Abstract
The vertebrate heart arises from distinct first and second heart fields. The latter also share a common origin with branchiomeric muscles in the pharyngeal mesoderm and transcription regulators, such as Nkx2–5, Tbx1 and Islet1. Despite significant progress, the complexity of vertebrate embryos has hindered the identification of multipotent cardiopharyngeal progenitors. Here, we summarize recent insights in cardiopharyngeal development gained from ascidian models, among the closest relatives to vertebrates. In a simplified cellular context, progressive fate specification of the ascidian cardiopharyngeal precursors presents striking similarities with their vertebrate counterparts. Multipotent cardiopharyngeal progenitors are primed to activate both the early cardiac and pharyngeal muscles programs, which segregate following asymmetric cells divisions as a result of regulatory cross-antagonisms involving Tbx1 and Nkx2–5 homologs. Activation of Ebf in pharyngeal muscle founder cells triggers both Myogenic Regulatory Factor-associated differentiation and Notch-mediated maintenance of an undifferentiated state in distinct precursors. Cross-species comparisons revealed the deep conservation of the cardiopharyngeal developmental sequence in spite of extreme genome sequence divergence, gene network rewiring and specific morphogenetic differences. Finally, analyses are beginning to uncover the influence of surrounding tissues in determining cardiopharyngeal cell identity and behavior. Thus, ascidian embryos offer a unique opportunity to study gene regulation and cell behaviors at the cellular level throughout cardiopharyngeal morphogenesis and evolution.
Introduction
The heart is a muscular organ that pumps blood through a circulatory system. In mammals, the embryonic heart tube is the first functional organ and further morphogenesis leads to a four-chambered organ. The frequency and diversity of congenital heart diseases reflects the complexity of cardiac morphogenesis. Discrete mesodermal cell populations, referred to as the first and second heart fields, are recruited sequentially to form the heart tube and progressively add tissue to the growing heart [1–5]. The first heart field (FHF) gives rise to the early embryonic heart tube, while the second heart field (SHF) later contributes to both arterial (e.g. outflow tract and right ventricle) and venous (e.g. right atrium) poles, (reviewed in [6]). Clonal analyses in the mouse demonstrated that cardiomyocytes of FHF and SHF origins derive from common progenitors that were initially thought to originate in the Mesp1+ mesoderm of the early embryo [7][8,9]. Further analyses revealed a common pharyngeal origin of the SHF and the branchiomeric/pharyngeal muscles [10–14]. Shared molecular determinants of the SHF and pharyngeal muscles have been identified and linked to the Cardio-Velo-Facial/DiGeorge syndrome, in which the 22q11.2 deletion removes the transcription factor TBX1 and is responsible for malformations of pharyngeal apparatus and cardiac outflow tract [15,16]. Developmental and genetics studies point to essential roles for the homeobox genes Islet1/Isl1 and Nkx2–5 alongside Tbx1 in the cardiopharyngeal mesoderm, the source of SHF and branchiomeric muscles progenitors [13,17–21]. Mammalian cardiopharyngeal progenitors and the mechanisms underlying early heart vs. branchiomeric muscle specification remain elusive due to the complexity and relative inaccessibility of the early embryos.
The ascidian Ciona intestinalis has emerged as a simple chordate model to study early cardiac development with cellular resolution [22–24]. As tunicates, ascidians are marine invertebrates among the closest living relatives of the vertebrates [25–27]. Tunicates and vertebrates form the clade Olfactores [27]. A model tunicate, Ciona uniquely combines genetic and cellular simplicity, experimental amenabilities and olfactores-specific traits, which are lacking in distant genetic models including flies or nematodes [28]. The adult Ciona heart consists of U-shaped tube comprising two monolayers of cells: an external pericardium surrounding a contractile myocardium, with no endocardium [29–31]. The ascidian heart derives from a single pair of bilateral blastomeres in the 110-cell stage embryo [31,32]. The B7.5 blastomeres, named after Conklin, and their daughter cells, the B8.9 and B8.10 founder cells, transiently express the Mesp/Mesogenin sole pro-ortholog [31]. As in vertebrates, early Mesp function is crucial for heart development in Ciona [31]. The founder cells then divide asymmetrically to produce two anterior tail muscle cells (ATMs) cells and their sister cells, the trunk ventral cells (TVCs), which migrate towards the ventral side of the trunk (Figure 1; [30,31,33]). TVC specification and migration are controlled by the sequential activation of the FGF-MAPK-Ets signaling pathway and the transcription factor FoxF ([34–36]; reviewed in [22,24]). Migrating TVCs activate conserved regulators of cardiac development including Nk4/Nkx2–5, Hand and Gata4/5/6 homologs [30,31]. The TVCs are common progenitors for the juvenile heart, atrial siphon muscles (ASM) and longitudinal body wall muscles (LoM) [32,37]. The latter muscle populations derive from Islet+ and Tbx1/10+ precursors, constituting likely homologs of the vertebrate branchiomeric muscles. Here, we review recent progress in characterizing the TVCs as multipotent cardiopharyngeal progenitors and the mechanisms underlying fate choices in their progeny. We present how the cardiopharyngeal ontogenetic motif is conserved amid specific developmental variations and gene regulatory drift in distantly related tunicates. Finally, we summarize recent insights into the crosstalk between extrinsic influences, intrinsic properties and cell behavior during TVC induction and collective migration.
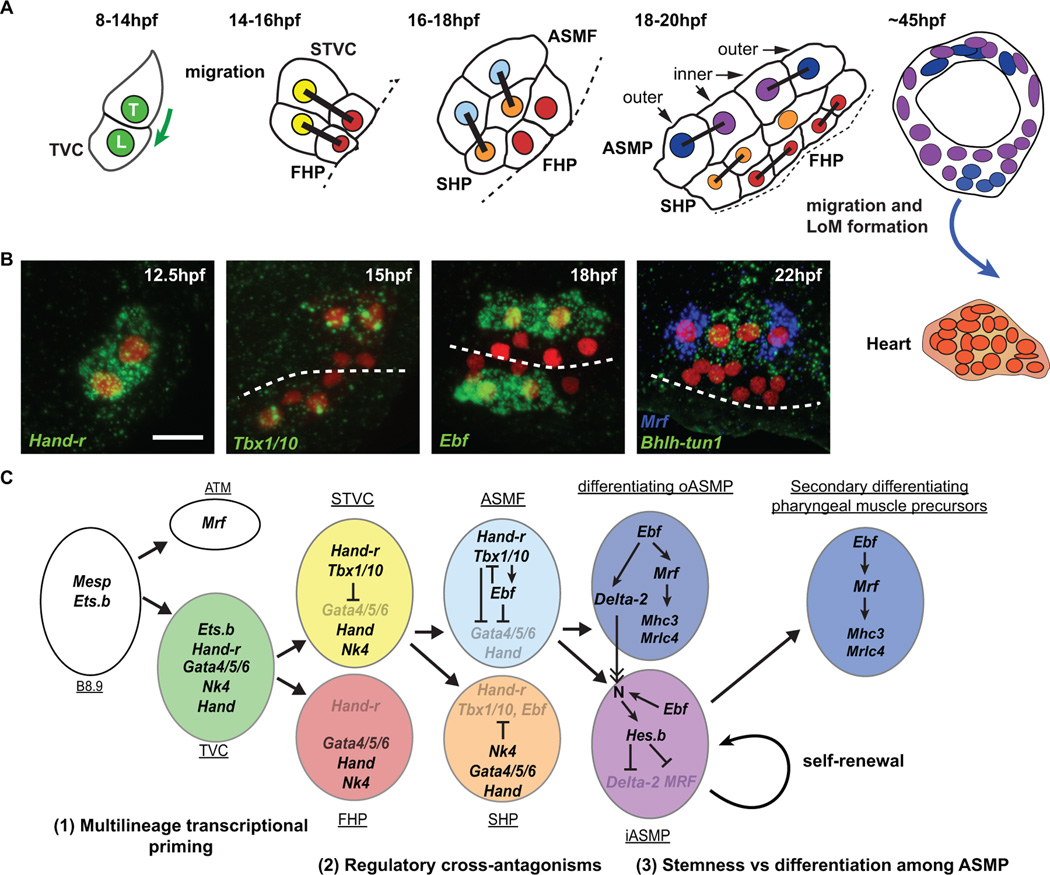
Figure 1. Schematic representation of cardio-pharyngeal development in Ciona intestinalis.
(A) Schematic showing the trunk ventral cell progeny with divisions and migrations (green and blue arrows), from initial tail bud stage (8 hpf) to metamorphosing juvenile (45 hpf). Trunk ventral cells (TVC, green), STVC (secondary TVC, yellow), first heart precursors (FHP, red), atrial siphon muscle founder cells (ASMF, light blue), second heart precursors (SHP, orange), inner and outer ASM precursors (ASMPs, dark blue and violet, respectively), heart (red/orange). The first longitudinal muscle (LoM) derives from the ASM ring. The left side only is presented, linked nuclei indicate sister cells, dashed lines represent the midline. (B) Fluorescent in situ hybridization of key markers of the TVC progeny from 12.5 hpf to 22 hpf. The B7.5 lineage (red) is marked with Mesp>NLS::lacZ revealed by anti β-galactosidase immunostaining. Scale bar, 10 µm. (C) Summary of the ontogenetic interactions of the B7.5 cardiopharyngeal lineage. The TVC are transcriptionally primed pluripotent progenitors (1). Heart (e.g. Gata4/5/6, Hand) and ASM (e.g. Hand-r, Tbx1/10) transcriptional regulatory programs are segregated through cross-antagonisms coupled to asymmetric divisions (2). A myogenic program associated with Mrf is deployed downstream of Ebf, which also promotes Notch-mediated lateral inhibition of Mrf and maintenance of a pool of stem cell-like muscle progenitors (3). Same color code as above.
TVC are multilineage-primed multipotent cardiopharyngeal progenitors
Following migration into the trunk and association with the ventral endoderm, each TVC divides asymmetrically into a small median first heart precursor (FHP) and a large lateral secondary TVC (STVC) [33,37]. The latter divide again asymmetrically into a small medial second heart precursor (SHP) and a large atrial siphon muscle founder cell (ASMF; Figure 1; [37–39]). ASMFs immediately activate the atypical helix-loop-helix transcription factor Ebf (previously referred to as COE, for Collier/Olf/EBF; [37,40]). Gain- and loss-of-function assays, using either a repressor form or lineage-specific CRISPR/Cas9 constructs, indicated that Ebf promotes ASM specification at the expense of the heart fate [37,41]. Thus, the mechanisms controlling ASMF-specific expression of Ebf determine the initial heart vs. ASM fate choice in the ascidian cardiopharyngeal mesoderm.
Tissue-specific transcription profiling using fluorescence activated cell sorting (FACS) and microarrays characterized the transcriptional dynamics underlying heart vs. ASM fate choice [38]. Time-series and Ebf-perturbations datasets suggested that Ebf-inhibited genes, viewed as candidate heart-specific genes, were first expressed in the TVC prior to asymmetric cell divisions. However, fluorescent in situ hybridizations assays revealed that asymmetric divisions are accompanied by progressive restriction of expression of distinct TVC genes into either the heart precursors or the STVCs and then ASMFs [38]. Among the TVC genes restricted to the STVCs and ASMFs, Hand-r (Hand-related, previously Hand-like/NoTrlc; [31,40,42]) is necessary for Ebf expression. On the other hand, Gata4/5/6 and Hand expressions become restricted to the heart precursors. Thus, the TVCs are transcriptionally primed for both pharyngeal and cardiac fate specification. Such multilineage transcriptional priming of multipotent progenitors is common in ascidians [43] and in vertebrate hematopoiesis but has not been documented in vertebrate cardiopharyngeal mesoderm [38]. Instead, studies using stem cell models for mammalian cardiogenesis revealed "chromatin priming", whereby cardiac enhancers are poised for future activation in mesoderm progenitors [44,45]. Future studies will determine whether late ASM- and/or heart-specific enhancers are also "primed" in multipotent cardiopharyngeal progenitors.
Regulatory cross-antagonisms segregate the early heart and ASM programs
Multilineage transcriptional priming of cardiopharyngeal progenitors begs the question as to how the segregation of heart and pharyngeal muscle programs is coordinated with asymmetric cell divisions. Among the primed transcriptional regulators, Tbx1/10 is first expressed in the bipotent STVCs and preferentially maintained in the ASMFs, where it is necessary for Ebf expression [39]. Tbx1/10 function is also necessary and sufficient to inhibit Gata4/5/6 expression and subsequent heart specification. Conversely, the Nkx2.5 homolog Nk4 is expressed throughout the cardiopharyngeal lineage and its repressor function is necessary to shut off Tbx1/10 expression and prevent ectopic Ebf activation in the SHPs. This data indicated that STVCs' multilineage transcriptional priming is resolved through regulatory cross-antagonisms between early cardiopharyngeal regulators, which segregate the ASM and heart programs to their corresponding fate-restricted precursors [39]. Similar regulatory antagonisms involving Nkx2.5, Tbx1 and Gata4/5/6 homologs contribute to delaying cardiac differentiation in the mouse SHF, thus permitting proliferation and growth of the developing heart [19,21,46].
Ebf promotes differentiation and stemness in distinct pharyngeal muscle precursors
Ebf promotes subsequent ASM development including collective migration of ASM precursors (ASMP) and muscle differentiation. Ebf activates a myogenic program associated with upregulation of the sole myogenic regulatory factor Mrf [38,47]. Following division of the ASMFs, Mrf expression becomes restricted to the anterior- and posterior-most "outer" ASMPs, which are the only cells to activate muscle differentiation genes (Figure 1; [38]). By contrast, Notch activation in the "inner" ASMP upregulates the repressor Hes-b, which inhibits Mrf expression and subsequent differentiation. Instead, "inner" ASMPs proliferate and later produce the full complement of ASMs and LoMs in juveniles (Figure 1; [38]). Thus, Tbx1/10 and subsequent Ebf activities govern both Mrf-associated differentiation and the Notch-dependent maintenance of an undifferentiated, stem cell-like, progenitor state among distinct pharyngeal muscle precursors. This developmental progression is reminiscent of the vertebrate situation where Tbx1 governs branchiomeric myogenesis by controlling MyoD expression [48–50]. In vertebrates, producing the full complement of muscle cells also requires Notch-mediated inhibition of precocious differentiation and proliferation [51,52]. Head muscle myofibers and associated stem cells also derive from common progenitors in the early embryo [53]. Ebf homologs act upstream of MyoD and Myf5 during pharyngeal and somitic myogenesis [54]. This suggests a conserved role for Ebf homologs in branchiomeric myogenesis [54–56]. Thus, ASM development in ascidians likely recapitulates key developmental transitions conserved with the branchiomeric muscles of vertebrates.
A cardiopharyngeal ontogenetic motif for chordate heart and head muscle development?
Successive gene expression profiles define transient cell identities in the cardiopharyngeal mesoderm. Together with a conserved clonal topology of the lineage, this invites a specific parallel between vertebrate and ascidian cardiopharyngeal development (Figure 1). Namely, early B-line blastomeres, TVCs, STVCs, FHPs, SHPs and ASMPs would be the ascidian counterparts of putative common Mesp1+ mesoderm progenitors, Nkx2.5+ pan-cardiopharyngeal progenitors, Nkx2.5+/Tbx1+ second cardiopharyngeal progenitors (common to the SHF and branchiomeric muscles), FHF precursors, SHF precursors and branchiomeric muscle precursors, respectively (Figure 1).
Recent lineage studies challenged the existence of common Mesp1+ pan-cardiopharyngeal progenitors in mice. Using distinct clonal analyses, both Lescroart et al. [57] and Devine et al. [58] conclude that cardiac progenitors are fate-restricted before they turn on Mesp1, which is first expressed in FHF and only later in the SHF, consistent with independent cardiac lineages deriving from distinct Mesp1+ progenitors. Vertebrate Mesp genes function in a context-dependent manner, integrating stage of differentiation and signaling environment in precursors that are committed to certain fates [59]. Nevertheless, the study by Lescroart et al. supports the existence of bipotent Mesp1+ progenitors for SHF cardiomyocytes and branchiomeric myocytes, in line with retrospective clonal analysis [12,60]. In Ciona, precocious expression of Mesp precedes cell fate diversification suggesting that gene network deployment can be evolutionarily decoupled from progressive fate restrictions. Similarly, although Tbx1/10 is activated in the common progenitors of the SHPs and ASMPs in Ciona, Tbx1 expression in vertebrates may start independently in distinct SHF and branchiomeric muscle progenitors [49,61,62].
Evolutionary changes in cardiopharyngeal development among distant ascidians
Tunicate and vertebrate embryos differ profoundly, rendering tentative comparisons between developmental processes arduous. In vertebrate development, cell-cell signaling is crucial to pattern plastic populations of thousands of progenitor cells. By contrast, the small number of progenitors in the ascidian embryo imposes hardwiring of every cell fate decision. These are thought to have fostered deep evolutionary conservation of early ascidian embryogenesis to the point that early cleavage patterns and cell lineages are perfectly conserved between the Stolfidobranchia and Phlebobranchia suborders of ascidians, despite over 500 million years of evolutionary divergence [63,64] (Figure 2A).
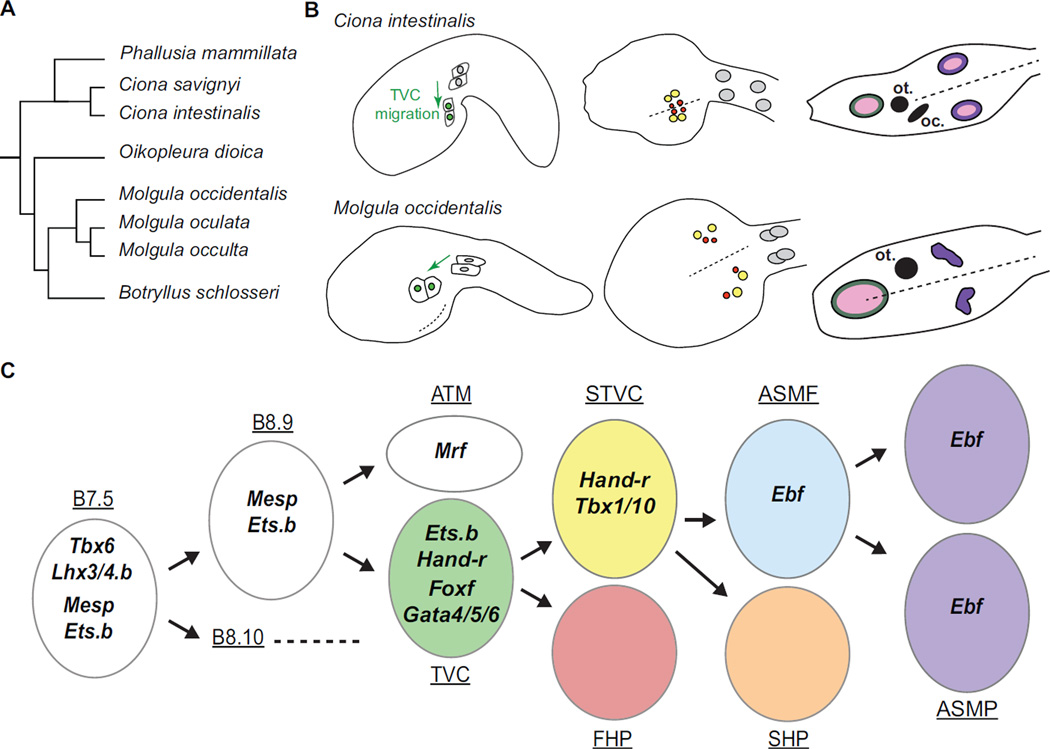
Figure 2. Variations on a conserved ontogenetic motif: comparison of the cardiopharyngeal development in Ciona intestinalis and Molgula occidentalis.
(A) Simplified evolutionary tree of Tunicates based on 18s phylogeny, adapted from [92]. The Ciona and Molgula genera are positioned on the two most divergent branches within Tunicates. (B) Comparative schematic showing the B7.5 lineage in Ciona intestinalis and Molgula occidentalis. In both species, the TVCs (green) migrate away from the ATMs. In Ciona intestinalis, the TVCs converge at the midline of the ventral trunk before they divide into STVCs (yellow) and FHPs (red). In Molgula occidentalis, the TVCs divide more laterally into STVCs and FHPs, resulting in distinct bilateral clusters of heart precursors. In both species, the oral siphon muscle precursors (dark green) derive from a different cell lineage and form a ring surrounding a single oral placode (pink). In Ciona intestinalis, 4 ASMPs migrate towards the atrial placode (pink), divide and from a rings of 8 cells (violet) on either side. In Molgula occidentalis, the absence bilateral atrial placodes causes the ASMPs to remain as two bilateral clusters of cells. (C) Summary of the ontogeny of the B7.5 cardiopharyngeal lineage of Molgula occidentalis. Conservation of the cell division patterns is coupled to conserved spatiotemporal expression patterns of the main markers of the B7.5 lineage. Same color code as above
This divergence is reflected in the genome sequences of three Stolidobranch species of the genus Molgula and the two Ciona species [65]. Yet, early Ciona and Molgula embryos are virtually indistinguishable. The successive gene expression, cell divisions and progressive fate specification events in the M. occidentalis B7.5 lineage are nearly identical to that in Ciona (Figure 2 B and C; [65]). These homologous developmental sequences demonstrate the ancestral origin of the ascidian cardiopharyngeal ontogenetic motif and allow to study how profoundly divergent genomes encode extremely conserved embryonic processes [64,66].
Functional B7.5- and TVC-specific enhancers for Molgula Mesp, FoxF and Hand-r were identified. These enhancer constructs fail to drive reporter gene expression when transfected in Ciona embryo, interpreted as an acute form of developmental system drift. The homologous Ciona enhancers showed variable degree of activity in Molgula embryos, revealing widespread interspecific unintelligibility of regulatory mechanisms controlling otherwise identical patterns of gene expression.
Interspecific differences in cell behavior were also apparent. In M. occidentalis, TVCs migrate into the trunk following a more lateral migration path than observed in Ciona. This causes M. occidentalis heart precursors to coalesce only later, during metamorphosis, and transiently display two disjointed clusters of cardioblasts in the trunk (Figure 2C). This condition has only been induced in Ciona by endoderm-specific disruption of Gata4/5/6 function [67]. Although midline convergence followed by asymmetric and oriented TVC divisions is a stereotyped cell behavior sequence in Ciona, its evolutionary remodeling in Molgula suggests that the ascidian cardiopharyngeal development is a modular assembly of specific morphogenetic events.
Another morphogenetic difference concerns the ASMPs. In Ciona larvae, bilateral 4-cell ASMP clusters migrate towards atrial siphon placodes (ASP), where they divide and form conspicuous 8-cell rings (Figure 2C; [37]). Only during metamorphosis do bilateral anlagen fuse to form a single atrial siphon [68]. By contrast, ASM ring formation is not observed in Molgula larvae, where the single ectodermal ASP is not yet specified. This presumably causes Molgula ASMP to migrate dorsally into a cluster without forming a ring, as observed in Hox1 mutant Ciona larvae lacking ASP [69]. These differences point the plasticity of cell-cell interactions that may have fostered diversification of cardiopharyngeal forms, while preserving ancestral patterns.
Intrinsic vs. extrinsic control of cardiopharyngeal cell fate and behavior
Ascidians offer a unique opportunity for high-resolution studies of chordate cardiopharyngeal development [22–24]. Transcriptional inputs from Mesp and FGF-MAPK-Ets govern TVC induction and their collective migratory behavior, in part through regulating FoxF [31,33,34,36]. Studies in vertebrates identified key roles for MESP1, ETS2, FoxF1 and FoxF2 in controlling specific aspects of mammalian heart specification and morphogenesis [70,71]. Whole genome studies of FACS-purified TVC cells identify distinct waves of transcriptional activation of genes in the TVCs prior to their divisions [35,72]. “Primary” TVC genes, such as FoxF and Hand-r are likely direct transcriptional targets of Ets.b as soon as the TVCs are born, while transcription of “secondary” TVC genes, such as the small GTPase-encoding RhoD/F, starts later in response to feed-forward inputs from MAPK-activated Ets.b and primary targets like FoxF [35,72].
Gene regulatory networks governing progressive cell fate specification also determine transient cell behaviors. Previous analyses identified numerous TVC-specific candidate migration effector genes involved in basic cellular processes, including actin dynamics, cell polarity and cell-matrix adhesion [35]. The abundance of genes encoding trans-membrane proteins and signaling molecules in the TVC transcriptome stresses the importance of the extracellular environment for progressive cardiopharyngeal fate specification and morphogenesis. Recent studies advanced our understanding of the roles of surrounding tissues in ascidian cardiopharyngeal development.
Asymmetric cell-matrix adhesion polarize MAPK activity and TVC induction
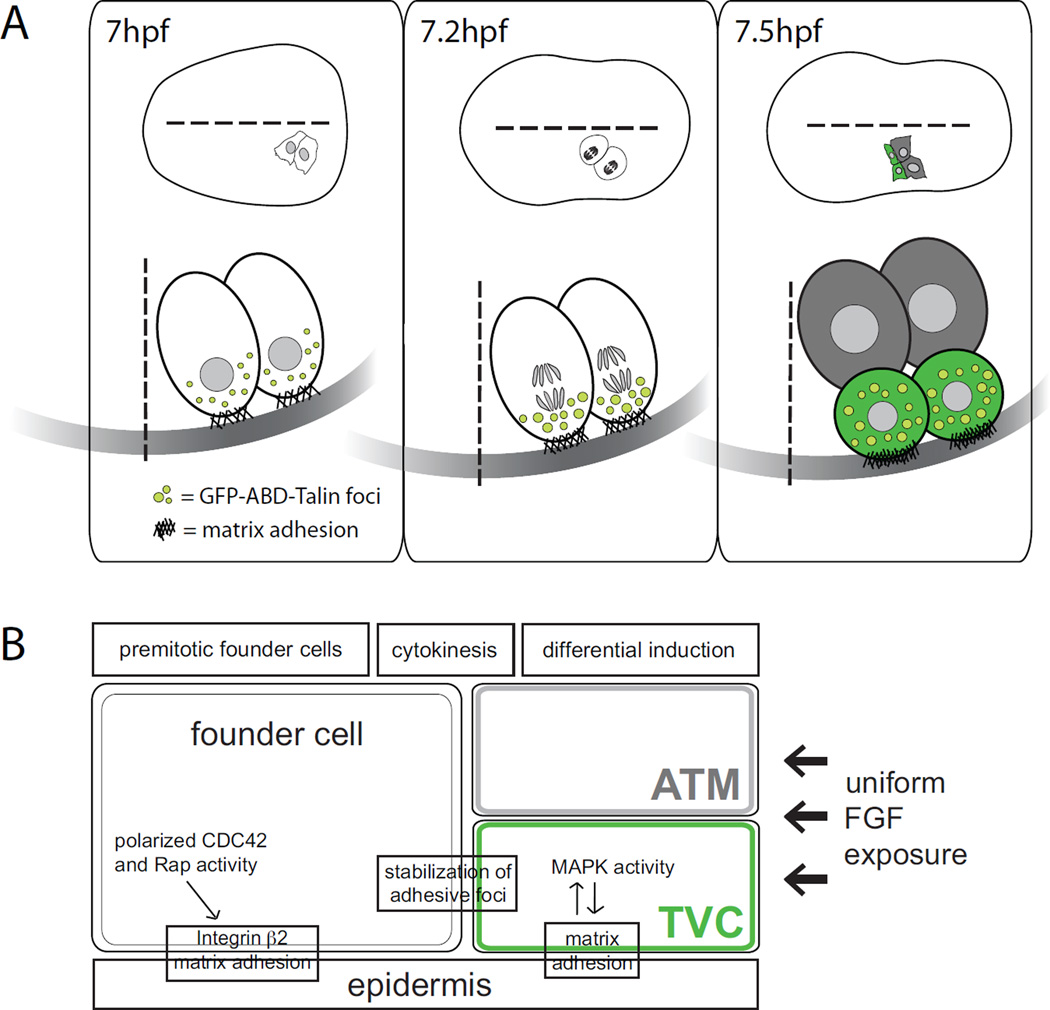
High-resolution analyses uncovered an essential role for cell-matrix adhesion in polarizing the signaling events controlling TVC fate specification [22,73,74]. Following asymmetric division of the B8.9 and B8.10 founder cells, FGF-MAPK signaling induces the TVCs [36]. MAPK activation triggers Ets-dependent transcription only in the two ventral/anterior-born cells from the asymmetric division (i.e. the prospective TVCs), even though founder cells seem to be uniformly exposed to an FGF9/16/20 ligand [74]. This differential induction of TVC fate is achieved by polarized matrix adhesion of the founder cells to the underlying epidermis [73,74]. A GFP-tagged talin actin-binding domain, reporting actin accumulation at adhesive foci, was enriched at the ventral founder cell membrane, re-distributed to the ventral edge throughout mitosis, and inherited by newborn TVCs after cytokinesis [73]. Cell-matrix adhesion requires Rap GTPase-dependent integrin β2 activation, which appeared necessary and sufficient to promote MAPK activation and TVC induction (Figure 3; [73]). This work parallels documented roles for integrin-mediated cell-matrix interactions during vertebrate cardiac development [75,76].
Figure 3. Model showing asymmetric induction of TVC fate by regional matrix adhesion.
(A) Dorsal (top) and lateral (bottom) views of schematic B8.9 and B8.10 founder cells at neurula stage (~7hpf). GFP-tagged actin binding domain of Talin labels actin at adhesive foci (light green), which localize ventrally in founder cells before they divide; black hashes represent matrix adhesion to the underlying epidermis at the ventral border of founder cells. During mitosis, GFP-ABD-Talin foci increase in size and intensity, which correlates with stronger adhesion to the underlying ECM. After cytokinesis (~7.5hpf), two different fates are specified: ATM (gray), and TVC (green). GFP-ABD-Talin foci are inherited by TVCs, and ventral edges of newly born TVCs appear to protrude into the underlying epidermis. (B) Model of regional MAPK activation and asymmetric TVC induction despite uniform FGF exposure. Polarized Rap activity in founder cells leads to regional stabilization of adhesion during cytokinesis at the presumptive TVC membrane in founder cells. Strong differential adhesion in newly born TVCs leads to differential MAPK activity and finally to TVC specification.
Surrounding tissues canalize cardiac progenitor migration
Following induction, TVCs collectively polarize and migrate in response to coordinated transcriptional inputs, extracellular cues and signaling events. Previous analyses revealed that Gata4/5/6 plays cell autonomous and non-cell autonomous roles during Ciona heart development [67], as is the case in vertebrates [77–83]. In Ciona TVCs, Gata4/5/6 function is required for FoxF, Nk4, and Bmp2/4 expression and migration [67]. Gata4/5/6 is also required in the adjacent endoderm for bilateral pairs of migrating TVCs to converge and meet at the midline [67]. The resulting ‘split heart’ phenotype is reminiscent of the vertebrate cardia bifida condition [79–83]. Thus, directional heart progenitor migration in vertebrates and ascidians requires interactions with the endoderm, among other surrounding tissues, for proper positioning.
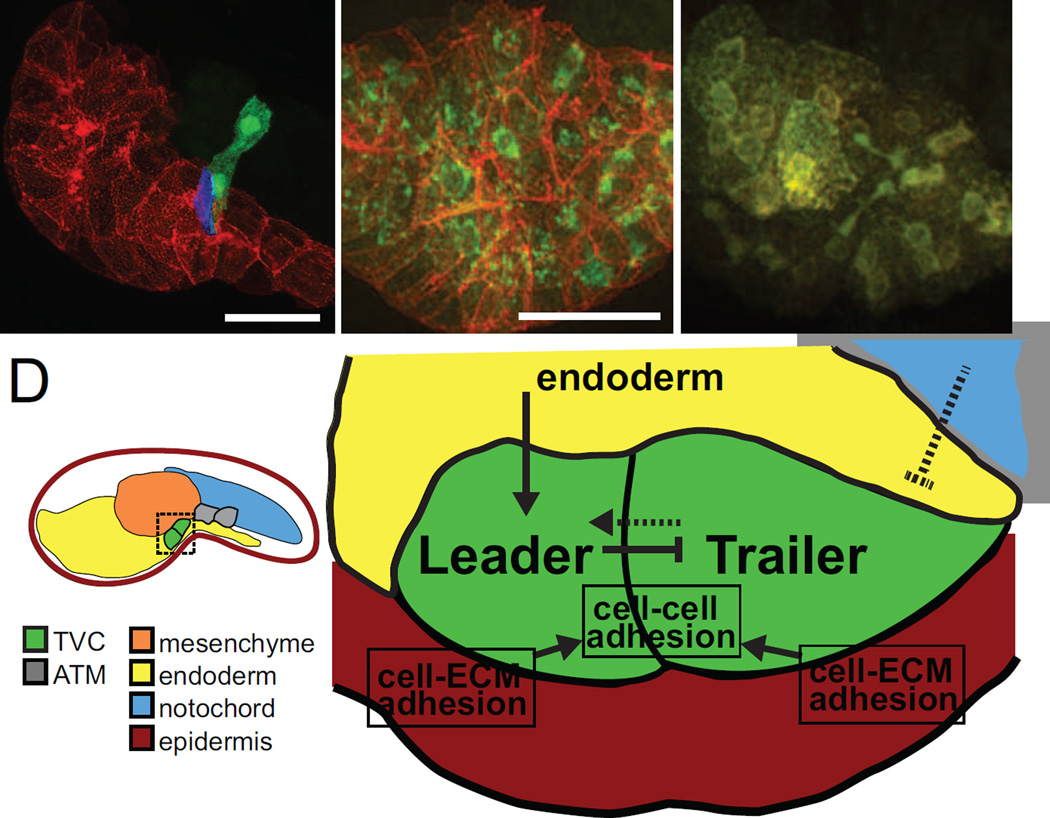
Systematic tissue-specific disruption of the secretory pathway and quantitative analyses probed the influence of surrounding tissues on collective TVC migration [84]. Quantification of the contact between TVC and surrounding tissues showed that the TVC are born touching each other, the mesenchyme, the epidermis, and their sister cells, the anterior tail muscle (Figure 4). The leader TVC usually arises for the anterior-most founder cell and is initially the only one to contact the trunk endoderm, but each TVC is capable of migrating individually, albeit imperfectly. The secretory pathway was inhibited in each TVC-surrounding tissue by expressing a dominant negative form of the small GTPase Sar1 to block endoplasmic reticulum to Golgi transport. Analyses revealed (1) a role for the mesenchyme in robust specification of the trailer TVC; (2) a role for the notochord in the timely initiation of TVC migration, possibly through chemorepulsion; (3) a role for the endoderm in the establishment and maintenance of leader-trailer polarity and (4) a role for the epidermis in TVC guidance and cell-cell adhesion. These data suggested that a combination of influences from surrounding tissues canalizes intrinsically motile TVCs towards stereotyped collective polarity and directed migration. Future studies will determine the signal and transduction pathways of TVC-specific interpretation of external cues.
Figure 4. Surrounding tissues canalize TVCs towards directional migration.
(A) Initial tailbud embryo expressing membrane-localized reporter hCD4::mCherry in the endoderm (red), and GFP in the B7.5 lineage (green). Leader and trailer TVCs are shown. Surface contact analysis between TVC and endoderm indicates that only the presumptive leader TVC contacts the endoderm (blue). Scale bar, 40 µm. (B) Localization of membrane localized protein hCD4::mCherry (red) at cell membranes and KDEL receptor KDELR::GFP (green) labelling the endoplasmic reticulum (ER) in control embryos. Scale bar, 40 µm. (C) In embryos expressing dnSar1, hCD4::mCherry accumulates in the ER and cannot be properly trafficked to the membrane, indicated by the co-localization of red and green at the ER, and the absence of red fluorescence at cell membranes. (D) Schematic mid-tailbud embryo showing TVCs (green) and ATMs (gray) and tissues analyzed by Gline et al. by tissue-specific disruption of the secretory pathway: epidermis (red), mesenchyme (orange), endoderm (yellow) and notochord (blue). Model for extrinsic tissue influences on TVCs and intercellular interactions during TVC migration. The endoderm (yellow) signals to the TVCs (green) to establish leader-trailer polarity and TVCs in turn signal to each other to maintain polarity during migration. The notochord (blue) possibly sends chemorepulsive signals to migrating TVCs. TVCs adhere to each other during migration and also to the underlying epidermis (red) through cell-ECM interactions.
Concluding remarks and future directions
Recent studies have furthered our understanding of conserved features of heart and pharyngeal muscle development between vertebrates and ascidians. Despite variations in cell topology due to larger cell populations in vertebrates, little doubt remains about the homology of vertebrate and ascidian cardiopharyngeal mesoderm [85]. Deployment of a core cardiopharyngeal gene regulatory network follows similar spatio-temporal sequences in both groups, from early mesoderm with Tbx6 homologs acting upstream of Mesp/Mesogenin genes [86–88], to muscle specification and differentiation controlled by Ebf, Mrf and Delta/Notch homologs.
Ebf function in pharyngeal myogenesis, first studied in Ciona, illustrates the relevance of ascidian models, building on their amenability for high-resolution analyses, to uncover the conserved molecular basis of cardiopharyngeal development in chordates. Outstanding questions remain about the polarized cell-cell signaling in asymmetrical fate choices; the chromatin dynamics underlying multilineage priming, early fates' segregation and commitment to either a cardiac or pharyngeal muscle identity; the molecular underpinnings of collective polarity and directed migration and the genetic changes underlying developmental system drift.
Computational approaches using whole genome data from an increasing number of ascidian species will empower gene regulatory network comparisons across ascidians and uncover fundamental rules governing cardiopharyngeal mesoderm evolution.
Future studies will dissect the intricate relationships between TVCs intrinsic transcriptional inputs and signaling pathways that interpret behavioral cues from the environment. They will benefit from the recent development of reliable methods for gene loss-of-function using electroporation, including short hairpin RNA-mediated RNAi [38,39,89] and tissue-specific targeted genome editing using TALEN and/or CRISPR/Cas9 technologies [41,90,91].
Acknowledgments
We are grateful to Christiaen lab members for discussions. N.K. was supported by the NIH 5 T32 HD007520-14 training program in developmental genetics. Our work is supported by NIH/NIGMS grant R01GM096032 and NIH/NHLBI grant R01HL108643 to L.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 2.Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol. 2013;5:a008292. doi: 10.1101/cshperspect.a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. 2010;107:1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 5.Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. doi: 10.1016/S0070-2153(10)90001-X. [DOI] [PubMed] [Google Scholar]
- 6.Kelly RG. The second heart field. Curr Top Dev Biol. 2012;100:33–65. doi: 10.1016/B978-0-12-387786-4.00002-6. [DOI] [PubMed] [Google Scholar]
- 7.Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell. 2004;6:685–698. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- 8.Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 9.Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med. 2000;10:345–352. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- 10.Harel I, Maezawa Y, Avraham R, Rinon A, Ma HY, Cross JW, Leviatan N, Hegesh J, Roy A, Jacob-Hirsch J, et al. Pharyngeal mesoderm regulatory network controls cardiac and head muscle morphogenesis. Proc Natl Acad Sci U S A. 2012;109:18839–18844. doi: 10.1073/pnas.1208690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lescroart F, Hamou W, Francou A, Theveniau-Ruissy M, Kelly RG, Buckingham M. Clonal analysis reveals a common origin between nonsomite-derived neck muscles and heart myocardium. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1424538112. ◦ Using retrospective clonal analysis, this paper shows that neck muscles have a branchiomeric origin and share a clonal relationship with cardiomyocytes of the venous pole, which derives from the posterior second heart field.
- 12.Lescroart F, Kelly RG, Le Garrec JF, Nicolas JF, Meilhac SM, Buckingham M. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development. 2010;137:3269–3279. doi: 10.1242/dev.050674. [DOI] [PubMed] [Google Scholar]
- 13.Nathan E, Monovich A, Tirosh-Finkel L, Harrelson Z, Rousso T, Rinon A, Harel I, Evans SM, Tzahor E. The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development. 2008;135:647–657. doi: 10.1242/dev.007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tirosh-Finkel L, Elhanany H, Rinon A, Tzahor E. Mesoderm progenitor cells of common origin contribute to the head musculature and the cardiac outflow tract. Development. 2006;133:1943–1953. doi: 10.1242/dev.02365. [DOI] [PubMed] [Google Scholar]
- 15.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 16.Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 17.Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 18.Golzio C, Havis E, Daubas P, Nuel G, Babarit C, Munnich A, Vekemans M, Zaffran S, Lyonnet S, Etchevers HC. ISL1 directly regulates FGF10 transcription during human cardiac outflow formation. PLoS One. 2012;7:e30677. doi: 10.1371/journal.pone.0030677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe Y, Zaffran S, Kuroiwa A, Higuchi H, Ogura T, Harvey RP, Kelly RG, Buckingham M. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2–5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc Natl Acad Sci U S A. 2012;109:18273–18280. doi: 10.1073/pnas.1215360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. An Nkx2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cota CD, Segade F, Davidson B. Heart genetics in a small package, exploiting the condensed genome of Ciona intestinalis. Brief Funct Genomics. 2014;13:3–14. doi: 10.1093/bfgp/elt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson B. Ciona intestinalis as a model for cardiac development. Semin Cell Dev Biol. 2007;18:16–26. doi: 10.1016/j.semcdb.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tolkin T, Christiaen L. Development and evolution of the ascidian cardiogenic mesoderm. Curr Top Dev Biol. 2012;100:107–142. doi: 10.1016/B978-0-12-387786-4.00011-7. [DOI] [PubMed] [Google Scholar]
- 25.Delsuc F, Tsagkogeorga G, Lartillot N, Philippe H. Additional molecular support for the new chordate phylogeny. Genesis. 2008;46:592–604. doi: 10.1002/dvg.20450. [DOI] [PubMed] [Google Scholar]
- 26.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 27.Satoh N, Rokhsar D, Nishikawa T. Chordate evolution and the three-phylum system. Proc Biol Sci. 2014;281:20141729. doi: 10.1098/rspb.2014.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stolfi A, Christiaen L. Genetic and genomic toolbox of the chordate Ciona intestinalis. Genetics. 2012;192:55–66. doi: 10.1534/genetics.112.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simoes-Costa MS, Vasconcelos M, Sampaio AC, Cravo RM, Linhares VL, Hochgreb T, Yan CY, Davidson B, Xavier-Neto J. The evolutionary origin of cardiac chambers. Dev Biol. 2005;277:1–15. doi: 10.1016/j.ydbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 30.Davidson B, Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc Natl Acad Sci U S A. 2003;100:11469–11473. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satou Y, Imai KS, Satoh N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131:2533–2541. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- 32.Hirano T, Nishida H. Developmental fates of larval tissues after metamorphosis in ascidian Halocynthia roretzi. I. Origin of mesodermal tissues of the juvenile. Dev Biol. 1997;192:199–210. doi: 10.1006/dbio.1997.8772. [DOI] [PubMed] [Google Scholar]
- 33.Davidson B, Shi W, Levine M. Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development. 2005;132:4811–4818. doi: 10.1242/dev.02051. [DOI] [PubMed] [Google Scholar]
- 34.Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134:3297–3305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- 35.Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–1352. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- 36.Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 2006;20:2728–2738. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stolfi A, Gainous TB, Young JJ, Mori A, Levine M, Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565–568. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Razy-Krajka F, Lam K, Wang W, Stolfi A, Joly M, Bonneau R, Christiaen L. Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors. Dev Cell. 2014;29:263–276. doi: 10.1016/j.devcel.2014.04.001. ◦◦ This paper offered the first description of multineage transcriptional priming in chordate cardiopharyngeal multipotent precursors. It also unveiled a role for EBF/COE in both differentiation of pharyngeal myoblasts and maintenance of their associated muscle stem cells through Notch/Delta lateral inhibition.
- 39. Wang W, Razy-Krajka F, Siu E, Ketcham A, Christiaen L. NK4 antagonizes Tbx1/10 to promote cardiac versus pharyngeal muscle fate in the ascidian second heart field. PLoS Biol. 2013;11:e1001725. doi: 10.1371/journal.pbio.1001725. ◦◦ This paper showed with cellular resolution how a simple regulatory cross antagonism between two transcription factors, Tbx1/10 and NK4, promotes cardiac versus branchiomeric cell fate decision in Ciona intestinalis.
- 40.Stolfi A, Sasakura Y, Chalopin D, Satou Y, Christiaen L, Dantec C, Endo T, Naville M, Nishida H, Swalla BJ, et al. Guidelines for the nomenclature of genetic elements in tunicate genomes. Genesis. 2014 doi: 10.1002/dvg.22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stolfi A, Gandhi S, Salek F, Christiaen L. Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9. Development. 2014;141:4115–4120. doi: 10.1242/dev.114488. ◦◦ This paper used CRISPR/Cas9 system to achieve targeted mutagenesis by inducing site-specific double stranded breaks in a gene of interest through electroporation of CRISPR/Cas9 components into Ciona embryos. The application of the CRISPR/Cas9 system for tissue-specific loss-of-function in Ciona makes this species an appropriate model for assessing gene function.
- 42.Imai KS, Satoh N, Satou Y. A Twist-like bHLH gene is a downstream factor of an endogenous FGF and determines mesenchymal fate in the ascidian embryos. Development. 2003;130:4461–4472. doi: 10.1242/dev.00652. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi K, Yamada L, Satou Y, Satoh N. Differential gene expression in notochord and nerve cord fate segregation in the Ciona intestinalis embryo. Genesis. 2013;51:647–659. doi: 10.1002/dvg.22413. [DOI] [PubMed] [Google Scholar]
- 44.Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao J, Aggarwal VS, Nowotschin S, Bondarev A, Lipner S, Morrow BE. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol. 2008;316:524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meedel TH, Chang P, Yasuo H. Muscle development in Ciona intestinalis requires the b-HLH myogenic regulatory factor gene Ci-MRF. Dev Biol. 2007;302:333–344. doi: 10.1016/j.ydbio.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grifone R, Jarry T, Dandonneau M, Grenier J, Duprez D, Kelly RG. Properties of branchiomeric and somite-derived muscle development in Tbx1 mutant embryos. Dev Dyn. 2008;237:3071–3078. doi: 10.1002/dvdy.21718. [DOI] [PubMed] [Google Scholar]
- 49.Kelly RG, Jerome-Majewska LA, Papaioannou VE. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum Mol Genet. 2004;13:2829–2840. doi: 10.1093/hmg/ddh304. [DOI] [PubMed] [Google Scholar]
- 50.Sambasivan R, Gayraud-Morel B, Dumas G, Cimper C, Paisant S, Kelly RG, Tajbakhsh S. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev Cell. 2009;16:810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 51.Mourikis P, Gopalakrishnan S, Sambasivan R, Tajbakhsh S. Cell-autonomous Notch activity maintains the temporal specification potential of skeletal muscle stem cells. Development. 2012;139:4536–4548. doi: 10.1242/dev.084756. [DOI] [PubMed] [Google Scholar]
- 52.Vasyutina E, Lenhard DC, Wende H, Erdmann B, Epstein JA, Birchmeier C. RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc Natl Acad Sci U S A. 2007;104:4443–4448. doi: 10.1073/pnas.0610647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harel I, Nathan E, Tirosh-Finkel L, Zigdon H, Guimaraes-Camboa N, Evans SM, Tzahor E. Distinct origins and genetic programs of head muscle satellite cells. Dev Cell. 2009;16:822–832. doi: 10.1016/j.devcel.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green YS, Vetter ML. EBF proteins participate in transcriptional regulation of Xenopus muscle development. Dev Biol. 2011;358:240–250. doi: 10.1016/j.ydbio.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. El-Magd MA, Allen S, McGonnell I, Otto A, Patel K. Bmp4 regulates chick Ebf2 and Ebf3 gene expression in somite development. Dev Growth Differ. 2013;55:710–722. doi: 10.1111/dgd.12077. ◦ This paper examined the regulation of EBF transcription factors by retinoic acid (RA) in somites and pharyngeal arches during chick embryonic development and found that the three chick EBF genes are regulated differently, either indirectly or directly, by RA in these tissues.
- 56.El-Magd MA, Saleh AA, El-Aziz RM, Salama MF. The effect of RA on the chick Ebf1–3 genes expression in somites and pharyngeal arches. Dev Genes Evol. 2014;224:245–253. doi: 10.1007/s00427-014-0483-y. [DOI] [PubMed] [Google Scholar]
- 57. Lescroart F, Chabab S, Lin X, Rulands S, Paulissen C, Rodolosse A, Auer H, Achouri Y, Dubois C, Bondue A, et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat Cell Biol. 2014;16:829–840. doi: 10.1038/ncb3024. ◦◦ This paper provides evidence that Mesp1 is expressed sequentially in two distinct pools of of cardiovascular progenitors. Those correspond to the FHF and to pharyngeal mesoderm (i.e. SHF and branchiomeric muscles), respectively. This paper thus refutes the existence of a Mesp1-expressing common progenitor for both FHF and SHF in mice.
- 58. Devine WP, Wythe JD, George M, Koshiba-Takeuchi K, Bruneau BG. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. Elife. 2014;3 doi: 10.7554/eLife.03848. ◦ This paper showed that Mesp-expressing cells in the early gastrulating embryo are assigned to distinct mesodermal fates, including, but not restricted to, cardiac compartiments. This study also identified the chromatin remodeling factor Smarcd3 as an early specific marker of specified cardiac progenitors within the gastrulating mesoderm
- 59.Chan SS, Shi X, Toyama A, Arpke RW, Dandapat A, Iacovino M, Kang J, Le G, Hagen HR, Garry DJ, et al. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. 2013;12:587–601. doi: 10.1016/j.stem.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lescroart F, Mohun T, Meilhac SM, Bennett M, Buckingham M. Lineage tree for the venous pole of the heart: clonal analysis clarifies controversial genealogy based on genetic tracing. Circ Res. 2012;111:1313–1322. doi: 10.1161/CIRCRESAHA.112.271064. [DOI] [PubMed] [Google Scholar]
- 61.Huynh T, Chen L, Terrell P, Baldini A. A fate map of Tbx1 expressing cells reveals heterogeneity in the second cardiac field. Genesis. 2007;45:470–475. doi: 10.1002/dvg.20317. [DOI] [PubMed] [Google Scholar]
- 62.Kong P, Racedo SE, Macchiarulo S, Hu Z, Carpenter C, Guo T, Wang T, Zheng D, Morrow BE. Tbx1 is required autonomously for cell survival and fate in the pharyngeal core mesoderm to form the muscles of mastication. Hum Mol Genet. 2014;23:4215–4231. doi: 10.1093/hmg/ddu140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen JY, Huang DY, Peng QQ, Chi HM, Wang XQ, Feng M. The first tunicate from the Early Cambrian of South China. Proc Natl Acad Sci U S A. 2003;100:8314–8318. doi: 10.1073/pnas.1431177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemaire P. Unfolding a chordate developmental program, one cell at a time: invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Dev Biol. 2009;332:48–60. doi: 10.1016/j.ydbio.2009.05.540. [DOI] [PubMed] [Google Scholar]
- 65. Stolfi A, Lowe EK, Racioppi C, Ristoratore F, Brown CT, Swalla BJ, Christiaen L. Divergent mechanisms regulate conserved cardiopharyngeal development and gene expression in distantly related ascidians. Elife. 2014;3 doi: 10.7554/eLife.03728. ◦◦ This paper dissected the cell division and gene expression patterns in the cardiopharyngeal lineage in Molgula, and found that sequential fate specification events are identical to the homologous B7.5 lineage in Ciona. While a conserved motif appears to underlie cardiopharyngeal development in these ascidians, this study also revealed widespread interspecific unintelligibility of regulatory mechanisms beyond highly divergent cis-regulatory sequences.
- 66.Lemaire P, Smith WC, Nishida H. Ascidians and the plasticity of the chordate developmental program. Curr Biol. 2008;18:R620–R631. doi: 10.1016/j.cub.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ragkousi K, Beh J, Sweeney S, Starobinska E, Davidson B. A single GATA factor plays discrete, lineage specific roles in ascidian heart development. Dev Biol. 2011;352:154–163. doi: 10.1016/j.ydbio.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiba S, Sasaki A, Nakayama A, Takamura K, Satoh N. Development of Ciona intestinalis juveniles (through 2nd ascidian stage) Zoolog Sci. 2004;21:285–298. doi: 10.2108/zsj.21.285. [DOI] [PubMed] [Google Scholar]
- 69.Sasakura Y, Kanda M, Ikeda T, Horie T, Kawai N, Ogura Y, Yoshida R, Hozumi A, Satoh N, Fujiwara S. Retinoic acid-driven Hox1 is required in the epidermis for forming the otic/atrial placodes during ascidian metamorphosis. Development. 2012;139:2156–2160. doi: 10.1242/dev.080234. [DOI] [PubMed] [Google Scholar]
- 70.Hoffmann AD, Yang XH, Burnicka-Turek O, Bosman JD, Ren X, Steimle JD, Vokes SA, McMahon AP, Kalinichenko VV, Moskowitz IP. Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS Genet. 2014;10:e1004604. doi: 10.1371/journal.pgen.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci U S A. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woznica A, Haeussler M, Starobinska E, Jemmett J, Li Y, Mount D, Davidson B. Initial deployment of the cardiogenic gene regulatory network in the basal chordate, Ciona intestinalis. Dev Biol. 2012;368:127–139. doi: 10.1016/j.ydbio.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Norton J, Cooley J, Islam AF, Cota CD, Davidson B. Matrix adhesion polarizes heart progenitor induction in the invertebrate chordate Ciona intestinalis. Development. 2013;140:1301–1311. doi: 10.1242/dev.085548. ◦ The above paper showed that pre-cardiac founder cells adhere to the underlying epidermal cell-matrix leading to a localized response to a uniform FGF ligand exposure, which is necessary and sufficient for regional heart progenitor induction.
- 74.Cooley J, Whitaker S, Sweeney S, Fraser S, Davidson B. Cytoskeletal polarity mediates localized induction of the heart progenitor lineage. Nat Cell Biol. 2011;13:952–957. doi: 10.1038/ncb2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bowers SLK, Baudino TA. Laying the Groundwork for Growth: Cell-Cell and Cell-ECM Interactions in Cardiovascular Development. Birth Defects Research Part C-Embryo Today-Reviews. 2010;90:1–7. doi: 10.1002/bdrc.20168. [DOI] [PubMed] [Google Scholar]
- 76.Ross RS, Borg TK. Integrins and the myocardium. Circulation Research. 2001;88:1112–1119. doi: 10.1161/hh1101.091862. [DOI] [PubMed] [Google Scholar]
- 77.Peterkin T, Gibson A, Patient R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev Biol. 2007;311:623–635. doi: 10.1016/j.ydbio.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao R, Watt AJ, Battle MA, Li JX, Bondow BJ, Duncan SA. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Developmental Biology. 2008;317:614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 80.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes & Development. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 81.Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DYR. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes & Development. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haworth KE, Kotecha S, Mohun TJ, Latinkic BV. GATA4 and GATA5 are essential for heart and liver development in Xenopus embryos. Bmc Developmental Biology. 2008;8 doi: 10.1186/1471-213X-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peterkin T, Gibson A, Patient R. Common genetic control of haemangioblast and cardiac development in zebrafish. Development. 2009;136:1465–1474. doi: 10.1242/dev.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gline S, Kaplan N, Bernadskaya Y, Abdu Y, Christiaen L. Surrounding tissues canalize motile cardiopharyngeal progenitors towards collective polarity and directed migration. Development. 2015;142:544–554. doi: 10.1242/dev.115444. ◦◦ By inhibiting the secretory pathway in a tissue-specific manner, this paper characterized the effects of surrounding tissues, notably the mesenchyme, endoderm, epidermis, and notochord, during the collective migration of bilateral pairs of cardiopharyngeal progenitors in Ciona intestinalis.
- 85.Diogo R, Kelly RG, Christiaen L, Levine M, Ziermann JM, Molnar J, Noden DM, Tzahor E. The Cardiopharyngeal Field and Vertebrate Evolution: A New Heart for a New Head. Nature. 2015 doi: 10.1038/nature14435. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Christiaen L, Stolfi A, Davidson B, Levine M. Spatio-temporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Dev Biol. 2009;328:552–560. doi: 10.1016/j.ydbio.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 87.Tazumi S, Yabe S, Uchiyama H. Paraxial T-box genes, Tbx6 and Tbx1, are required for cranial chondrogenesis and myogenesis. Dev Biol. 2010;346:170–180. doi: 10.1016/j.ydbio.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 88.Tazumi S, Yabe S, Yokoyama J, Aihara Y, Uchiyama H. PMesogenin1 and 2 function directly downstream of Xtbx6 in Xenopus somitogenesis and myogenesis. Dev Dyn. 2008;237:3749–3761. doi: 10.1002/dvdy.21791. [DOI] [PubMed] [Google Scholar]
- 89.Nishiyama A, Fujiwara S. RNA interference by expressing short hairpin RNA in the Ciona intestinalis embryo. Dev Growth Differ. 2008;50:521–529. doi: 10.1111/j.1440-169X.2008.01039.x. [DOI] [PubMed] [Google Scholar]
- 90. Sasaki H, Yoshida K, Hozumi A, Sasakura Y. CRISPR/Cas9-mediated gene knockout in the ascidian Ciona intestinalis. Dev Growth Differ. 2014;56:499–510. doi: 10.1111/dgd.12149. ◦ This paper provides the first evidence for CRISPR/Cas9-induced mutations upon transient expression of Cas9 and single guide RNAs (sgRNA) in Ciona embryos.
- 91. Treen N, Yoshida K, Sakuma T, Sasaki H, Kawai N, Yamamoto T, Sasakura Y. Tissue-specific and ubiquitous gene knockouts by TALEN electroporation provide new approaches to investigating gene function in Ciona. Development. 2014;141:481–487. doi: 10.1242/dev.099572. ◦ This paper shows that custom-design TALE nucleases can be used for targeted mutagenesis and functional studies by transient transfection in Ciona embryos.
- 92.Tsagkogeorga G, Turon X, Hopcroft RR, Tilak MK, Feldstein T, Shenkar N, Loya Y, Huchon D, Douzery EJ, Delsuc F. An updated 18S rRNA phylogeny of tunicates based on mixture and secondary structure models. BMC Evol Biol. 2009;9:187. doi: 10.1186/1471-2148-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]