1, glutathione S-transferase pi 1; GSTA2, glutathione S-transferase alpha 2; GSTK1, glutathione S-transferase kappa 1; GGT5, gamma-glutamyltransferase 5; GABA, gamma-aminobutyric acid; α-KG, α-ketoglutarate; GLS, glutaminase; ACY1, aminoacylase 1; ASS1, argininosuccinate synthase 1.
To validate this pathway, we examined the GSH pathway as a function of glutamine in vitro; cell viability assay results showed that RCC cell viability was decreased in both cell lines dose-dependently with glutamine depletion (Supplemental Fig. 2B). To further analyze the function of glutamine as a source for the GSH pathway, absolute quantitative GSH and GSSG levels and GSH/GSSG ratio were measured in cells grown in with and without glutamine. Levels of both GSH and GSSG and GSH/GSSG ratios decreased significantly in both cell lines in glutamine-depleted media (Supplemental Fig. 2C), consistent with our postulated role of glutamine in the tumoral anti-oxidant response system. To confirm that the effect of glutamine depletion correlated with a change in redox state, we evaluated the NADPH/NADP+ ratio in the glutamine-depleted media and found that the ratio was significantly decreased in the glutamine-depleted state in both cell lines (Supplemental Fig. 2D), further supporting an increased signature of oxidative stress in the absence of glutamine. Taken together, these findings confirm the necessity of glutamine for attenuating oxidative stress by kidney tumors.
The TCA cycle is down-regulated as a function of tumor grade
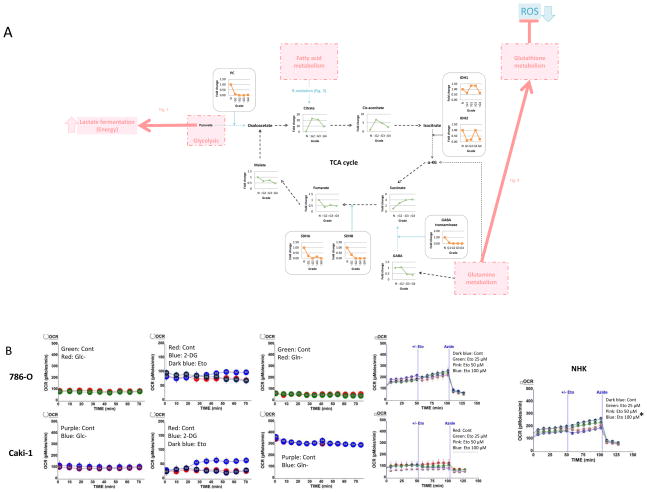
PC, one of the main routes by which pyruvate enters the TCA cycle, was decreased in RCC (Figs. 1 and 5A; Supplemental Table 4), and several enzymes regulating the “input” pathways to the TCA cycle, such as enzymes involved in β-oxidation and gamma-aminobutyric acid (GABA) transaminase were down-regulated as a function of grade (Figs. 3, 4, and 5A). Furthermore, the increased acetylcarnitine level suggested that acetyl-CoA was not being used to feed the TCA cycle (Fig. 3). Taken together, these findings suggest that three major inputs to the TCA cycle (glycolysis, fatty acid metabolism, and the glutamine pathway) are reprogrammed in RCC. To confirm this finding, we measured OCR, as a proxy for oxidative metabolism, in RCC cell lines treated with glucose depletion, etomoxir (inhibitor of CPT1), or glutamine depletion. OCR was not affected by inhibition of glycolysis, fatty acid metabolism, or glutamine metabolism in RCC cells (786-O and Caki-1) while NHK cells had a significantly decreased OCR with etomoxir treatment (Fig. 5B), indicating that these major energy pathways, all converging on the TCA cycle, were not significantly contributing to oxidative metabolism in two RCC cell lines while normal cells did rely more heavily on this pathway. These findings, in light of the above data indicating an increased transit through the aerobic glycolysis pathway, suggest that RCC cells do not rely on the TCA cycle but utilize lactate fermentation as their major energy source.
Figure 5. 5. The TCA cycle is not fed by glycolysis, the FA pathway, or the glutamine pathway in RCC.
(A) Combined proteomics and metabolomics data of human RCC tissue was overlayed onto a stylized KEGG-based pathway diagram. Green, metabolite; orange, enzyme; black dot arrow, metabolism; red arrow, upregulated pathway; blue arrow, downregulated pathway; ROS, reactive oxygen species; PC, pyruvate carboxylase; IDH, isocitrate dehydrogenase; GABA, gamma-aminobutyric acid; SDHA, succinate dehydrogenase complex subunit A; SDHB, succinate dehydrogenase complex subunit B.,
(B) Oxygen consumption rates (OCR) were measured with a Seahorse XF24 Analyzer in 786-O (RCC; top panels), Caki-1 (RCC; lower panels), and NHK (right panel) cells maintained in their growth media. Cells were treated 30 minutes prior to OCR measurement with glucose depleted media (Glc-; left panel), glutamine depleted media (Gln-; right panel), etomoxir (Eto) 50 μM, or 2-DG (middle panel) 5 mM unless stated otherwise. DMSO was used as the vehicle solution for etomoxir and 2-DG treatments. Data are the average OCR from six wells per group and error bars are the standard error of the mean. The data are representative of three repeats.
The tryptophan pathway results in indoleamine 2,3-dioxygenase (IDO)-dependent production of anti-inflammatory metabolites
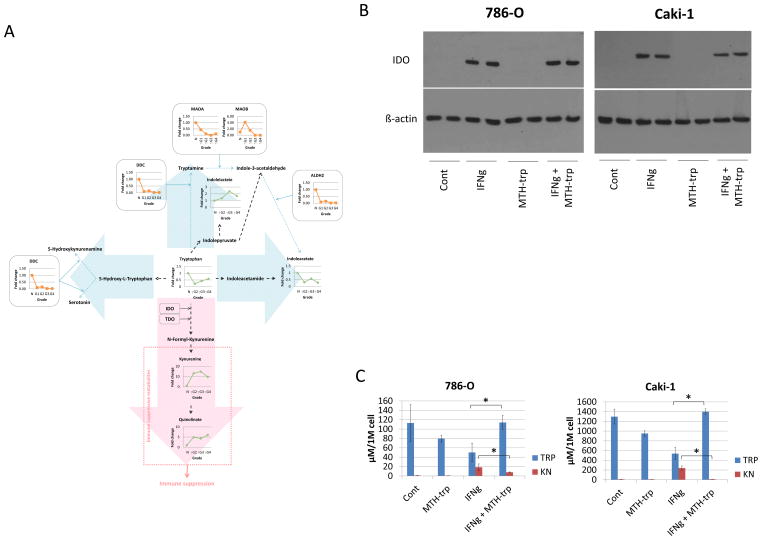
Tryptophan is an essential amino acid which undergoes multiple pathways of metabolism in normal and cancerous tissue, resulting in immunosuppressive and serotoninergic properties (32). Based on previously published work from our laboratory demonstrating high levels of immunosuppressive tryptophan metabolites in human RCC urine (7) and in human RCC xenograft mice urine and serum (6), we next asked whether, and in what manner, the tryptophan metabolic pathway is upregulated as a function of RCC grade. When tryptophan pathways were examined by combined metabolomic and proteomic analyses, the immunosuppressive metabolite kynurenine increased in tumors compared to normal tissues, the immunosuppressive metabolite quinolinate increased in grade 2 tumors compared to nor, and the activity of enzymes which catalyze tryptophan to the other known arms of tryptophan metabolism (including serotonin and indoleacetate) were decreased inversely proportional to tumor grade (Fig. 6A), consistent with our previously published data.
Figure 6. 6. Tryptophan metabolism favors a grade-dependent increase in immune suppressive metabolites.
(A) Combined proteomics and metabolomics data of human RCC tissue was overlayed onto a stylized KEGG-based pathway diagram. Green, metabolite; orange, enzyme; black dot arrow, metabolism; red arrow, upregulated pathway; blue arrow, downregulated pathway; MAO, monoamine oxidase; DDC, dopa decarboxylase; ALDH, aldehyde dehydrogenase; IDO, indoleamine 2,3-dioxygenase; TDO, tryptophan 2,3-dioxygenase.
(B) 786-O (VHL-mut) and Caki-1 (VHL-wt) cells were plated in 6-well plates the day before treating with either human IFN-g (50 ng/ml) and/or MTH-trp (100 μM) for four days and immunoblotted with the antibodies indicated.
(C) 786-O (VHL-mut) and Caki-1 (VHL-wt) cells were plated in 6-well plates the day before treating with either human IFN-g (50 ng/ml) and/or MTH-trp (100 μM) for three days. The conditioned media was harvested and tryptophan (TRP) and kynurenine (KN) were measured by HPLC and normalized for cell number counted using the cell viability assay kit (EMD Millipore, Billerica, MA) on a MUSE (EMD Millipore, Billerica, MA). Data shown are means (n=3) and standard deviations. *P < 0.05.
The cytokines IFNγ, IFNα, and IFNβ stimulate the transcription of IDO, the primary enzyme in the tryptophan pathways that produce immunomodulatory metabolites, and this finding may explain the relative lack of success when interferons are used clinically to treat advanced RCC. To validate the tryptophan pathway data, we showed that IFNγ increases IDO levels (Fig. 6B) and inhibition of IDO by MTH-trp results in increased tryptophan and decreased kynurenine levels in the culture media (Fig. 6C). The fact that an IDO inhibitor, 1-D-MT, is available and currently in clinical trials for other cancers (Clinicaltrials.gov NCT00567931), presents an incentive to evaluate IDO as a novel RCC therapeutic target, which work is underway in our laboratory.
Strengths and weaknesses of current study
A strength of our study is that it demonstrates that each Fuhrman grade of tissue possesses distinct biochemical characteristics with a specific magnitude of metabolic reprogramming, a phenomenon which is logical given the markedly disparate clinical behavior of tumors of different grades. In addition, despite tumor heterogeneity observed in RCC (33), it would appear that in this era of personalized medicine, and based on the differences which we have observed, it may now be more appropriate to evaluate patient responses to drug therapies with knowledge of their initial tumor grade being part of the clinical decision-making process.
There exist potential criticisms of this study, the most significant being that the same tissues were not used for both platforms (measurement of protein and metabolite levels). This was necessary given the nature of the study in which several investigators in different institutions collaborated on the project, and for this reason the human samples were collected separately. However, given the universally established criteria for Fuhrman grading and the use of experienced pathologists for determination of such, we have attempted to minimize potential inconsistencies from this issue. While there exists variability in any omics experiment, this possibility was minimized in this work by utilizing laboratories with abundant experience in non-targeted metabolomics and proteomics, both of which were based on mass spectrometry. To further minimize potential errors from this issue, validation of findings from each pathway was performed in vitro.
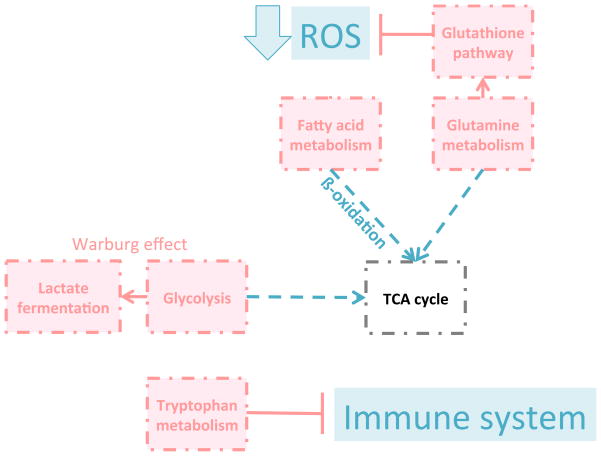
While some of our findings, such as enhancement of the Warburg effect as a function of increasing tumor grade, were not surprising given the known activation of hypoxia pathways in the majority of RCCs, other pathway alterations were quite unexpected and represent novel potential therapeutic targets for further research (Fig. 7). The overarching theme of our findings regarding energy metabolism are that (1) the Warburg effect seemed to be of prime importance for these tissues especially of high grades, at the expense of oxidative metabolism including fatty acid oxidation and transit through the TCA cycle, (2) the glutamine pathway functions to attenuate the high levels of ROS which have long been known to be present in RCC (34), and (3) tryptophan metabolism was upregulated leading to the generation of immune suppressive metabolites. These findings lead to obvious issues relevant to tumor targeting which in theory could be designed to defeat these mechanisms.
Figure 7. 7. Summary of grade-dependent metabolic pathway alterations in RCC.
With higher grade, glycolysis was directed towards lactate metabolism at the expense of TCA cycle intermediaries. Fatty acid β-oxidation was decreased, while the glutamine pathway served to attenuate oxidative stress thereby increasing cancer cell survival. Tryptophan was metabolized preferentially to immunosuppressive compounds. Red: increased; blue: decreased.
In summary, we have for the first time utilized data from a combination of metabolomics and proteomics studies to dissect energy and immune-relevant metabolic reprogramming of RCC to a high resolution and in a grade-dependent manner. These data show markedly different biochemistry of higher grade tumors, which suggests that stratification in the clinical setting is likely to be advantageous. In addition, the changes in energy metabolism that favor alterations of the oxidative stress and aerobic metabolism pathways, as well as generation of immune-suppressive tryptophan metabolites, have profound implications for designing new targets for this disease.
Supplementary Material
Acknowledgments
We thank Dr. Alan Buckpitt and Mr. Ken Chmiel for their assistance with this work.
Grant support
This work was supported by NIH grants 5UO1CA86402 (Early Detection Research Network), 1R01CA135401-01A1, and 1R01DK082690-01A1, and the Medical Service of the US Department of Veterans’ Affairs (all to R.H.W.), the LLNL-UCDCC Fitzpatrick Award (to H.I.W.), and grants from the Paula Moss Trust for the research into the cure and treatment of kidney cancer and the J. Randall & Kathleen L. MacDonald Research Fund in Honor of Louis V. Gerstner (to J.J.H.).
Reference List
- 1.Baker M. Big biology: The ‘omes puzzle. Nature. 2013;494:416–9. doi: 10.1038/494416a. [DOI] [PubMed] [Google Scholar]
- 2.Weiss RH, Kim K. Metabolomics in the study of kidney diseases. Nat Rev Nephrology. 2011;8:22–33. doi: 10.1038/nrneph.2011.152. [DOI] [PubMed] [Google Scholar]
- 3.Aboud OA, Weiss RH. New opportunities from the cancer metabolome. Clinical Chemistry. 2013;59:138–46. doi: 10.1373/clinchem.2012.184598. [DOI] [PubMed] [Google Scholar]
- 4.Perroud B, Ishimaru T, Borowsky AD, Weiss RH. Grade-dependent proteomics characterization of kidney cancer. Mol Cell Proteomics. 2008;8:971–85. doi: 10.1074/mcp.M800252-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perroud B, Lee J, Valkova N, Dhirapong A, Lin PY, Fiehn O, et al. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol Cancer. 2006;5:64. doi: 10.1186/1476-4598-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganti S, Taylor SL, Abu AO, Yang J, Evans C, Osier MV, et al. Kidney tumor biomarkers revealed by simultaneous multiple matrix metabolomics analysis. Cancer Res. 2012;72:3471–9. doi: 10.1158/0008-5472.CAN-11-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim K, Taylor SL, Ganti S, Guo L, Osier MV, Weiss RH. Urine Metabolomic Analysis Identifies Potential Biomarkers and Pathogenic Pathways in Kidney Cancer. OMICS. 2011;15:293–303. doi: 10.1089/omi.2010.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, et al. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics. 2009;8:558–70. doi: 10.1074/mcp.M800165-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchi C, Bombelli S, Raimondo F, Torsello B, Angeloni V, Ferrero S, et al. Primary cell cultures from human renal cortex and renal-cell carcinoma evidence a differential expression of two spliced isoforms of Annexin A3. Am J Pathol. 2010;176:1660–70. doi: 10.2353/ajpath.2010.090402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villas-Boas SG, Hojer-Pedersen J, Akesson M, Smedsgaard J, Nielsen J. Global metabolite analysis of yeast: evaluation of sample preparation methods. Yeast. 2005;22:1155–69. doi: 10.1002/yea.1308. [DOI] [PubMed] [Google Scholar]
- 11.Laich A, Neurauter G, Widner B, Fuchs D. More rapid method for simultaneous measurement of tryptophan and kynurenine by HPLC. Clin Chem. 2002;48:579–81. [PubMed] [Google Scholar]
- 12.Inoue H, Hwang SH, Wecksler AT, Hammock BD, Weiss RH. Sorafenib attenuates p21 in kidney cancer cells and augments cell death in combination with DNA-damaging chemotherapy. Cancer Biol Ther. 2011:12. doi: 10.4161/cbt.12.9.17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105–10. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120:142–56. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merrill CL, Ni H, Yoon LW, Tirmenstein MA, Narayanan P, Benavides GR, et al. Etomoxir-induced oxidative stress in HepG2 cells detected by differential gene expression is confirmed biochemically. Toxicol Sci. 2002;68:93–101. doi: 10.1093/toxsci/68.1.93. [DOI] [PubMed] [Google Scholar]
- 16.Fong MY, McDunn J, Kakar SS. Metabolomic profiling of ovarian carcinomas using mass spectrometry. Methods Mol Biol. 2013;1049:239–53. doi: 10.1007/978-1-62703-547-7_18. [DOI] [PubMed] [Google Scholar]
- 17.Evans AM, Dehaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 18.WARBURG O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 19.Unwin RD, Craven RA, Harnden P, Hanrahan S, Totty N, Knowles M, et al. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics. 2003;3:1620–32. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- 20.Kim JW, Dang CV. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–30. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 21.Firth JD, Ebert BL, Ratcliffe PJ. Hypoxic regulation of lactate dehydrogenase A. Interaction between hypoxia-inducible factor 1 and cAMP response elements. J Biol Chem. 1995;270:21021–7. doi: 10.1074/jbc.270.36.21021. [DOI] [PubMed] [Google Scholar]
- 22.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–34. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227–32. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreurs M, Kuipers F, van der Leij FR. Regulatory enzymes of mitochondrial beta-oxidation as targets for treatment of the metabolic syndrome. Obes Rev. 2010;11:380–8. doi: 10.1111/j.1467-789X.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 25.Ganti S, Taylor S, Kim K, Hoppel CL, Guo L, Yang J, et al. Urinary acylcarnitines are altered in kidney cancer. Int J Cancer. 2012;130:2791–800. doi: 10.1002/ijc.26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, et al. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5:e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paillasse MR, de MP, Amouroux G, Mhamdi L, Poirot M, Silvente-Poirot S. Signaling through cholesterol esterification: a new pathway for the cholecystokinin 2 receptor involved in cell growth and invasion. J Lipid Res. 2009;50:2203–11. doi: 10.1194/jlr.M800668-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koochekpour S, Majumdar S, Azabdaftari G, Attwood K, Scioneaux R, Subramani D, et al. Serum glutamate levels correlate with Gleason score and glutamate blockade decreases proliferation, migration, and invasion and induces apoptosis in prostate cancer cells. Clin Cancer Res. 2012;18:5888–901. doi: 10.1158/1078-0432.CCR-12-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martino JJ, Wall BA, Mastrantoni E, Wilimczyk BJ, La Cava SN, Degenhardt K, et al. Metabotropic glutamate receptor 1 (Grm1) is an oncogene in epithelial cells. Oncogene. 2013;32:4366–76. doi: 10.1038/onc.2012.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gameiro PA, Yang J, Metelo AM, Perez-Carro R, Baker R, Wang Z, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17:372–85. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adam W, Simpson DP. Glutamine transport in rat kidney mitochondria in metabolic acidosis. J Clin Invest. 1974;54:165–74. doi: 10.1172/JCI107738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung KT, Gadupudi GS. Possible roles of excess tryptophan metabolites in cancer. Environ Mol Mutagen. 2011;52:81–104. doi: 10.1002/em.20588. [DOI] [PubMed] [Google Scholar]
- 33.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto K, Toyokuni S, Kim WJ, Ogawa O, Kakehi Y, Arao S, et al. Overexpression of human mutT homologue gene messenger RNA in renal-cell carcinoma: evidence of persistent oxidative stress in cancer. Int J Cancer. 1996;65:437–41. doi: 10.1002/(SICI)1097-0215(19960208)65:4<437::AID-IJC7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.