Abstract
The interface between viruses and their hosts’ are hot spots for biological and biotechnological innovation. Bacteria use restriction endonucleases to destroy invading DNA, and industry has exploited these enzymes for molecular cut-and-paste reactions that are central to many recombinant DNA technologies. Today, another class of nucleases central to adaptive immune systems that protect bacteria and archaea from invading viruses and plasmids are blazing a similar path from basic science to profound biomedical and industrial applications.
In retrospect, we probably should have anticipated that bacteria and archaea would have sophisticated immune systems. After all, viruses are the most abundant biological agents on the planet, causing roughly 1023 infections every second [1–3]. The selective pressures imposed by viral predation have resulted in the evolution of numerous phage defense systems, but it was only recently that sophisticated adaptive defense systems were identified in both bacteria and archaea [4–7].
Initial indications that Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) were part of an adaptive defense system came from a series of bioinformatics observations revealing that the short spacer sequences embedded in CRISPRs were sometimes identical to sequences found in phages and plasmids [8–10]. These observations led to the hypothesis that CRISPRs are central components of an adaptive immune system, and in 2007 Barrangou et. al. provided the first demonstration of adaptive immunity in bacteria by monitoring CRISPR loci in phage-challenged cultures of Streptococcus thermophilus [11]. This paper showed that CRISPRs evolve by acquiring short fragments of phage-derived DNA and strains with new spacers are resistant to these phages. It was immediately clear that this paper would serve as a foundation for an emerging team of scientists interested in understanding the mechanisms of adaptive defense systems in bacteria and archaea, but few of us anticipated the broader impacts of these discoveries for new applications in genome engineering.
Building on this initial foundation, a series of mechanistic studies showed that CRISPR loci are transcribed and processed into a library of small CRISPR derived RNAs (crRNAs) that guide dedicated nucleases to complementary nucleic acid targets [5–7,12,13]. In nature, these RNA-guided nucleases provide bacteria and archaea with sequence specific resistance to previously encountered genetic parasites. However, sequence specific nucleases have considerable value in biotechnology and one of these CRISPR-associated nucleases (i.e. Cas9) has recently been co-opted for new applications in biomedical, bioenergy, and agricultural sciences [14–17].
A goldmine for biotechnology
The molecular interface between a parasite and its host is a hot spot for innovation. A resistant host has a competitive advantage over a susceptible host, but an obligatory parasite faces extinction unless it is able to subvert host defense mechanisms. This conflict results in an accelerated rate of evolution that stimulates genetic innovation on both sides of this molecular arms race.
Genes at the interface of a genetic conflict have proven to be a goldmine for enzymes with activities that can be creatively repurposed for applications in biotechnology. In the 1970s, basic research on bacteriophages led to the discovery of DNA restriction endonucleases, which transformed molecular biology by making it possible to cleave specific DNA sequences [18]. The discovery of these enzymes paved the way for the emergence of recombinant DNA technologies, and in 1978 Werner Arber, Daniel Nathans, and Hamilton Smith shared the Nobel Prize in Physiology or Medicine "for the discovery of restriction enzymes and their application to problems of molecular genetics" [19]. Identification and application of type II restriction enzymes, which are integral to almost every aspect of DNA manipulation, effectively triggered the emergence of a global biotech industry.
Like restriction enzymes, CRISPR systems evolved as components of a prokaryotic defense system. However the mechanisms of sequence recognition by these enzymes are fundamentally different. Unlike DNA restriction enzymes, which typically rely on protein mediated recognition of 4 to 8 base pairs; CRISPR-associated nucleases are guided by base pairing between an RNA-guide and a complementary target. The implications of this targeting mechanism have triggered a sea-change in biology and now the historical precedent of nucleases in biotechnology seems poised to repeat itself.
Why all the fuss?
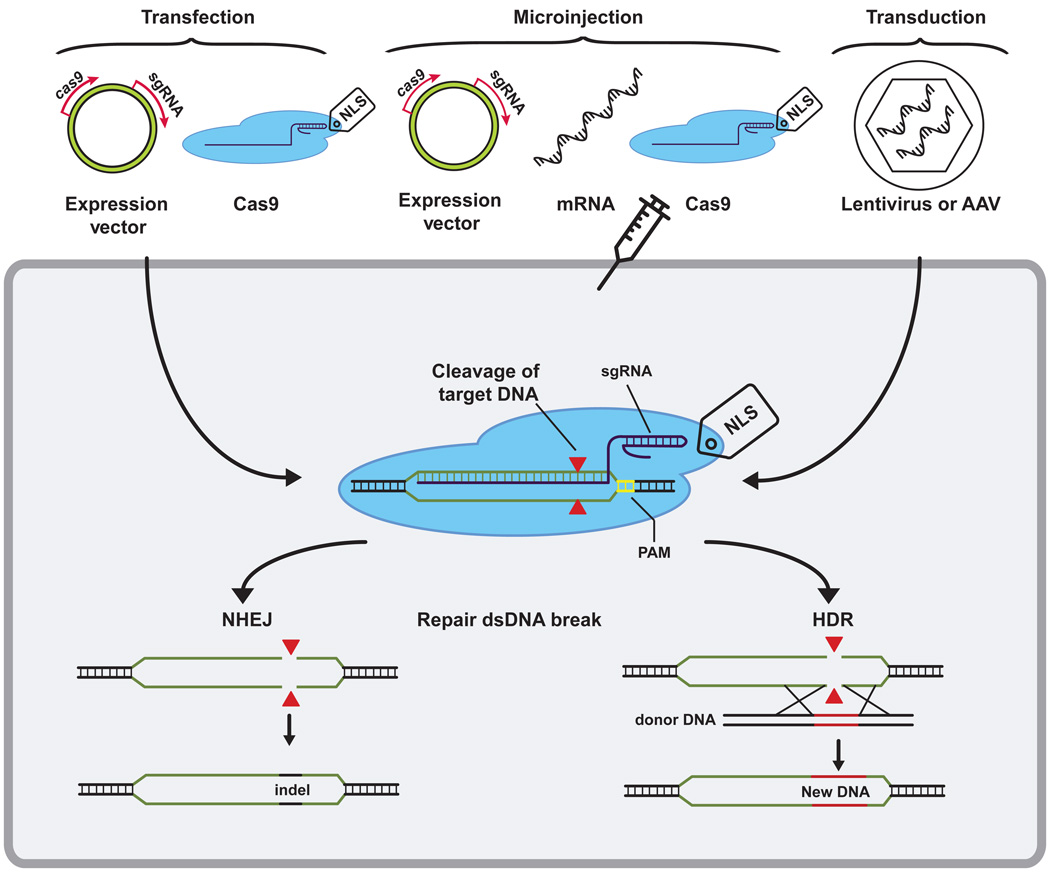
Reverse genetics is a powerful method used to determine the biological function of a specific gene. This approach is used routinely to determine gene function in organisms with simple genomes, but existing techniques are not applicable for high-throughput genetic screens in organisms with large genomes and multiple chromosomes. However, the recently discovered mechanism of DNA cleavage by the CRISPR RNA-guided nuclease Cas9 [20], has transformed the field of genetics by allowing efficient and precise genetic manipulation of diverse eukaryotic genomes, including human cells [14–17]. To repurpose the Cas9 nuclease for targeted genome editing, the cas9 gene has been codon optimized for expression in eukaryotic systems and tagged with a nuclear localization signal (NLS) [21–23]. Cas9 and the guide RNA have been delivered to eukaryotic cells by transient transfections with expression vectors [24] or purified Cas9/sgRNA [25,26], viral transduction using Lentiviruses [27–29] or Adeno-Associated Virus [30–33], and cytoplasmic or nuclear injections [34–37] (Figure 1). In each case, RNA-guided targeting of Cas9 to a complementary DNA target results in a double-stranded DNA break (DSB) at the target site. These lesions are typically repaired by non-homologous end joining (NHEJ), which is an error-prone process that is often accompanied by insertion or deletion of nucleotides (indels) at the targeted site, resulting in a genetic knock-out of the targeted gene due to a frameshift mutation. Alternatively, DSBs are repaired by homology directed repair (HDR). In most systems, HDR is an inefficient process that requires a DNA donor with sequence homology to regions of the genome that flank the DSB [38]. Using Cas9 in combination with a DNA donor provides a method to target cleavage and repair of naturally occurring genetic defects, add foreign DNA encoding genes with new functions to specific locations, or precisely excise defined fragments of DNA [14–16,39].
Figure 1. Cas9 delivery and repair of the targeted DNA.
Cas9 and a single-guide RNA (sgRNA) have been delivered to eukaryotic cells by several methods: transient transfections (expression vectors or purified Cas9/sgRNA complex), cytoplasmic or nuclear injections (expression vectors, mRNA or purified Cas9/sgRNA) and by transduction (Lentiviruses or Adeno-Associated Virus). Cas9 identifies its target by protein mediated PAM (yellow) recognition and base pairing between the sgRNA and the DNA target. Target recognition activates the nuclease sites (red triangles), resulting in double stranded breaks (DSBs) 3–4 nucleotides downstream from the PAM. DSBs can be repaired by non-homologous end joining (NHEJ) or homology directed repair (HDR). NHEJ results in insertion or deletions (indels), which often results in a frameshift mutation. HDR relies on homologous recombination with a donor DNA molecule. This donor DNA can be used to specifically insert desired sequences.
Cas9 is not the first programmable nuclease developed for engineering eukaryotic genomes [40]. Some of the earliest methods for introducing targeted genome modifications relied on meganucleases (e.g. HO and I-SceI), which are endonucleases that have long recognition sequences (12 to 40 base pairs) [41]. The enhanced specificity of these nucleases, as compared to the 4 to 8 base pairs recognized by most restriction enzymes, presents an opportunity to target specific locations in large eukaryotic genomes. However, these enzymes rely on protein-mediated recognition of the target DNA, and reprogramming these proteins to target new DNA sequences has been challenging due to the integrated nature of the DNA binding and nuclease domains of these proteins. To address this issue, non-natural chimeric nucleases composed of distinct DNA binding and nuclease domains have been engineered. The most celebrated examples of these are Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs).
Zinc fingers (ZFs) are sequence specific DNA binding domains found in eukaryotic transcription factors and transcription activator-like effectors (TALEs) are modular DNA binding proteins made by bacterial pathogens that infect plants. The mechanism of DNA recognition by these proteins is well understood and the modular nature of these interactions has been exploited for reprogramming. While the success of these proteins for targeted genome engineering cannot be overstated, they both rely on protein-mediated recognition of the DNA, which means that every new target requires the engineering of new proteins. In contrast to these techniques, Cas9 is an RNA-guided nuclease that relies on complementary base pairing and protein mediated recognition of an adjacent short sequence motif, commonly referred to as the PAM (protospacer adjacent motif) [42]. PAMs are typically 2 to 5 basepairs in length, which means that PAMs occur at high frequency and are rarely a limitation when designing RNA guides to specific target sequences. However, Cas9 proteins from different organisms often recognize different PAM sequences, so in rare instances where a target sequence does not contain a PAM recognized by one Cas9 (i.e. Cas9 from Streptococcus pyogenes recognizes a 5’-NGG PAM), then another Cas9 with a distinct PAM recognition sequence may be used (i.e. Cas9 from Neisseria meningitides recognizes a 5′-NNNNGATT PAM) [43]. The diversity of Cas9 proteins and the simplicity of RNA-guided programing abrogates the need for sophisticated protein engineering, and affords rapid generation of designer nucleases. In fact, guide RNAs that target Cas9 to as many as 20,000 different genes and 1,800 microRNAs in the human genome have been generated in a single experiment [28,29,44]. These whole-genome knockout techniques are transforming functional genomics and redefining the possibilities of reverse genetics.
The Emerging Market
CRISPR applications span almost every industry that involves biological systems (Table 1). Danisco (DuPont) was an initial pioneer of commercial use of CRISPR technology to enhance viral immunity in bacteria used to make yogurts and cheese, but other markets have been rapidly emerging. Applications in agriculture have lower regulatory hurdles than biomedical application and some of these markets are anticipated to produce earlier returns on investments. Dow Agrosciences has co-developed intellectual property (IP) with Sangamo Biosciences for developing genetically modified crops using Cas9, and Cellectis Plant Sciences is leveraging its relationship with parent Cellectis SA to move the technology into crops. Similarly, Recombinetics Inc. is using TALENs, ZFNs and Cas9 to enhance productivity in the livestock industry [45]. While the USDA has yet to decide on how it will treat genomes edited using Cas9, it has already ruled that ZFNs and TALENs do not fall under their governance [46]. This saves an average 5.5 years and $35 million in related regulatory costs for bringing a product to market [47]. Similar treatment of CRISPR-based genome editing may stimulate economic activity around the development of new agricultural and industrial products.
Table 1. Industry interests in Cas9-based technologies.
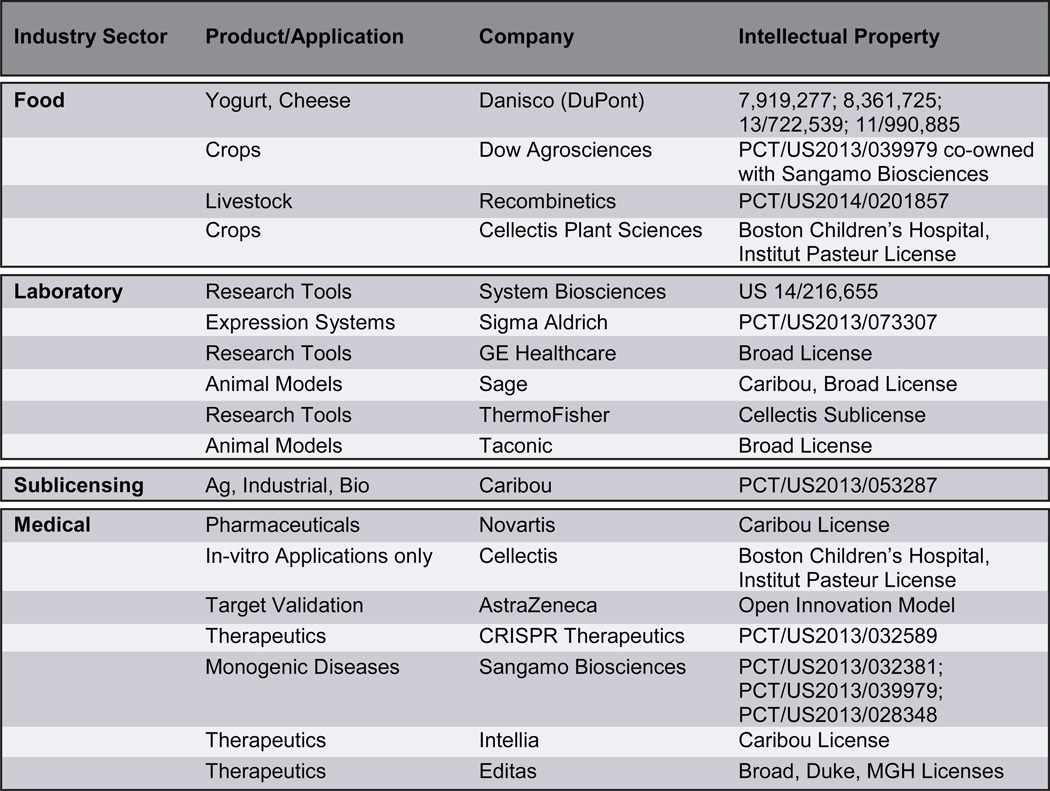 |
In addition to Cas9-based applications in the agricultural industry, market segments for Cas9 endonucleases within the human health sectors are experiencing frenetic growth. These markets include: gene-, cell-, and immunotherapy, fast and efficient development of transgenic research animals, drug discovery, as well as target validation and screening. It is difficult to accurately estimate the value of the nascent market for CRISPR RNA-guided nucleases in the biomed sector, but documents from the initial public offering (IPO) of Horizon Discovery Group, plc., which has in-licensed Cas9 IP, indicate a market size of $46 billion [48] and recent private equity financings of Cas9-based genome engineering companies include: Caribou Biosciences, (undisclosed venture estimated at $2.9 million from Novartis), CRISPR Therapeutics, ($25 million), Recombinetics, Inc. ($5 million), Intellia Therapeutics ($15 million), and Editas Medicine ($43 million). In total, companies with an interest in using Cas9 for applications related to gene therapy have raised over $600 million in venture capital and public markets since the beginning of 2013. The pace of this activity is remarkable given that the first granted patent for the use of CRSIPR technology in eukaryotic cells was issued April 15, 2014.
Commercial interest in Cas9 IP has not escaped the interest of big pharmaceuticals. Novartis has partnered with a tier I private equity firm, Atlas Ventures to commit $15 million to kick start Intellia Therapeutics, and Pfizer partner Cellectis SA will be using Cas9-based technologies to make T-cells with chimeric antigen receptors. In January of this year, AstraZeneca announced four partnerships with academia around the use of Cas9 nucleases to validate new drug targets.
In addition to the agricultural and biomedical sectors, the research tools market is also embracing CRISPR-based technology. Sigma-Aldrich has in-licensed technology in order to make, use, and distribute tools for the generation of modified plant and animal models, custom cell line creation and for pooled genetic screening. Perhaps the financial activity in each of these market sectors is heightened by over-exuberance common to the early market development. However, given the scope of current applications across multiple industries, we see no limits to research or financial commitments in this space.
Highlights.
Genes involved in genetic conflict are a source for biological and biotechnological innovation.
Bacteria and archaea have evolved adaptive immune systems that rely on RNA-guided nucleases.
Cas9 is an RNA-guided endonuclease that has been repurposed for genome engineering.
Market valuations estimate Cas9 related technologies in the billions.
Acknowledgements
We are grateful to Jennifer A. Doudna for valuable feedback on this manuscript. Research in the Wiedenheft lab is supported by the National Institutes of Health (P20GM103500 and R01GM108888), the National Science Foundation EPSCoR (EPS-110134), the M.J. Murdock Charitable Trust, and the Montana State University Agricultural Experimental Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A. Explaining microbial population genomics through phage predation. Nature Reviews Microbiology. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 2.Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 3.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 5.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 6.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrangou R, Marraffini LA. CRISPR-Cas Systems: Prokaryotes Upgrade to Adaptive Immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolotin A, Ouinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 9.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 10.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 11. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. ** This paper provides the first direct evidence for adaptive immunity in bacteria.
- 12.Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. Biochem J. 2013;453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson R, Wiedenheft B. A CRISPR method for genome engineering. F1000Prime Rep. 2014;6:3. doi: 10.12703/P6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 17.Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 18.Roberts RJ. How restriction enzymes became the workhorses of molecular biology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5905–5908. doi: 10.1073/pnas.0500923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobel Media, editor. 2015. The Nobel Prize in Physiology or Medicine 1978. [Google Scholar]
- 20. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. ** This paper provides mechanistic insights that explain how Cas9 can be used as a programmable nuclease for genome engineering.
- 21. Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. **This is one of the first demonstations that Cas9 can be used for precise multiplexed genome engineering in mouse and human cells.
- 22. Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. **This is one of the first demonstations that Cas9 can be used for precise genome engineering in human cells.
- 23. Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. **This is one of the first demonstations that Cas9 can be used for precise genome engineering in human cells, including induced pluripotent stem cells.
- 24. https://http://www.addgene.org/CRISPR/. [Google Scholar]
- 25.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9- based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. *This paper demonstates how lentiviral delivery of sgRNAs can be used to generate human genome knockout libraries for genome-scale screening.
- 29. Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science. 2014;343:80–84. doi: 10.1126/science.1246981. *This paper demonstates how lentiviral delivery systems can be used to generate human genome knockout libraries.
- 30.Cheng R, Peng J, Yan Y, Cao P, Wang J, Qiu C, Tang L, Liu D, Tang L, Jin J, et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014;588:3954–3958. doi: 10.1016/j.febslet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maggio I, Holkers M, Liu J, Janssen JM, Chen X, Goncalves MA. Adenoviral vector delivery of RNA-guided CRISPR/Cas9 nuclease complexes induces targeted mutagenesis in a diverse array of human cells. Sci Rep. 2014;4:5105. doi: 10.1038/srep05105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holkers M, Maggio I, Henriques SF, Janssen JM, Cathomen T, Goncalves MA. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat Methods. 2014;11:1051–1057. doi: 10.1038/nmeth.3075. [DOI] [PubMed] [Google Scholar]
- 34.Horii T, Arai Y, Yamazaki M, Morita S, Kimura M, Itoh M, Abe Y, Hatada I. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep. 2014;4:4513. doi: 10.1038/srep04513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terns RM, Terns MP. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet. 2014;30:111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carroll D. Genome Editing by Targeted Chromosomal Mutagenesis. Methods in Molecular Biology. 2014;1239:1–13. doi: 10.1007/978-1-4939-1862-1_1. [DOI] [PubMed] [Google Scholar]
- 41.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Research. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAMdependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou Z, Zhang Y, Propson NE, Howden SE, Chu L-F, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, Fahrenkrug SC. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. PNAS. 2013;110:16526–16531. doi: 10.1073/pnas.1310478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glorikian H. Gene Editing Will Change Everything—Just Not All at One Time. 2015 Edited by. [Google Scholar]
- 47.Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 2014;12:e1001877. doi: 10.1371/journal.pbio.1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horizon Discovery Group, Admission Document. 2015 [Google Scholar]