Abstract
Animal motility varies with genotype, disease, aging, and environmental conditions. In many studies, it is desirable to carry out high throughput motility-based sorting to isolate rare animals for, among other things, forward genetic screens to identify genetic pathways that regulate phenotypes of interest. Many commonly used screening processes are labor-intensive, lack sensitivity, and require extensive investigator training. Here, we describe a sensitive, high throughput, automated, motility-based method for sorting nematodes. Our method is implemented in a simple microfluidic device capable of sorting thousands of animals per hour per module, and is amenable to parallelism. The device successfully enriches for known C. elegans motility mutants. Furthermore, using this device, we isolate low-abundance mutants capable of suppressing the somnogenic effects of the flp-13 gene, which regulates C. elegans sleep. By performing genetic complementation tests, we demonstrate that our motility-based sorting device efficiently isolates mutants for the same gene identified by tedious visual inspection of behavior on an agar surface. Therefore, our motility-based sorter is capable of performing high throughput gene discovery approaches to investigate fundamental biological processes.
Introduction
In 1974, Sidney Brenner proposed using the nematode Caenorhabditis elegans as an animal model to understand nervous system function1. In his classic publication, Brenner described the results of a random mutagenesis screen for mutants with motility defects. In the ensuing forty years, numerous additional genetic screens have been performed for such mutants, often referred to as “uncoordinated”. In a typical genetic screen, animals are observed under the microscope and the investigator selects animals that qualitatively move differently than the norm. This simple strategy has proven powerful in the identification of over a hundred genes that, when mutated, affect animal locomotion.
Notwithstanding the success of this type of screen for locomotion-defective animals, there are limitations. First, the screen is laborious since each animal must be individually inspected. Since mutants of interest are rare, with a typical gene being meaningfully-mutated in less than one in a thousand animals1, the number of mutants identified is limited by investigator time, vigilance, and competence. Second, prior screens for mutants that affect locomotion have required that the phenotype be sufficiently severe to be qualitatively detectable by the observer. In fact, there are mutants that appear normal to the casual observer, yet have locomotion defects when analyzed with sensitive machine vision methods2-4. Ideally, one would want to develop quantitative methods capable of identifying even subtle, gene-induced variations in locomotion. Additionally, one would want to identify chemicals that affect locomotion, as such studies may assist in developing drugs and identifying hazardous chemicals, which later may be translated to human-based studies. Finally, one would want to also identify genetic and chemical perturbations that improve locomotion parameters. Such screens are not yet easily feasible with direct observation strategies.
Importantly, until recently, identifying the molecular lesion responsible for a phenotype was a challenging endeavor that could take years to complete using meiotic recombination genetic mapping strategies. An easily identified phenotype was crucial to the success of such genetic mapping experiments. However, now, with the ability to carry out whole genome sequencing (WGS)5-7, the approach for identifying molecular mechanisms of behavioral phenotypes has changed. WGS allows one to forgo the need for laborious genetic mapping, which, in turn, obviates the need for a strong mutant phenotype. In the new era of WGS, the isolation of candidate mutants has become the rate limiting step in many forward genetic screens.
To keep up with advances in genotyping technologies, machine vision methods, microfluidic platforms, and automated worm-handling systems have been developed for high-throughput phenotyping and sorting of worms2-4, 8-30. Sophisticated, sensitive machine vision programs are enabling the identification of subtle phenotypes that are not easily detectable with human eyes2, 3. Miniaturized microfluidic platforms are facilitating simultaneous monitoring of many animals9, 11, 14-16, 18, 22-27. Device miniaturization is particularly crucial when each individual animal needs to be monitored for a prolonged period of time as in C. elegans sleep and aging studies14, 22. Various microfluidic modules have been developed for on-chip assays, including wells and capsules11, 14-16, 18, 22-27, worm traps9, 12, 13, 31, and electrophysiological measurement modules21. Sophisticated automated worm handling systems, such as fluorescence-activated sorters like the COPAS Biosort machine,32, 33 enable the isolation of mutants with altered fluorescent protein expression10, 19. High-throughput, size-based microfluidic sorting device has also been developed recently28. However, while these methods are powerful for carrying out size/morphology/fluorescent label-based sorting, they are not capable of sorting based on motility.
Recently, researchers20, 34 have utilized electrotaxis, the tendency of nematodes to migrate towards the negative pole of an electric field35, 36, to direct the motion of animals. By collecting animals at a predetermined location, based on their arrival time, one can achieve motility-based separation. The reported method suffers, however, from a relatively low throughput and is limited to strains that exhibit electrotaxis. Furthermore, prolonged exposure to electric field may adversely impact animals’ motility.
As an alternative, we describe here a simple, high throughput, motility-based sorter that separates out animals whose propulsive power exceeds a preset (controllable) threshold. The sorter isolates animals capable of swimming upstream, against a fluid flow. A single module of our sorter can process hundreds to thousands of animals per hour. Multiple modules can operate in parallel. To demonstrate the efficacy of our motility-based sorter, we separate motility mutants from non-mutant (wild-type) C. elegans animals. We then use our device to carry out a large-scale genetic screen to identify rare mutants that suppress the locomotion-impairment conferred by over-expression of the gene flp-13, which regulates C. elegans sleep37
Results
Device Description
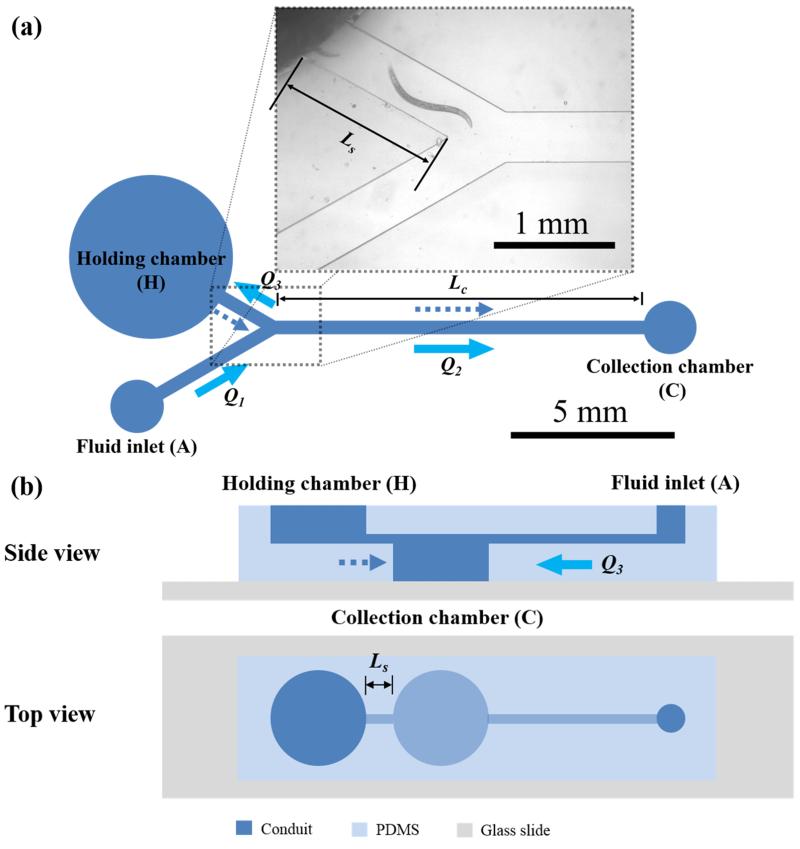
A device for high-throughput, motility-based sorting must rapidly and selectively isolate animals whose propulsive power differs from a pre-set threshold. We achieve this objective with a device comprised of a circular holding chamber (4 mm in diameter × 5 mm deep) connected to a separation conduit (Fig. 1). The holding chamber can house thousands of adult C. elegans, each measuring ~1 mm in length and ~69 m in diameter. The controllable flow velocity in the separation conduit (length Ls) is directed towards the holding chamber. The animals are loaded into the holding chamber. Animals occasionally enter the separation conduit. Only the ones with a sufficient propulsive thrust to overcome the adverse flow progress upstream (Video S1); the others remain in the holding chamber. Fig. 1 depicts two embodiments of the sorter, differing in the method by which the escaping animals are collected.
Fig. 1.
A schematic depiction of the sorting devices. (A) The Y sorter – a photograph and top view. The fluid inlet is connected to a syringe pump, which controls the flow rate Q1. The collection chamber is connected to a second syringe pump that operates in suction mode and controls the flow rate Q2. Q3 = Q1-Q2. Solid arrows and dashed arrows denote, respectively, flow direction and able animal direction of movement. (B) The L sorter: Side cross-section (top) and top view (bottom). The fluid inlet was connected to a syringe pump, which controlled the flow rate Q3 in the separation conduit (Ls). Animals that moved with sufficient velocity to escape the length Ls of the sorting conduit sank to the bottom of the collection chamber and were thus isolated.
Fig. 1a depicts schematically the top view of a single separation module, which we dub the Y sorter, and shows a photograph of the device’s bifurcation region. The Y module is comprised of a holding chamber, a “Y” – shaped conduit, and a collection chamber. One leg of the “Y” is connected to the holding chamber and the other to a syringe pump. The stalk of the “Y” is connected to a collection chamber and farther downstream to a second syringe pump that operates in a suction mode. Although the device can be operated with a single syringe pump, we elected to use here two pumps to ease flow control. The positive pressure pump (A) supplies flow rate Q1. At the bifurcation, the flow splits into two streams. One stream, with flow rate Q2 (collecting flow) proceeds to the collection chamber. The other stream, with flow rate Q3 (separation flow), goes to the holding chamber. Q1=Q2+Q3. If only a positive pressure pump were used, the fraction of the flow Q3/Q1 would equal to R2/(R3+ R2), where R2 and R3 are, respectively, the hydraulic resistances of the collecting and separating conduits. The separation flow Q3 was selected so that the average fluid velocity (u3) in the separation conduit is lower than the swimming velocity of the animals to be sorted (us) and higher than the normal velocity of animals to be retained in the holding chamber. The velocities u1 and u2 associated, respectively, with the positive pressure pump flow rate Q1 and the collecting pump flow rate Q2 are sufficiently high to preclude the sorted animals from progressing upstream. u3<us<u1 and us<u2.
Since the nematode’s normal speed (us) is not significantly affected by the background flow, the animals’ velocity in the laboratory frame of reference is the superposition of (1) the animal’s velocity when the medium is stagnant and (2) the medium’s velocity. In other words, the absolute animal’s velocity in the sorting conduit is us-u3. In operation, animals are loaded into the holding chamber. The able animals enter the sorting conduit and swim upstream while the weaker animals remain in the holding chamber. Once an animal arrives at the bifurcation, it is carried by the collecting flow into the collection chamber (C).
The second embodiment (Fig. 1b), which we dub the linear (L) sorter, is comprised of a single conduit leading into the holding chamber, and is operated with a single syringe pump. The collection chamber is located upstream of the holding chamber beneath the conduit’s level. Since C. elegans is denser than the carrier water38, when the animal arrives at the collection chamber, it sinks to the chamber’s bottom, unable to escape. Although not attempted here, one can envision expanding the L sorter to include a cascade of collection chambers, each doubling as a holding chamber with a judiciously designed separation conduit to sort animals with various motilities.
Both devices were fabricated with polydimethylsiloxane (PDMS) using soft lithography. The PDMS cast was then bonded to a glass slide. The width and height of the sorting conduit were sufficiently large to accommodate uninhibited swimming of individual adult animals with typical body diameter of 69 μm and gait amplitude of 340 μm, but sufficiently small to prevent two animals from concurrently occupying any cross-section of the sorting conduit, thus minimizing jamming and/or interference. Unless otherwise stated, we used a 91μ m deep, 400μm wide separation conduit in the Y sorter and a 91μ m deep, 600μ m wide separation conduit in the L sorter. In most experiments, only one animal occupied any cross-section at any given time.
Another factor to consider is the fluid flow’s effects on the animal’s orientation. If the animals were to orient with the direction of the flow (negative rheotaxis), the operation of the separation conduit would be compromised. Fortunately, this is not the case. In fact, we have recently demonstrated that C. elegans exhibits hydrodynamically–induced, non-deliberate tendency to swim towards boundaries39 and, when sufficient space available, to orient against the flow (positive rheotaxis)40. The width of the separation conduit in our experiments is smaller than the length of a typical young adult animal - too narrow for the reorientation of animals by hydrodynamic forces.
During sorter’s operation, animals would enter the separation conduit by chance. Once in the separation conduit, most animals remained oriented against the flow direction. Only infrequently, did animals change their direction of motion by deliberately bending their body into the shape of the Greek letter omega (omega turns) and were washed back into the holding chamber to await a second attempt at escape. These sporadic changes in swimming direction are common to nematodes, such as C. elegans41. The time interval between successive omega turns ranges from a few to tens of seconds41, though the probability of making an omega turn may vary by genotype42. In our devices, the separation conduit and the animal’s residence time in it were short enough to render these events sufficiently rare as not to significantly impair device performance. Although we did not carry out an optimization study for the separation conduit’s length Ls, we found that a separation conduit length on the order of one animal body length, 1 mm<Ls<1.5 mm, to be adequate.
Multiple modules of either Y type (Fig. 1a) and/or L type (Fig. 1b) sorters can be accommodated on a single substrate to operate in parallel with the same or different motility thresholds to increase throughput. Also, a number of modules can be connected in series to refine the separation process. Below, we characterize the sorter. Then, to demonstrate the utility of our sorters, we describe two experiments. In the first experiment, we sort known mutants and use this sorting experiment as a proof of concept for our device. The second experiment serves an actual research purpose - a genetic screen for a yet unidentified gene.
Device Characterization: Probability of an able animal (us>u3) escaping the holding chamber
To quantify the sorter’s operation, we constructed a simple mathematical model. We assume that the probability τ-1 (s−1) of escaping from the holding chamber is independent of the number of able animals Na(t) present in the holding chamber at any instance t. This approximation is likely to be valid when the animals in the holding chamber are sufficiently dilute as not to interact significantly with each other and jamming at the separation conduit’s inlet occurs rarely. Since we often start the sorting with the holding chamber tightly packed with animals, the model will likely go into effect only after the holding chamber has been partially emptied.
We define able animals as the animals that we wish to isolate and that have a swimming velocity us greater than the sorting velocity u3. The rate of change in the number of able animals in the holding chamber . Thus, , where τ can be viewed as the time constant whose magnitude depends on the motility of the able animals (us), the dimensions of the holding chamber, the average opposing fluid velocity in the separation conduit (u3), the length of the separation conduit (Ls), and the likelihood of making an omega turn.
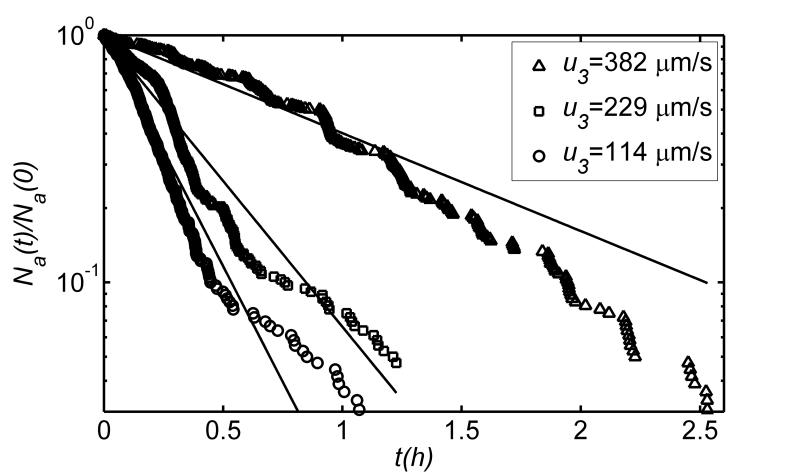
To test this model, we carried out a set of experiments with the Y sorter. We loaded the holding chamber with wild-type animals, adjusted the opposing flow velocity in the separation conduit, and monitored the number of animals in the holding chamber as a function of time. Fig. 2 depicts Na(t)/Na(t0) as a function time when the adverse flow velocities are 114μ m/s (15 μL/h), 229 μm/s (30 μL/h), and 382 μm/s (50 μL/h). In all three cases, Na(t0)=360. The symbols and solid lines correspond, respectively, to experimental data and best fits. As predicted, the number of animals in the holding chamber decayed exponentially with the time constants τ= 0.2h, 0.4h, and 1.1h when u3 = 114 μm/s, 229 μm/s, and 382 μm/s, respectively. The time constant τ increased with the average opposing flow velocity (u3) in the separation conduit. As u3 increases, the upstream swimming velocity of the animals (us-u3) in the separation conduit (in the laboratory frame of reference) decreases. As a result, it takes longer for the animals to pass through the separation conduit. We hypothesize that this increased residence time in the separation conduit has two effects. First, and most important, it takes the animal longer to clear the separation conduit to make room for the next animal’s entry into the separation conduit. Second, and perhaps less significantly, the longer residence time in the separation conduit increases the likelihood of the animal making an omega turn, and returning into the holding chamber, which would reduce, on average, the rate at which able animals escape from the holding chamber.
Fig. 2.
The normalized number of fit animals Na(t)/Na(t0) in the holding chamber as a function of time when the average, adverse fluid velocity in the separation conduit is 114 μm/s (circles), 229 μm/s (squares), and 382 μm/s (triangles). The symbols and solid lines correspond, respectively, to experimental data and best fits. The experiment was carried out with the Y sorter.
Motility-Based Phenotyping
As a proof of concept, we used the Y module (Fig. 1a) to separate binary mixtures, each containing two different genotypes in equal portions: In each separation experiment, we mixed wild-type C. elegans expressing the mCherry fluorescent protein (FP) with unlabeled animals carrying a mutation in one of the three genes unc-63, dys-1, or lev-10. These three genes are known to be required for normal locomotion3. The FP allowed us to readily distinguish between the wild types and the mutants.
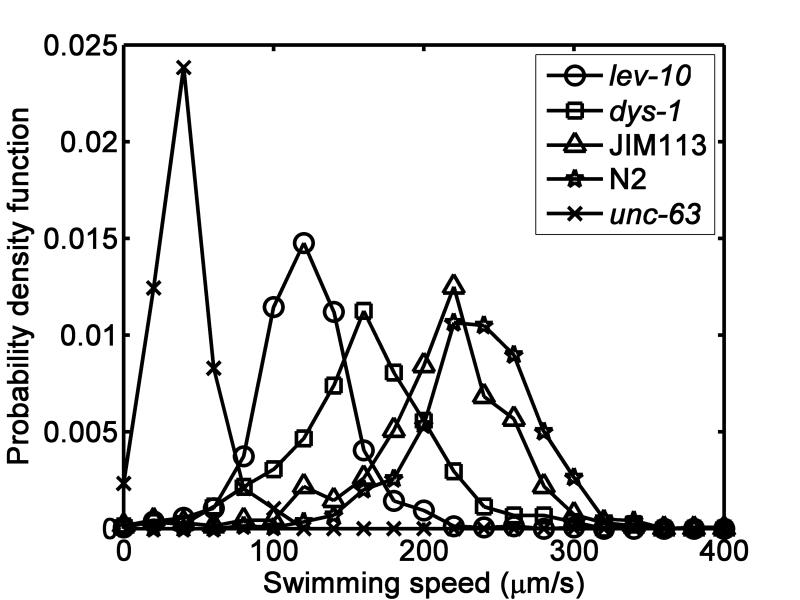
Before embarking on the sorting experiments, we assessed the motility of the various strains used in our experiments under temperature and liquid composition similar to the ones in our sorting process. Young adult wild type expressing FP (WT-FP, n=344), wild type without FP (WT, n=719), unc-63 (UNC, n= 193), dys-1 (DYS, n=440), and lev-10 (LEV, n=803) were suspended, one strain at a time, in water confined between two glass slides, spaced 130 μm to 160 μm apart and tracked with a video camera. The maximum velocities of the animals were deduced with the wrMTrck ImageJ plug-in. Fig. 3 depicts the probability density function of the measured velocities of the various strains. The averages of the maximum velocities of the strains were us(WT)=235 μm/s ± 40 μm/s, us(WT-FP)= 208 μm/s ± 50 μm/s, us(DYS)= 160 μm/s ± 49 μm/s, us(LEV)= 121 μm/s ± 32 μm/s, and us(UNC)= 40 μm/s ± 20 μm/s. The wild type expressing FP were slightly slower than the wild type without FP (P=10−24), suggesting that either the expression of FP itself or the process of generating this strain compromises propulsive power somewhat. Our data is consistent with prior reports3. Fig. 3 indicates overlaps in the various strains’ velocity distributions. Thus, we can expect the sorter to only significantly enrich, but not completely purify, the sorted populations.
Fig. 3.
The probability density functions of the maximum swimming speeds of animals of the genotypes: wild-type (N2, stars), wild-type expressing fluorescent protein (JIM113, triangles), lev-10 (circles), dys-1 (squares), and unc-63 (crosses).
Table 1 records our experimental conditions and the sorting results. In each experiment, wild-type animals expressing FP were mixed with one of the mutants unc-63, dys-1, or lev-10 in equal proportions. The sorter’s objective is to separate the more motile wild-type animals out of the mixture. The average liquid velocity in the separation conduit (u3) was adjusted to the values indicated in the table. At the conclusion of the sorting operation, we counted the FP(+) and the FP(−) animals in both the holding as well as in the collection chambers. In all cases, the sorter enriched the fractions of the slower mutants in the holding chamber from 50% to over 90%. The fraction of the slowest mutants in the collection chamber ranged from 0.7% to 11.7%, with higher percentages (i.e. poorer enrichment) of mutants whose distribution of swimming velocities showed significant overlap with that of wild-type animals. Since there is very little overlap in the velocity distributions of the wild-type (FP) animals and unc–63 mutants, very few unc-63 animals ended in the collection chamber (0.7%). This corresponds to 71.4 fold enrichment, where we define enrichment as the percentage of unable animals before sorting (50) divided by the percentage of unable animals after sorting (0.7). The corresponding enrichments for the mixtures with dys-1 and lev-10 were, respectively, 4.3 and 4.7. Although we have not yet done so, the sorted animals can be resorted to achieve even higher levels of enrichment. Resorting can be accomplished either by loading the sorted animals back into the holding chamber or by using an L sorter with a cascade design as discussed above.
Table 1.
Throughput and sensitivity of the sorter
| Genotype |
us (μm/s) |
u3 (μm/s) |
Total number of worms |
% in initial mix |
Sorting time (h) |
% in holding chamber |
% in collection chamber |
Throughput (#/h) |
|---|---|---|---|---|---|---|---|---|
| unc-63 | 40 | 165 | 496 | 50 | 1.6 | 90.9 | 0.7 | 317 |
| dys-1 | 160 | 204 | 991 | 50 | 1.1 | 90.0 | 11.7 | 901 |
| lev-10 | 121 | 127 | 1036 | 50 | 0.4 | 96.0 | 10.7 | 2486 |
| Average | 92.3 | 7.7 | 1235 |
The % are for the slow mutants, which lacked fluorescent protein expression.
us: Swimming velocityu3: Fluid velocity in the separation conduit
Forward Genetic Screen to Isolate Animals with flp-13 Suppressors
Next, we demonstrate the utility of the sorter to enrich the fraction of rare (low abundance) mutants in an unbiased forward genetic screen. Here, we put the sorter to use to assist us in an actual research carried out in our lab. The gene flp-13, which encodes neuropeptides and is expressed in the sleep-promoting ALA neuron, has recently been shown to regulate sleep-like, quiescent behavior in C. eleagns37. The flp-13 transcript and hence protein can be over-expressed by making a transgene of the gene under the control of a heat-inducible promoter37. When flp-13 is over expressed, the majority (~90%) of the animals cease feeding and moving on agar surfaces37. To understand the mechanism by which flp-13 confers its somnogenic effects, we performed a random mutagenesis suppressor screen of the flp-13 induced sleepy behavior. By identifying genes that, when mutated, suppress the flp-13 over-expression phenotype, we hope to gain insights into the mechanism of flp-13 somnogenic action.
To identity genetic pathways involved in regulating quiescent behavior in response to flp-13, we mutagenized the flp-13 over-expressing transgenic animals, and isolated their granddaughters filial 2 (F2) animals that remained active after heat shock–induced, over-expression of flp-13. See the methods section for the protocol. We use the L sorter (Fig. 1b) to enrich for the rare mutated animals that remained active post heat shock. To further enrich for true suppressor mutations, we subjected the sorted animals to a second heat shock the next day. Animals that remained motile through these two heat shock treatments were individually cultured and their progenies were retested for post heat shock activity during the animals’ first day of adulthood. The progenies that retain their activity post heat shock were candidates for genetic sequencing.
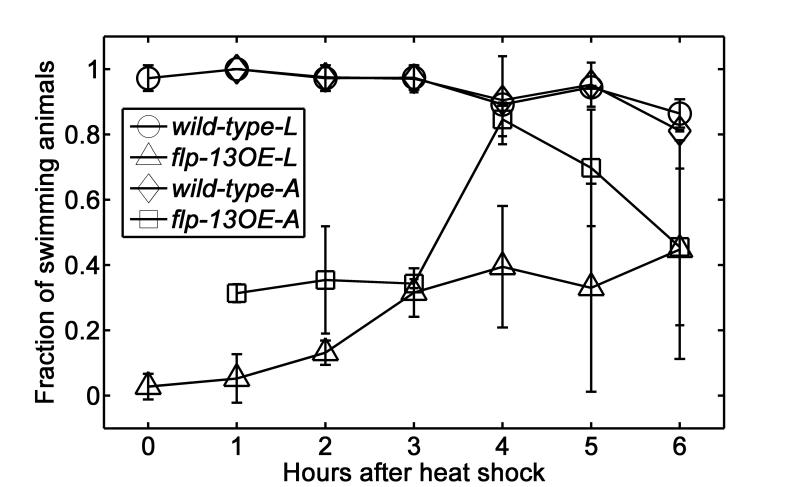
Since our prior heat shock induced flp-13 over-expression (OE) experiments were carried out with animals on agar and not in solution, we first assessed the response of animals suspended in solution to heat shock. Two sets of experiments were carried out. In the first set of experiments, animals were heat-shocked while on agar, washed, and then suspended in M9 buffer. In the second set of experiments, the animals were heat-shocked while suspended in M9 buffer. In both cases, the animals’ activity was monitored while suspended in solution. Animals with average body bending frequency exceeding 0.25Hz over a 20 second time interval were classified as active. The experiments were carried out both with wild type animals (control) and the flp-13 OE strain. Fig. 4 depicts the fractions of active animals as functions of time after a 30-minute heat shock. The circles and diamonds correspond, respectively, to wild-type animals that were heat shocked on agar (n=2 trials with 16 animals in one trial and 20 animals in the other) and in suspension (n=2 trials with 18 animals in one trial and 19 animals in the other trial). As expected, the heat shock had no effect on the wild type animals’ activity. The slight decline in activity after about 4 hours can be attributed to quiescence that is occasionally exhibited by suspended, wild type animals43. The squares correspond to flp-13 OE strain (n=2 trials with 17 animals in one trial and 21 animals in the other) heat shocked on agar and then suspended in buffer. Relatively high fraction of these animals (>30%) remained active. We hypothesize that the process of transferring the animals from agar to suspension stimulated the animals and counteracted, to a degree, the soporific effects of over-expressing flp-13. The triangles correspond to the flp-13 OE strain (n=2 trials with 18 animals in each trial) heat shocked while in solution. The heat shocked animals appear to maintain their quiescent state for about two hours after heat shock and then gradually regained their activity. The experiments shown in Fig. 4 teaches us two things. First, the animals to be sorted must be heat-shocked while in M9 buffer since transfer of animals from agar surfaces to M9 buffer may counteract the heat shock effects. Second, the sorting experiment must be completed during the first two hours after heat shock, when the effects of over-expression are the strongest.
Fig. 4.
The fraction of active animals in M9 buffer as functions of time after a 30-minute heat exposure by immersion in a 33°C water bath. Circles and diamonds represent data of wild-type animals heat shocked in M9 buffer and on agar, respectively and then suspended in M9 buffer for observation. Triangles and squares represent data of flp-13 over-expressing animals, heat shocked in M9 buffer and on agar, respectively, and then suspended in M9 buffer for observation. Error bars correspond to one standard deviation. N= 2 trials for each condition. The number of animals in each trial ranged from 16 to 21.
The granddaughters (F2s) of mutagenized, flp-13(OE) animals were screened for actively moving animals during the first day after adulthood. The F2 animals, suspended in M9 buffer, were heat shocked and subsequently transferred into the holding chambers of twenty L sorting modules, operating in parallel. Each sorting process lasted one hour. Eight sorting processes were carried out with each of the twenty L-type sorters over three days to sort the progeny of approximately 10,000 F1 animals, a total of 201,800 animals. The numbers of animals to be sorted were counted by random sampling. Three runs were carried out in the first day (76,150 animals), three in the second day (80,500 animals), and two in the third day (45,150 animals). Animals screened on each day were the progeny of different F1 animals. The average liquid velocity in the separation conduit (u3) was 254 μm/s. A total of 4,022 (725 in the 1st day, 2051 in the 2nd day, and 1246 in the 3rd day) animals that remained active after the first heat shock (2%) were sorted out.
The sorted animals were transferred from the collection chamber to an agar surface (with bacteria lawn) and subjected to a second heat shock on the following day. Of the 4,022 animals collected from the initial liquid sorting experiment, a total of 42 animals (15 from the first day’s experiment, 12 from the second day’s experiment, and 15 from the third day’s experiment) remained active after heat shock on an agar surface. These 42 animals were cultured individually. The progeny from these 42 animals were tested on an agar surface using direct investigator observation of behavior for heat-shock induced quiescence. Of these 42 candidate suppressors, the progeny of 12 (6 from the first day’s experiment, 3 from the second day’s experiment, and 3 from the last day’s experiment) remained active after heat shock on agar. These 12 animals are considered to be true suppressors and candidates for sequencing. The occurrence rate of true suppressors in the pre-sorted population was 6×10−5 ± 2×10−5. The sorter had enriched the true suppressors sixty-seven fold to 0.004 ± 0.004. The mean and standard deviation are calculated from the data of each of three days.
In parallel to the automated screening, we carried out a conventional genetic screen by manual inspection of animal behavior on an agar surface. Approximately 20 hours of observations were performed by four investigators, one (DMR) with extensive experience, observing C. elegans behavior and the other three with minimal experience. We examined 60,500 F2 progeny of heat-shocked, mutagenized flp-13(OE) transgenic animals for rare sleeping-defective mutants that moved and fed two hours after heat shock on an agar surface. These F2s were the progeny of approximately 3000 F1 animals. We identified two sleeping-defective mutants, which were obtained from independent mutagenesis experiments. One of these two suppressors was identified by the experienced observer (DMR). Based on the results of this conventional assay, the occurrence rate of true suppressors in the mutagenized population was estimated as 3.3×10−5, which is in reasonable agreement with the results obtained with our automated sorter. Hence, the sorter offers significant advantages in terms of throughput, and reduces the need for experienced observers of behavior. Of course, the sorter is scalable and its throughput can be greatly increased.
To determine whether the sleeping-defective mutant animals identified by our motility-based sorter carried loss-of-function mutations in the same gene identified by the manual screening of behavior on an agar surface, we performed genetic complementation tests. We first established that the mutations conferring the sleeping-defective phenotype were recessive to the wild-type chromosome (see Methods), suggesting that they reduce or eliminate gene function. We then compared the post-heat shock behavioral quiescence phenotype of the progeny of a genetic cross between pairs of mutants to that of qnIs303 control animals. Failure of complementation was indicated when cross-progeny animals showed significantly greater movement and feeding than the qnIs303 control animals. The results of the pair-wise complementation tests are shown in Table 2 and the detailed data is provided in Supplementary Tables 1 and 2. Of the nine mutants tested, seven showed a failure to complement in pair-wise testing. These seven mutants thus define a single locus, which we name slep-1 (SLEePing defective 1). Of the seven mutants, two, qn40 and qn44, were isolated by manual screening of behavior on an agar surface and the remaining five were isolated by the motility-based sorter. We conclude that the motility-based sorter is capable of isolating mutations in the same genes identified by conventional mutant screens; furthermore, our analysis indicates that the motility-based sorter is more efficient in the isolation of mutants than the manual inspection. Therefore, high throughput motility-based screening combined with whole genome sequencing holds great promise as a tool for identifying new genes.
Table 2.
Results of the pair-wise complementation tests
| Male parent | Hermaphrodites parent | Outcome | Interpretation |
|---|---|---|---|
| qn52 | qn51 | Failed to complement | Same gene |
| qn52 | qn53 | Failed to complement | Same gene |
| qn44 | qn52 | Failed to complement | Same gene |
| qn44 | qn40 | Failed to complement | Same gene |
| qn44 | qn45 | Failed to complement | Same gene |
| qn44 | qn49 | Failed to complement | Same gene |
| qn44 | qn54 | Complemented | Different gene |
| qn44 | qn46 | Complemented | Different gene |
In addition to the mutant allele shown, which was homozygous, each strain also contained a homozygous copy of the integrated transgene qnls303[Phsp-16.2:flp13; Phsp-16.2:gfp; Prab-3mCherry].
Discussion
We describe a simple, low cost, motility-based sorter for nematodes and demonstrate its utility for both genotyping and forward genetic screening to identify rare mutants. Two different embodiments of the sorter are described, both capable of high throughput operation, enabling the sorting of hundreds of thousands of animals. Many modules can be operated in parallel to further increase throughput. Identical sorting modules can be connected in series to improve enrichment efficiency. It is also possible to form a cascade of modules designed to isolate animals with different motilities.
While we used in our experiments syringe pumps to induce adverse flow and thus select for able swimmers, it is possible to achieve a similar selective effect without the aid of adverse flow by simply tilting the L sorter at an angle and using gravity as the escape barrier. Our preliminary observations (see supplement) suggest that such a separation method works, allowing for reduced cost and even greater economy of scale. Such a sorter assumes that the animals have a narrow density (mass per unit volume) distribution as indicated by a recent study38.
Our proof-of-principle genetic screen was designed in a fashion that allows us to select animals with enhanced motility. Yet the screen can easily be modified to select for animals with reduced motility, as demonstrated by our analysis of the mobility-defective mutants unc-63, lev-10, and dys-1. Such motility defective mutants remain in the holding chamber and fail to escape via the separation conduit.
Our device sorts animals based on their swimming motility in liquids. While the mutants we chose to validate our device behaved similarly while swimming as they did while crawling on an agar surface, it is conceivable that some mutations would affect swimming behavior without affecting crawling behavior, or vice versa. Indeed, mutants with medium-dependent locomotion defects have been described42.
Although our experiments focus on C. elegans, the same device design can be used with other types of worms, including parasitic worms to identify drug resistance, and potentially with other motile cells and organisms, including sperm, bacteria, and zebrafish.
Methods
Device fabrication
A master mold for the conduits was made using standard photolithography with negative photoresist (SU8 2025, Microchem). A three inch wafer (EI-Cat Inc.) was rinsed with acetone, isopropyl alcohol (IPA), and de-ionized water; heated to 65°C on a hot plate (Torrey Pines Sci.); and loaded on a spinner (WS-650S-6NPP/LITE, Laurell). About 5 mL of SU8 2025 photoresist was poured onto the center of the wafer. The wafer was then spun at 500 rpm for 5s and then at 800 rpm for 25s. Next, the wafer was baked at 95°C on a hot plate for two hours. Once cooled, the wafer was exposed to a 365nm wavelength light at 3.3 mW/cm2 power through a transparency mask (designed with LayoutEditor software and printed by Photo Plot Store) for 140s. Then, the wafer was baked at 65°C on a hot plate for 10 minutes and at 95°C for 60 minutes. The wafer was allowed to cool at room temperature for five minutes. Then, the wafer was immersed in SU8 developer (Microchem) for 110 minutes after which the wafer was rinsed with fresh SU8 developer and IPA. The height of the conduit’s mold was measured with a profilometer (Alpha step 200, Tencor) to be 91 μm.
Polydimethylsiloxane (PDMS, Sylgard 184, Ellsworth Adhesives), a pre-polymer and a cure agent in the ratio of 1:10, was cast on the master mold, and cured at room temperature for 24 hours to form a 5 mm thick PDMS slab. The PDMS replica was then peeled off from the master mold and cut into modules containing individual conduits. A 2.90 mm OD hole-puncher (15077, Harris Uni-Core, Ted Pella, Inc.) was used to puncture holes for fluid inlet and outlet. A 4.39 mm OD hole-puncher (15080, Harris Uni-Core, Ted Pella, Inc.) was used to puncture holes for the loading well and worm reservoir. The PDMS piece was then permanently bonded to a glass slide (plain microscopic slide, 76.2×25.4×1 mm, Fisher Scientific). Both the PDMS piece and glass slide were treated with oxygen plasma prior to bonding.
Animal Cultivation and Strains Used
Prior to the experiments, animals were cultivated on the surfaces of NGM agar 1, fed the bacterial strain DA837 44, and kept in a constant temperature, 20°C incubator. The wild-type strain used was N2, variety Bristol 1. Other strains used were LS292 dys-1(cx18) I, ZZ17 lev-10, ZZ37 unc-63(x37) I and JIM113 ujIs113[pie-1::mCherry::Histone H2B; Pnhr-2::mCherry::HIS-24; unc-119(+)] II 45. Strains generated in this study were: NQ793 qnIs303[Phs:flp-13; Phs:gfp; Prab-3:mCherry]; slep-1(qn40), NQ51 qnIs303; slep-1(qn40), NQ52 qnIs303; slep-1(qn52), NQ53 qnIs303; slep-1(qn53), NQ54 qnIs303; qn54, NQ56 qnIs303; qn56, NQ57 qnIs303; qn57, NQ58 qnIs303; qn58, NQ59 qnIs303; qn59, NQ60 qnIs303; qn60, NQ792 qnIs303, slep-1(qn45), NQ810 qnIs303; slep-1(qn44), NQ814 qnIs303; slep-1(qn49). Unless stated otherwise, experiments were performed on hermaphrodites. Experiments were performed on well-fed young adult animals, which were staged based on developmental time (3-4 days after feeding L1-arreseted animals) or by selecting for L4 animals the day prior to the experiment and then aging at 20 degrees for one day.
Mutagenesis of C. elegans
EMS mutagenesis was performed according to standard procedure 46. Large quantities of staged L4 animals were obtained using the alkaline bleach method 47 and were incubated in 4 mL of 50 mM Ethyl methanesulfonate (EMS) solution in a 15 mL conical tube for 4 hours at room temperature (21-23 degrees) on a nutator mixer. Then, the EMS solution in the conical tube was separated from the animals using a centrifuge and replaced by M9 buffer. After five washes to remove residual EMS, the animals were plated on the surfaces of NGM agar with bacteria, and kept in a constant temperature, 20 degree incubator for 3-4 days. Eggs of the F1 generation of these animals were isolated using the alkaline bleach method 47 and were suspended in about 7 mL M9 buffer in a 15 mL conical tube for 18-24 hours. The L1 stage F2 animals that hatched from these eggs were then plated on the surfaces of NGM agar pre-seeded with a lawn of bacteria. The L1 stage animals were plated at a density of 800 animals per plate. The agar plates had a diameter of 5.5 cm and contained a volume of 11 mL of NGM agar. After 3 days at 20 degrees, the F2 animals were heat shocked by submersion in a 33°C water bath for 30 minutes and subsequently screened either manually or using the sorters during their young adulthood. In the manual screens, the animals were heat shocked on agar plates. In the automated screens, the animals were heat shocked in M9 buffer in a conical tube and subsequently transferred to the holding chambers of the sorters. In some experiments assessing the effect of heat-shock methods (results of which are shown in figure 4), the animals were first transferred from an agar surface to a M9 buffer-filled plastic petri-dish and then heat-shocked by submersing the petri-dish into a 33°C water bath for 30 minutes.
Genetic complementation testing
To determine whether or not the sleeping-defective mutants isolated contained loss-of-function mutations in the same gene, we performed pair-wise genetic complementation tests. We first established the recessive nature of each mutation by comparing the behavioral quiescence phenotype two hours after 33° heat shock of male progeny from a cross between qnIs303 males and mutant hermaphrodites to the phenotype of homozygous mutant males. Recessive mutants were those in which the sleeping-defective phenotype was observed in homozygous mutant males but not in heterozygous mutant males. For example, two hours after heat shock-based activation of flp-13 expression, the male progeny of NQ570 males crossed by NQ810 hermaphrodites were far less active than NQ810 males, which were homozygous for the qn44 mutation.
We then compared the post-heat shock behavioral quiescence phenotype of the progeny of a cross between pairs of mutants to that of qnIs303 males. Failure of complementation was indicated when cross-progeny animals showed significantly greater movement and feeding than the qnIs303 control males. The behavior of the animals was assessed during their first day of adulthood. In seven crosses (Table S1), we tested male progeny and were therefore certain that the animals tested were cross-progeny and not self-progeny. In the remaining four crosses (Table S2), in which we tested hermaphrodite progeny, we selected the progeny from among cohorts containing approximately 50% males, suggesting that the mating was highly efficient.
Supplementary Material
Acknowledgments
This research was supported, in part, by National Institutes of Health (NIH) NIA Grant 5R03AG042690-02 and by the National Science Foundation NSEC DMR08-32802 to the University of Pennsylvania. D.M.R. was supported by NIH R01NS064030, R01NS088432, and R21NS091500. We are grateful to Gregg Artiushin, who had manually isolated the flp-13(OE) suppressor and demonstrated the feasibility of the genetic screen used in our work. Kun He Lee assisted with the mutagenesis of C. elegans. The strains of C. elegans used in this study were obtained from the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
References
- 1.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yemini E, Jucikas T, Grundy LJ, Brown AE, Schafer WR. Nature methods. 2013;10:877–879. doi: 10.1038/nmeth.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krajacic P, Shen X, Purohit PK, Arratia P, Lamitina T. Genetics. 2012;191:1015–1021. doi: 10.1534/genetics.112.141176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swierczek NA, Giles AC, Rankin CH, Kerr RA. Nature methods. 2011;8:592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayden EC. Nature. 2014;507:294–295. doi: 10.1038/507294a. [DOI] [PubMed] [Google Scholar]
- 6.Sarin S, Prabhu S, O’Meara MM, Pe’er I, Hobert O. Nature methods. 2008;5:865–867. doi: 10.1038/nmeth.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang W, Magrini VJ, Richt RJ, Sander SN, Stewart DA, Stromberg M, Tsung EF, Wylie T, Schedl T, Wilson RK, Mardis ER. Nature methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 8.Chronis N, Zimmer M, Bargmann CI. Nature methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- 9.Hulme SE, Shevkoplyas SS, Apfeld J, Fontana W, Whitesides GM. Lab Chip. 2007;7:1515–1523. doi: 10.1039/b707861g. [DOI] [PubMed] [Google Scholar]
- 10.Chung K, Crane MM, Lu H. Nature methods. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 11.Shi W, Qin J, Ye N, Lin B. Lab Chip. 2008;8:1432–1435. doi: 10.1039/b808753a. [DOI] [PubMed] [Google Scholar]
- 12.Chokshi TV, Ben-Yakar A, Chronis N. Lab Chip. 2009;9:151–157. doi: 10.1039/b807345g. [DOI] [PubMed] [Google Scholar]
- 13.Gilleland CL, Rohde CB, Zeng F, Yanik MF. Nature protocols. 2010;5:1888–1902. doi: 10.1038/nprot.2010.143. [DOI] [PubMed] [Google Scholar]
- 14.Hulme SE, Shevkoplyas SS, McGuigan AP, Apfeld J, Fontana W, Whitesides GM. Lab Chip. 2010;10:589–597. doi: 10.1039/b919265d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krajniak J, Lu H. Lab Chip. 2010;10:1862–1868. doi: 10.1039/c001986k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi W, Wen H, Lu Y, Shi Y, Lin B, Qin J. Lab Chip. 2010;10:2855–2863. doi: 10.1039/c0lc00256a. [DOI] [PubMed] [Google Scholar]
- 17.Albrecht DR, Bargmann CI. Nature methods. 2011;8:599–605. doi: 10.1038/nmeth.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung K, Zhan M, Srinivasan J, Sternberg PW, Gong E, Schroeder FC, Lu H. Lab Chip. 2011;11:3689–3697. doi: 10.1039/c1lc20400a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crane MM, Stirman JN, Ou CY, Kurshan PT, Rehg JM, Shen K, Lu H. Nature methods. 2012;9:977–980. doi: 10.1038/nmeth.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han B, Kim D, Ko UH, Shin JH. Lab Chip. 2012;12:4128–4134. doi: 10.1039/c2lc40209b. [DOI] [PubMed] [Google Scholar]
- 21.Lockery SR, Hulme SE, Roberts WM, Robinson KJ, Laromaine A, Lindsay TH, Whitesides GM, Weeks JC. Lab Chip. 2012;12:2211–2220. doi: 10.1039/c2lc00001f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belfer SJ, Chuang HS, Freedman BL, Yuan J, Norton M, Bau HH, Raizen DM. Sleep. 2013;36:689–698G. doi: 10.5665/sleep.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krajniak J, Hao Y, Mak HY, Lu H. Lab Chip. 2013;13:2963–2971. doi: 10.1039/c3lc50300c. [DOI] [PubMed] [Google Scholar]
- 24.Lee H, Kim SA, Coakley S, Mugno P, Hammarlund M, Hilliard MA, Lu H. Lab Chip. 2014;14:4513–4522. doi: 10.1039/c4lc00789a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu CC, Raizen DM, Fang-Yen C. Journal of neuroscience methods. 2014;223:35–39. doi: 10.1016/j.jneumeth.2013.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Stone HA, Murphy CT. Lab Chip. 2015;15:524–531. doi: 10.1039/c4lc01028k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen H, Yu Y, Zhu G, Jiang L, Qin J. Lab Chip. 2015 doi: 10.1039/c4lc01377h. DOI: 10.1039/c4lc01377h. [DOI] [PubMed] [Google Scholar]
- 28.Ai X, Zhuo W, Liang Q, McGrath PT, Lu H. Lab Chip. 2014;14:1746–1752. doi: 10.1039/c3lc51334c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P, Martin RJ, Dong L. Lab Chip. 2013;13:650–661. doi: 10.1039/c2lc41174a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johari S, Nock V, Alkaisi MM, Wang W. Lab Chip. 2013;13:1699–1707. doi: 10.1039/c3lc41403e. [DOI] [PubMed] [Google Scholar]
- 31.Chuang HS, Raizen DM, Lamb A, Dabbish N, Bau HH. Lab Chip. 2011;11:599–604. doi: 10.1039/c0lc00532k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doitsidou M, Poole RJ, Sarin S, Bigelow H, Hobert O. PloS one. 2010;5:e15435. doi: 10.1371/journal.pone.0015435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doitsidou M, Flames N, Lee AC, Boyanov A, Hobert O. Nature methods. 2008;5:869–872. doi: 10.1038/nmeth.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maniere X, Lebois F, Matic I, Ladoux B, Di Meglio JM, Hersen P. PloS one. 2011;6:e16637. doi: 10.1371/journal.pone.0016637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rezai P, Siddiqui A, Selvaganapathy PR, Gupta BP. Lab Chip. 2010;10:220–226. doi: 10.1039/b917486a. [DOI] [PubMed] [Google Scholar]
- 36.Sukul NC, Croll NA. Journal of nematology. 1978;10:314–317. [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson MD, Lee KH, Churgin MA, Hill AJ, Van Buskirk C, Fang-Yen C, Raizen DM. Current biology: CB. 2014;24:2406–2410. doi: 10.1016/j.cub.2014.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reina A, Subramaniam AB, Laromaine A, Samuel AD, Whitesides GM. PloS one. 2013;8:e69651. doi: 10.1371/journal.pone.0069651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan J, Raizen DM, Bau HH. 2015 in review. [Google Scholar]
- 40.Yuan J, Raizen DM, Bau HH. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1424962112. DOI: 10.1073/pnas.1424962112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava N, Clark DA, Samuel AD. Journal of neurophysiology. 2009;102:1172–1179. doi: 10.1152/jn.90952.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray JM, Hill JJ, Bargmann CI. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh R, Emmons SW. The Journal of experimental biology. 2008;211:3703–3711. doi: 10.1242/jeb.023606. [DOI] [PubMed] [Google Scholar]
- 44.Davis MW, Somerville D, Lee RY, Lockery S, Avery L, Fambrough DM. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1995;15:8408–8418. doi: 10.1523/JNEUROSCI.15-12-08408.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walton T, Preston E, Nair G, Zacharias AL, Raj A, Murray JI. PLoS genetics. 2015;11:e1005003. doi: 10.1371/journal.pgen.1005003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kutscher LM, Shaham S. In: WormBook. T. C. e. R. Community, editor. WormBook; DOI: 10.1895/wormbook.1.167.1. [Google Scholar]
- 47.Stiernagle T. In: WormBook. T. C. e. R. Community, editor. WormBook; DOI: 10.1895/wormbook.1.101.1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.