Abstract
Frontotemporal lobar degeneration with TAR DNA binding protein 43 inclusions (FTLD-TDP) is the most common pathology associated with frontotemporal dementia (FTD). Repeat expansions in chromosome 9 open reading frame 72 (C9ORF72) and mutations in progranulin (GRN) are the major known genetic causes of FTLD-TDP; however, the genetic etiology in the majority of FTLD-TDP remains unexplained.
In this study, we performed whole-genome sequencing in 104 pathologically confirmed FTLD-TDP patients from the Mayo Clinic brain bank negative for C9ORF72 and GRN mutations and report on the contribution of rare single nucleotide and copy-number variants in 21 known neurodegenerative disease genes. Interestingly, we identified 5 patients (4.8%) with variants in optineurin (OPTN) and TANK-binding kinase 1 (TBK1) that are predicted to be highly pathogenic, including two double mutants. Case A was a compound heterozygote for mutations in OPTN, carrying the p.Q235* nonsense and p.A481V missense mutation in trans, while case B carried a deletion of OPTN exons 13–15 (p.Gly538Glufs*27) and a loss-of-function mutation (p.Arg117*) in TBK1. Cases C–E carried heterozygous missense mutations in TBK1, including the p.Glu696Lys mutation which was previously reported in two amyotrophic lateral sclerosis (ALS) patients and is located in the OPTN binding domain. Quantitative mRNA expression and protein analysis in cerebellar tissue showed a striking reduction of OPTN and/or TBK1 expression in 4 out of 5 patients supporting pathogenicity in these specific patients and suggesting a loss-of-function disease mechanism. Importantly, neuropathologic examination showed FTLD-TDP type A in the absence of motor neuron disease in 3 pathogenic mutation carriers.
In conclusion, we highlight TBK1 as an important cause of pure FTLD-TDP, identify the first OPTN mutations in FTLD-TDP, and suggest a potential oligogenic basis for at least a subset of FTLD-TDP patients. Our data further adds to the growing body of evidence linking ALS and FTD and suggests a key role for the OPTN/TBK1 pathway in these diseases.
Keywords: Whole-genome sequencing, FTLD-TDP, OPTN, TBK1, oligogenic mechanism
INTRODUCTION
Frontotemporal lobar dementia (FTD) represents a collection of neurodegenerative diseases accounting for 5–10% of all dementia patients and 10–20% of patients who develop dementia before 65 years [22]. With 25–50% of patients reporting at least one other relative with a neurodegenerative disorder, it is clear that FTD has a strong genetic basis. Causal FTD mutations with high penetrance have been described in the microtubule associated protein tau gene (MAPT) [26,27], progranulin gene (GRN) [4,12] and chromosome 9 open reading frame 72 (C9ORF72) [14,45]; while mutations in a range of other genes have been found to cause FTD on rare occasions [49]. Despite recent advances, the genetic defect in a significant subset of familial and most sporadic FTD patients currently remains unknown.
FTD is a clinical syndrome that is characterized by progressive deterioration in behavior, personality and/or language, with relative preservation of memory [22]. Based on the symptomatology clinical subtypes of FTD are defined: behavioral variant (bvFTD), and two types of primary progressive aphasia: progressive nonfluent/agrammatic variant (agPPA) and semantic variant (svPPA) dementia. The term frontotemporal lobar degeneration (FTLD) is often used to describe the pathology associated with clinical FTD, and refers to the selective degeneration of the frontal and temporal lobes. FTLD is also heterogeneous with cases being classified into subtypes based on the main component(s) of pathologic protein aggregates. FTLD with aggregates of the TAR DNA-binding protein 43 (FTLD-TDP) is the most common pathologic subtype of FTLD accounting for more than 50% of the FTLD patients [47,33]. FTLD-TDP cases with moderate to numerous TDP-43-immunoreactive neuronal cytoplasmic inclusions (NCIs) and short dystrophic neurites (DNs) predominantly in the upper cortical layers II/III are designated type A; cases with moderate to numerous TDP-43-immunoreactive NCIs and sparse DNs across all cortical layers are designated type B; and cases in which long dystrophic neurites are present predominantly in the upper cortices and NCIs are rare are assigned a type C. The relevance of the FTLD-TDP heterogeneity and types A–C classification system is supported by clinical and genetic correlations, as well as by evidence for distinct biochemical properties of TDP-43 in the different subtypes [44]. For example, all patients carrying loss-of-function mutations in GRN are characterized by FTLD-TDP type A, while the majority of C9ORF72 mutation carriers have FTLD-TDP type B.
In recent years, clinical, pathologic and genetic studies have contributed to an emerging theme that FTD and amyotrophic lateral sclerosis (ALS) are part of a disease spectrum with a common underlying pathogenesis [6,23]. Fifteen percent of ALS patients have cognitive and behavioral impairment and 15% of FTD patients meet ALS criteria [31]. Moreover, TDP-43, fused in sarcoma (FUS) and p62 inclusions are found in post-mortem tissues of both ALS and FTD cases [2,42,1,3,28,41,57]. The most striking evidence, however, came from the identification of repeat expansions in C9ORF72 as a common cause of FTD, ALS and mixed FTD-ALS patients, which firmly established a genetic link between these disorders [14,45].
Despite a strong family history in up to 50% of patients, a substantial proportion of FTD patients are sporadic. While currently unknown environmental factors may contribute to the disease in these individuals, the presence of incomplete penetrance, compound heterozygous variants or genetic variants in multiple genes, i.e. oligogenic disease mechanism, could be contributing to the disease in these patients. In fact, recently several reports suggested an oligogenic mechanism as the basis for ALS in sporadic patients but also in ALS families [32,56,10]. An oligogenic basis for FTD however has not yet been systematically assessed [54].
To date, oligogenic studies have been largely based on the screening of candidate genes in large pedigrees or cohorts of unrelated individuals. Recent genetic discoveries, however, continuously increase the number of candidate genes to look at, diminishing the cost-effectiveness of this approach. The use of next-generation sequencing now allows sequencing the whole exome or genome at once, followed by a selection of candidate genes to study in detail. Whole exome and genome sequencing are thus likely to enable and accelerate discoveries of oligogenic mechanisms in human diseases.
In this study we present the first whole-genome sequencing study in FTLD-TDP patients, using a unique FTLD-TDP cohort from the Mayo Clinic Florida brain bank in which mutations in GRN and C9ORF72 were excluded. In a first attempt to genetically characterize this cohort, we hypothesized that rare variants in causal genes previously implicated in neurodegenerative disorders could be responsible (in part) for pathologically confirmed FTLD-TDP. The use of whole-genome sequencing data further allowed us to incorporate both single nucleotide variants (SNVs) and copy number variants (CNVs) into our analysis. Importantly, we identified potentially causative variants (CNV and SNVs) in optineurin (OPTN) and TANK-binding kinase 1 (TBK1), two functionally related genes previously associated with ALS, and provide additional support for an oligogenic basis of FTLD-TDP.
MATERIAL AND METHODS
Patient cohort
Neuropathologic and immunohistological records of the Mayo Clinic Florida brain bank were reviewed by the end of 2013 and 168 patients with a pathologic diagnosis of FTLD-TDP with frozen tissue availability were identified. Most patients were referred to our brain bank as part of the Mayo Clinic Alzheimer’s disease Research Center, the State of Florida Alzheimer’s Disease Initiative, the Mayo Clinic Study of Aging and The Cure PSP brain bank. Four pathologically confirmed FTLD-TDP patients without frozen tissue for which a blood sample was available as a source of DNA were also identified. Out of these 172 patients, previous genetic analyses identified 33 patients (19%) with pathogenic C9ORF72 repeat expansions, 33 (19%) patients with loss-of-function mutations in GRN, one patient (0.5%) with a valosin containing protein (VCP) mutation and one patient (0.5%) with a leucine rich repeat kinase 2 (LRRK2) mutation, resulting in 104 FTLD-TDP patients (60%) with unknown genetic cause. Among these 104 patients, 42% had FTLD-TDP type A, 27% had FTLD-TDP type B and 28% had FTLD-TDP type C. The average age at diagnosis was 66.8 years (range 38–86 years) and average age at death was 74.7 years (range 41–97 years). Two patients were Asian-American and two patients were Hispanic, all others were non-Hispanic whites. Information on family history was available for 82% (85/104) of the FTLD-TDP patients and 50.5% of these reported a family history of neurodegenerative disease. Limited ante-mortem clinical information was available on most patients and revealed diagnoses within the dementia spectrum, most commonly FTD and Alzheimer’s disease (AD).
Whole-Genome Sequencing and Bioinformatic analysis
Whole-genome sequencing was performed by Complete Genomics (Mountain View, CA, USA) using unchained combinatorial probe anchor ligation sequencing [16] in 104 pathologically confirmed FTLD-TDP patients with unknown genetic cause and 3 GRN mutation carriers (as internal positive controls). Sequence reads were mapped to the reference genome (GRCh37) and variants were called by local de novo assembly according to the methods previously described [16]. All samples passed internal Complete Genomics quality control parameters and a gender check. For additional quality control metrics Complete Genomics calculated the ratio of variants reported in dbSNPv137 to all variants identified (on average 95.6% of all called variants and 98.5% of high confidence variants). We generated 164Gb of sequence per individual covering 96% of the entire genome and 98% of the exome. Eighty-five percent of exonic high quality reads had a read depth higher than 30×. On average 3,349,512 high quality variants were identified per individual and 21,171 were exonic. Two different bioinformatics pipelines were then used to prioritize variants. First, variants identified by Complete Genomics in the “var” file were submitted to an in-house bioinformatics pipeline [20]. This tool includes variation information on their genomic position (exon, splice site, intron, untranslated region (UTR), promoter region), their predicted effect on protein level, and their frequency in different databases (dbSNPv37; Exome variant server (ESP), and the genome of the Netherlands (GoNL) [7]. In addition, it contains variant frequency data from two datasets generated using the same Complete Genomics sequencing technology, namely the Wellderly dataset [37] as well as 100 neurologically healthy parents of patients with severe intellectual disability [20]. Filtering was performed following strict criteria and consisted of removing any low confidence variants and excluding variants with a minor allele frequency (MAF) ≥0.01% in ESP and 1000 Genomes as well as variants present in control cohorts. Only missense, nonsense, stop-loss, indels and splice variants were considered.
In parallel, we uploaded the MasterVar file and the high Confidence Junctions Beta files into QIAGEN’s Ingenuity® Variant Analysis™ software (www.qiagen.com/ingenuity) from QIAGEN Redwood City. Similarly to the first pipeline, only high confidence variants were selected (variant quality score >50; read depth >10). We then selected only variants with a MAF<0.01% in ESP and 1000 Genomes, and only variants absent in 155 Harvard Personal Genome Project controls. Only missense, nonsense, stop-loss, indels and splice variants were selected.
In addition to the SNVs analysis, Complete Genomics also calls CNVs based on coverage determined in sliding windows of 2kb intervals. The presence of such structural and copy number variations in our FTLD-TDP cohort was assessed by using the Ingenuity Variant Analysis software (variant quality score>30; read depth>10). On average 242 CNVs were detected per individual. Manual examination of selected CNVs was performed using high Confidence SvEvents Beta files from Complete Genomics to confirm the coordinates and the all Junctions Beta files to examine the precise sequence of the event. All SNVs and CNVs detected in known neurodegenerative disease genes using either pipeline (n=13) were confirmed by Sanger sequencing.
OPTN copy number assay
To validate OPTN copy number variations duplex real-time quantitative PCR assays were performed on an ABI7900 using TaqMan copy number assay Hs02993339 _cn (OPTN, exon 14) from Life Technology (Life Technologies, Grand Island, NY, USA). For each assay, RNaseP was used as the reference (Life Technologies, Grand Island, NY, USA). To confirm the presence of a partial genomic deletion of OPTN and to confirm the deletion breakpoint, a PCR-based assay was also developed. PCR primers were designed on either side of the deletion breakpoint based on the exact deletion coordinates obtained from whole-genome sequencing analysis of one case by Complete Genomics (Forward: ATTTGGGACCCTGGAATCAT; Reverse: TGTTTTTGGAGGCAACCTTT; approximately 500-bp amplification product).
OPTN and TBK1 mRNA expression analysis
Total RNA was extracted from cerebellar tissue from FTLD-TDP cases A and B carrying TBK1/OPTN double mutations, from 6 FTLD-TDP patients without mutations and 4 control brains using the RNeasy Plus Mini Kit (Qiagen, Redwood City, CA, USA), and its quality was assessed on an Agilent 2100 Bioanalyzer. RNA concentration was determined using Nanodrop 8000 (Thermo Fisher Scientific, Walthman, MA, USA). A reverse transcription reaction was performed using the Superscript III system (Invitrogen™, Thermo Fisher Scientific, Walthman, MA, USA) with 400ng of RNA as template and a 1:1 mix of random hexamer and Oligo dT primers. Real-time quantitative PCR using an ABI7900HT PCR System (Invitrogen™, Thermo Fisher Scientific, Walthman, MA, USA) was performed in triplicate for each sample using Taqman gene expression probes for the targets OPTN (Hs00184221_m1 - exons 6–7) and TBK1 (Hs00975474_m1 - exons 3–4), and for endogenous control RPLP0 (large ribosomal protein, Hs00420895_gh). Results were analyzed using SDS software version 2.2.2 and relative quantification of OPTN was determined using the ΔΔCt method. A Mann-Whitney test was performed to assess the difference of RNA expression between FTLD-TDP patients without mutations and control brains using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, CA, USA)
OPTN and TBK1 western blotting
For Western blotting experiments, approximately 100 mg of cerebellar tissue from cases A and B, 6 FTLD-TDP patients without mutations and 6 control brains were homogenized in 400µl RIPA buffer (Boston BioProducts, Ashland, MA, USA) supplemented with protease/phosphatase inhibitors (Thermo Fisher Scientific, Waltham, MA, USA). Two independent tissue preps were performed for each cerebellar sample. Samples were subjected to centrifugation at 4°C for 5 minutes at 14,000rpm (rotation per minute) and the supernatant was transferred to a fresh tube. Protein concentrations were determined by the PierceTM Bicinchoninic Acid Protein Assay (Thermo Fisher Scientific, Waltham, MA, USA) per the manufacturer’s instructions using known concentrations of bovine serum albumin as standards. Lysates were prepared at a concentration of 1mg/ml protein in Novex sample buffer (Invitrogen™, Thermo Fisher Scientific, Walthman, MA, USA) supplemented to 5% with β-mercaptoethanol. Samples were denatured by heating for 5 minutes at 95°C before loading into SDS-polyacrylamide gels (Life Technologies, Grand Island, NY, USA). 30µg of protein for each sample was loaded in a 4–12% gel for migration. Gels were then transferred to Immobilon membranes (EMD Millipore, Darmstadt, Germany), and immunoblotted with the corresponding primary antibody overnight (anti-OPTN: ab79110 1/1,000 – anti-TBK1: ab40676 1/10,000 (Abcam, Cambridge, UK) – anti-GAPDH (Glyceraldehyde 3-phosphate dehydrogenase): H86504M 1/500,000 (Meridian Life Science, Cincinnati, OH, USA). The next day, blots were incubated with an HRP-conjugated secondary antibody (Promega, Madison, WI, USA) and bands were detected by enhanced chemiluminescence using Western Lightning Plus-ECL reagents (Perkin Elmer, Waltham, MA, USA). Densitometric analyses were performed using ImageJ software [48]. Relative quantification was carried out after normalization to the band intensities of 6 control brains using two independent protein extractions. A Mann-Whitney test was performed to assess the difference of protein expression between FTLD-TDP patients without mutations and control brains.
OPTN and TBK1 immunohistochemistry
All cases included in this study had been previously diagnosed as FTLD-TDP by a single, experienced neuropathologist (DWD) and had available hematoxylin and eosin as well as Thioflavin-S stains on a number of cortical and subcortical brain regions. For this study, immunohistochemistry was performed on formalin-fixed paraffin-embedded tissue from the frontal cortex, primary motor cortex, temporal cortex (with hippocampus), and medulla of FTLD-TDP cases A–E with potential pathogenic mutations in OPTN and TBK1, 6 FTLD-TDP cases without any causative mutations (matched for FTLD-TDP Type, age and sex), and 4 neuropathologically normal controls (matched for age and sex). Five micron-thick sections were deparaffinized and steamed in water for 30 minutes for antigen retrieval (pH 6 citrate buffer for tissue immunostained with TBK1). Immunohistochemistry was performed using Dako EnVision™+ reagents on a Dako Autostainer (Dako North America, Inc., Carpinteria, CA, USA). Primary antibodies included anti-optineurin (rabbit polyclonal, ab79110, 1:100, Abcam Inc., Cambridge, MA, USA), anti-p62/sequestosome-1 (mouse monoclonal, p62-lck, 1:250, BD Biosciences, Franklin Lakes, NJ, USA), anti-TBK1 (mouse monoclonal, LS-B1317, 1:200, LifeSpan BioSciences, Inc., Seattle, WA, USA) and anti-phosphorylated TDP-43 (mouse monoclonal, pS409/410, 1:5,000, Cosmo Bio Co., Tokyo, Japan). Subsequently, slides were counterstained with hematoxylin, dehydrated and coverslipped. Slides were viewed on an Olympus BX40 microscope.
Additional immunohistochemistry with p62 antibodies was performed on the frontal cortex of another 9 FTLD-TDP cases without causative mutations (matched for FTLD-TDP Type, age and sex; 2 cases with known C9ORF72 mutations and 2 cases with known GRN mutations). Immunostained slides of the FTLD-TDP cases A–E and 15 FTLD-TDP cases without causative mutations in OPTN and TBK1 were imaged using an Aperio AT2 bright field scanner (Leica Microsystems, Buffalo Grove, IL, USA). Using ImageScope software, gray matter (spanning all cortical laminae) and white matter tissue was analyzed using a color deconvolution computer algorithm. The pathologic burden was measured as the percentage of strong positive staining in the given region of interest for each of the 20 cases.
RESULTS
OPTN and TBK1 recurrently harbor highly damaging rare coding variants in FTLD-TDP
Whole-genome sequencing of 107 FTLD-TDP patients followed by filtering of rare coding single nucleotide variants (SNVs) through the two bioinformatic pipelines as described in the material and methods section resulted in a total of 22,121 variants. To determine the contribution of mutations in known neurodegenerative disease genes to FTLD-TDP, we next focused on 21 major disease genes previously implicated in AD, Parkinson’s disease, FTD, ALS, Creutzfeldt Jacob’s disease and hereditary diffuse leukoencephalopathy with axonal spheroids (Online Resource 1). This reduced the number to only 13 variants, including 3 different GRN mutations (one missense, one small deletion and one splice-site mutation) (Table 1). The 3 patients carrying GRN mutations were included in our cohort and blindly analyzed in order to assess the efficiency of our pipeline to detect different types of known pathogenic mutations, and all were successfully identified. Out of the remaining 10 variants, 4 were detected in TANK-binding kinase 1 (TBK1), two in leucine rich repeat kinase 2 (LRRK2), one in optineurin (OPTN), one in fused in sarcoma (FUS), one in profilin 1 (PFN1) and one in the colony stimulating factor 1 receptor (CSF1R). Importantly, when we sorted these 10 remaining variants by pathogenicity score based on CADD_Phred score, all 4 TBK1 variants and the OPTN variant had scores higher than 20, meaning that those substitutions are predicted to be among the 1% most deleterious substitutions in the human genome (Table 1; Figure 1a). Cases A and B carried nonsense mutations in OPTN (NM_001008211.1:c.703C>T; p.Gln235*), and TBK1 (NM_013254.3:c.349C>T; p.Arg117*) respectively; while the other 3 TBK1 mutations observed in cases C–E were missense changes. Importantly, one of the TBK1 missense changes (NM_013254.3:c.2086G>A; p.Glu696Lys; case C) was recently reported in two Swedish ALS patients and was shown to impair the binding of TBK1 to OPTN in vitro [18]. The 5 variants with CADD_Phred scores below 20 are less likely to be pathogenic and were located outside the previously reported domains implicated in disease (CSF1R) [43], previously seen in controls (PFN1) [61,53], or when identified in a family, did not segregate with the disease (FUS) [25].
Table 1.
Rare coding variants identified in FTLD-TDP patients in 21 known neurodegenerative disease genes.
| Gene name |
Case ID |
cDNA change |
Protein change |
Domain | CADD_PHRED | SNP id | ExAC N (frequency) |
Reference |
|---|---|---|---|---|---|---|---|---|
| OPTN | case A | NM_001008211.1: c.703C>T | p.Gln235* | Coil-Coiled domain | 39 | New | Cady et al., 2015 [8] | |
| TBK1 | case B | NM_013254.3: c.349C>T | p.Arg117* | Kinase domain | 37 | 1 (0.000008259) | Novel | |
| TBK1 | case C | NM_013254.3: c.2086G>A | p.Glu696Lys | 31 | 2 (0.00001654) | Freischmidt et al., 2015 [18] | ||
| TBK1 | case D | NM_013254.3: c.916C>A | p.Leu306Ile | Ubiquitin-like domain | 28.4 | rs201970436 | 36 (0.00001654) | Novel |
| TBK1 | case E | NM_013254.3: c.1201A>G | p.Lys401Glu | 22.9 | 1 (0.00001227) | Novel | ||
| LRRK2 | case F | NM_198578.3: c.776C>A | p.Ala259Glu | ARM | 18.11 | rs199557665 | New | Novel |
| PFN1 | case G | NM_005022.3: c.350_351delinsGT | p.Glu117Gly | 17.52 | rs140547520 rs151313170 | 50 (0.0004120) | Wu et al., 2012 – van Blitterswijk et al., 2013 [61,53] | |
| GRN | case H | NM_002087.2: c.26C>A | p.Ala9Asp | Signal peptide | 17.52 | rs63751243 | New | Mukheerje et al., 2006 [40] |
| CSF1R | case I | NM_005211.3: c.533G>A | p.Ser178Asn | Ig-like C2-type 2 | 16.05 | New | Novel | |
| LRRK2 | case J | NM_198578.3: c.3087G>T | p.Gln1029His | Leucine-rich repeat | 15.23 | New | Novel | |
| FUS | case K | NM_004960.3: c.537A>C | p.Gln179His | RGG1 | 13.05 | New | Huey et al., 2012 [25] | |
| GRN | case L | NM_002087.2: c.709-2A>G | p.Ala237Trpfs*4 | Granulin domain B | 12.51 | rs63750548 | New | Leverenz et al., 2007 [29] |
| GRN | case M | NM_002087.2: c.1145del | p.Thr382Serfs*30 | Granulin domain C | 0.103 | rs63750805 | New | Baker et al., 2006 [4] |
CADD_PHRED: Combined Annotation Dependent Depletion scaled score, ARM: Armadillo repeat domain
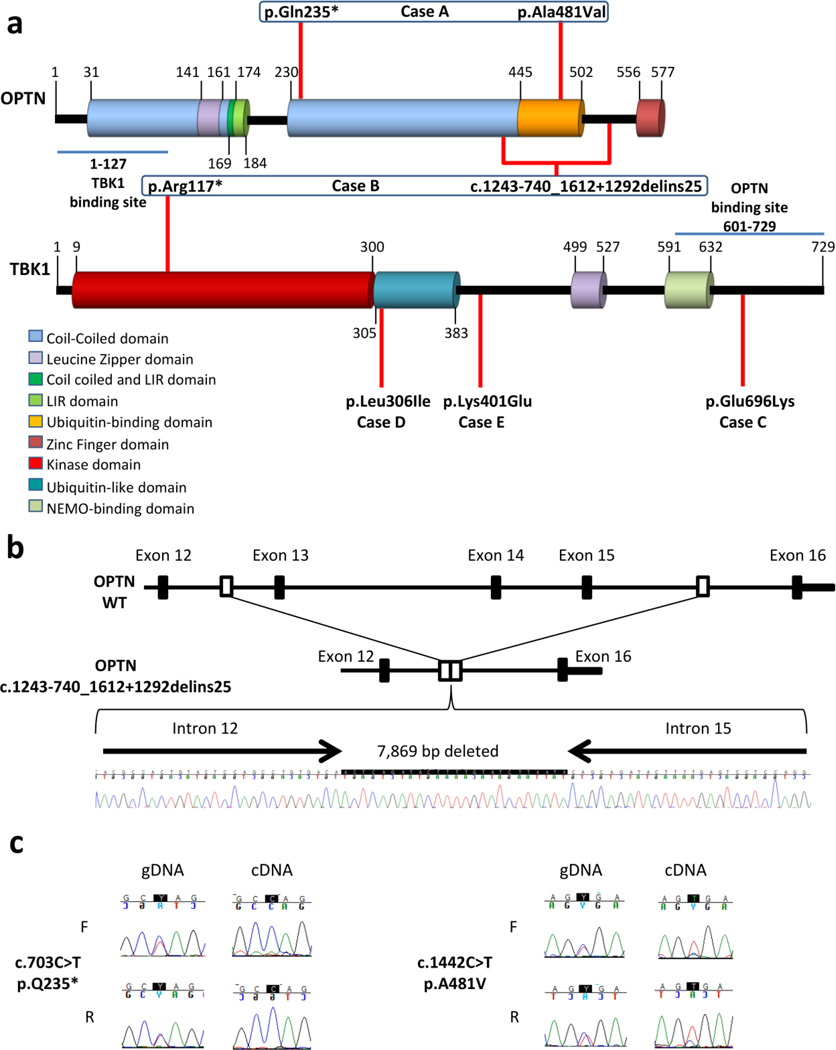
Figure 1. OPTN and TBK1 mutations in FTLD-TDP cases.
(a) Schematic representation of OPTN and TBK1 protein with their corresponding domains and the position of rare variants (red lines) identified in the study. Domain boundaries are shown with a vertical black line associated with the corresponding amino acid number. Each mutation has been found in a single patient which is indicated below. Mutations found in the same patient are indicated in a blue square. Binding sites to OPTN or TBK1 are indicated with a blue line. (b) Schematic representation of the partial deletion of exons 13 to 15 of OPTN (OPTN c.1243-740_1612+1292delins25) compared to wild-type OPTN (OPTN WT) along with the genomic DNA sequence chromatogram of the deletion observed in case B. (c) Genomic DNA (gDNA) and complementary DNA (cDNA) sequence chromatogram of exon 8 (right panel) demonstrating loss of the mutant (T) allele c.703C>T in cDNA for case A. gDNA and cDNA sequence chromatograms of exon14 (left panel) demonstrating the predominant presence of the mutant (T) allele c.1442C>T in cDNA sequence for case A. LIR: LC3-interacting region.
CNV analysis of the 21 neurodegenerative disease genes using Ingenuity Variant Analysis software further identified one patient with a partial deletion of OPTN (NM_001008211.1:c.1243-740_1612+1292delins25; p.Gly538Glufs27). The whole-genome sequence data identified a deleted region on chromosome 10, spanning 7,869 bp including the complete deletion of OPTN exons 13, 14 and 15. The deletion breakpoint was confirmed by PCR using primers that align to the non-deleted genomic sequence surrounding the deleted region and subsequent sequencing of the PCR product (Figure 1b). This partial OPTN deletion was absent from a cohort of 284 healthy subjects from Mayo Clinic Jacksonville as determined by a genomic Taqman assay designed to OPTN exon 14. Interestingly, the FTLD-TDP patient carrying the OPTN deletion is case B, who also carries the TBK1 p.Arg117* nonsense mutation, suggesting an oligogenic disease mechanism.
Prompted by the idea of an oligogenic mechanism of disease we further looked at the presence of more frequent variants (MAF <0.1%) in individuals already harboring extremely rare variants in OPTN and TBK1 and noted that case A carrying the p.Gln235* nonsense variant in OPTN, is compound heterozygote for mutations in OPTN as it also carries the rare variant p.Ala481Val (NM_001008211.1:c.1442C>T) in OPTN (MAF=0.0116% in ESP – CADD_Phred score: 34). In order to evaluate the likelihood of OPTN and TBK1 to harbor rare compound heterozygous variants or double mutations, we applied the same stringent filters that we used for our FTLD-TDP cases to our control dataset (155 Harvard PGP controls and 100 parents of intellectual disability patients). Using these filtering settings, no variants in OPTN or TBK1 were detected in any of our control datasets which emphasizes that the presence of rare double hits in our FTLD-TDP cohort is unlikely to have occurred by chance alone. Comparison of sequence traces of OPTN exon 8 (harboring the p.Gln235* mutation) in gDNA and mRNA prepared from cerebellar cortex of case A showed the absence of mutant RNA (T-allele) suggesting the degradation of mutant RNA by nonsense mediated decay (Figure 1c). A similar analysis of OPTN exon 14 (harboring the p.Ala481Val) mutation showed significantly reduced amounts of the wild-type (C-allele) in the cDNA sequence suggesting that the missense variant was inherited in trans with respect to the OPTN nonsense mutation (Figure 1c).
OPTN and TBK1 mutations reduce RNA and protein expression in cerebellar tissue
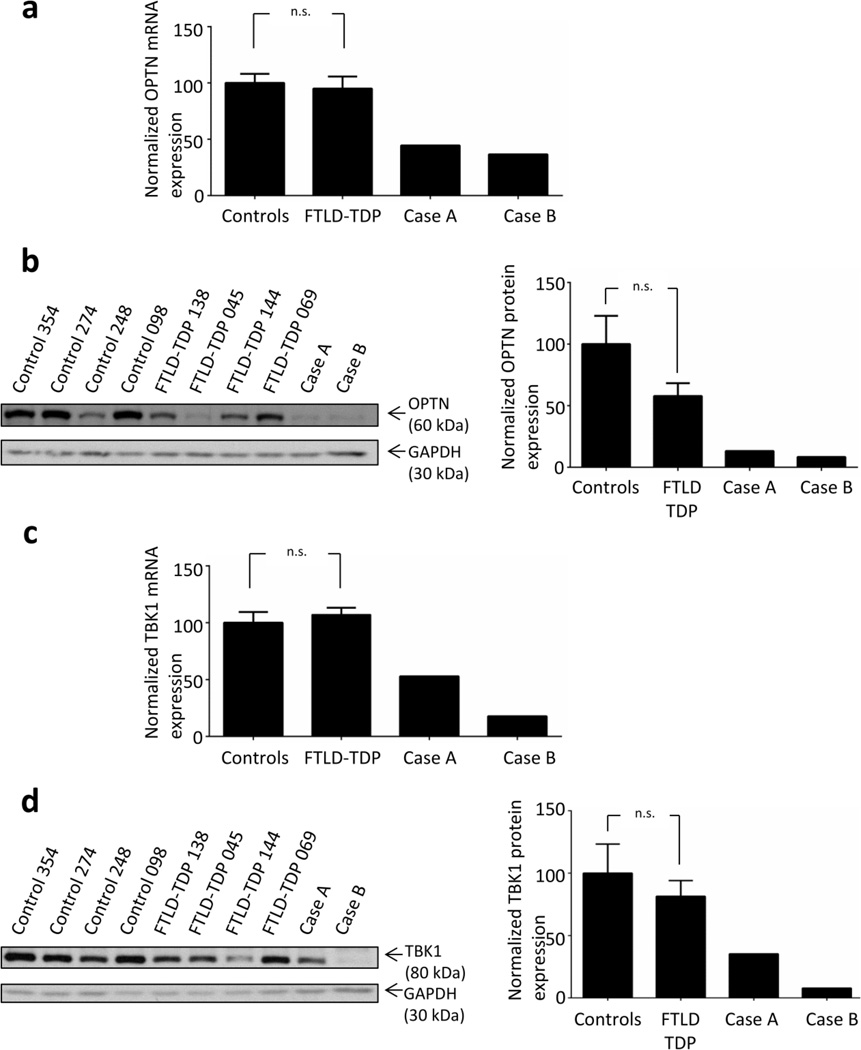
To provide further support for a pathogenic role of the OPTN/TBK1 pathway in FTLD-TDP, we performed quantitative mRNA expression and protein analysis of the OPTN compound heterozygous mutant (case A) and the OPTN/TBK1 double mutant (case B). To avoid overestimation of the RNA and protein levels due to neuronal loss, we focused on the cerebellum, a region of the brain less affected by neuronal loss. Despite variable expression, most likely resulting from the use of post-mortem tissues, OPTN mRNA and protein expression were dramatically reduced in cases A and B as compared to 6 FTLD-TDP patients without OPTN and TBK1 variants and 4 normal brains (mRNA relative expression levels of 0.5 and 0.4 respectively; Figure 2a–b). Of note, while as a group the FTLD-TDP patients without mutations in OPTN/TBK1 did not show significantly reduced OPTN protein levels compared to the normal controls (p=0.12), one case in particular (FTLD-TDP 045, Figure 2b) did show consistent lower levels of OPTN in two separate protein extractions, comparable to the level observed in cases A and B and suggesting it may have an as yet unidentified mutation affecting this pathway. TBK1 mRNA and protein analysis also showed reduced expression of TBK1 in case B, (the TBK1/OPTN double mutant); and, unexpectedly, TBK1 mRNA and protein levels were also reduced in case A despite two functional copies of the TBK1 gene as compared to normal controls (Figure 2 c–d).
Figure 2. Loss of function variants in OPTN and TBK1 reduce mRNA and protein expression.
(a) Relative OPTN mRNA expression levels normalized to RPLPO for cases A and B and 6 FTLD-TDP cases without mutations in OPTN and TBK1 are represented relatively to 4 control brains. (b) Representative western blot showing OPTN and GAPDH protein levels (left panel) in cases A and B, 4 FTLD-TDP cases without mutations in OPTN and TBK1 are 4 normal control brains. The relative quantification of OPTN protein (from 2 independent protein extractions) for cases A and B as well as FTLD-TDP cases without mutations in OPTN and TBK1 (n=6) and controls (n=6) is shown in right panel. (c) Relative TBK1 mRNA expression levels normalized to RPLPO for cases A and B and 6 FTLD-TDP cases without mutations in OPTN and TBK1 are represented relatively to 4 control brains. (d) Representative western blot showing TBK1 and GAPDH (left panel) for cases A and B, 4 FTLD-TDP cases without mutations in OPTN and TBK1 and 4 normal control brains. The relative quantification of TBK1 protein (from 2 independent protein extractions) for cases A and B as well as for 6 FTLD-TDP cases without mutations in OPTN and TBK1 (n=6) and controls (n=6) is shown in right panel. For all panels, the error bar +/− SEM is shown. n.s.: not significant after a Mann-Whitney test.
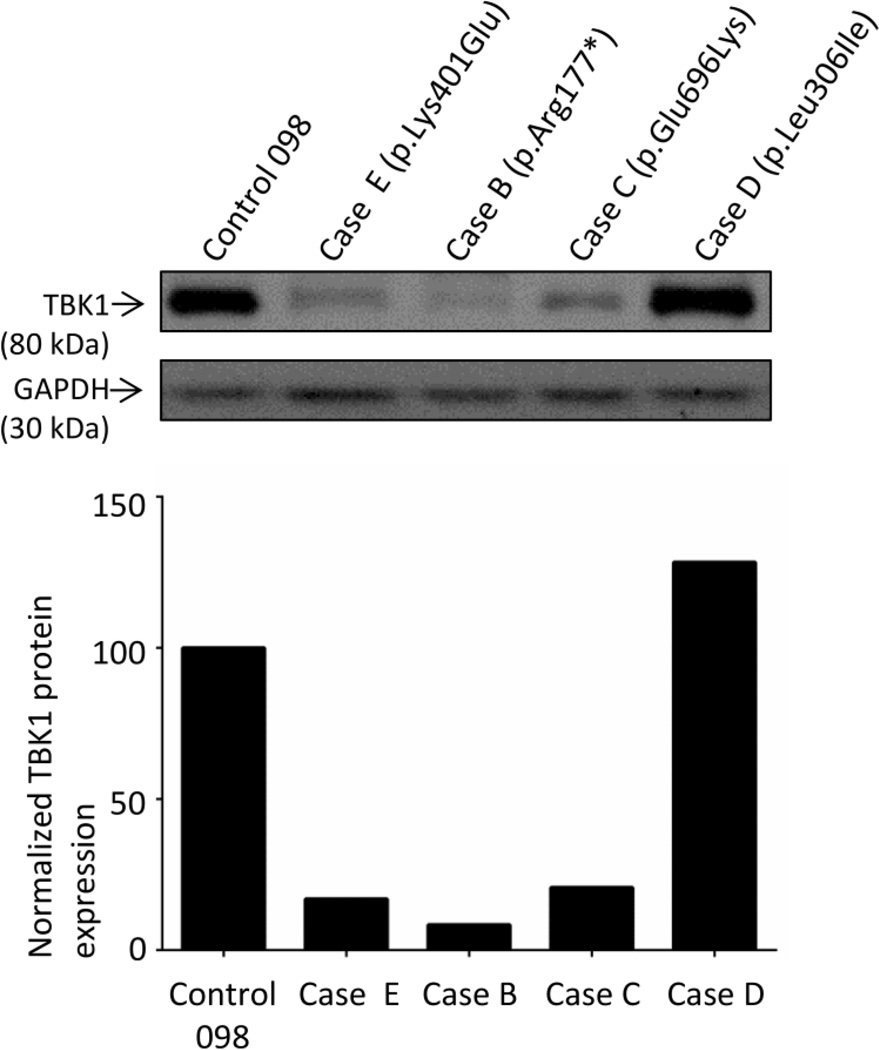
Encouraged by the ability to observe reduced levels of TBK1 protein in post-mortem tissue from a TBK1 loss-of-function mutation carrier, we next studied TBK1 protein expression in cerebellar tissue from cases C–E carrying TBK1 missense mutations with unknown pathogenicity. Interestingly, case C (NM_013254.3:c.2086G>A ; p.Glu696Lys) and case E (NM_013254.3:c.1201A>G; p.Lys401Glu) showed greatly reduced TBK1 protein levels, comparable to case B, while in case D (NM_013254.3:c.916C>A; p.Leu306Ile) TBK1 levels were normal (Figure 3).
Figure 3. TBK1 protein expression of TBK1 missense variants.
A western blot for TBK1 and GAPDH is presented (upper panel) for cases E, B, C and D as well as a representative control Control 098. Relative quantification of TBK1 protein for each case relatively to Control 098 is shown in the lower panel.
OPTN and TBK1 double mutations are associated with FTLD pathology without clinical motor neuron disease
Clinical and pathological features of cases A–E with possible pathogenic mutations in TBK1/OPTN are summarized in Table 2. Detailed case reports of the TBK1 and OPTN double mutants are included below:
Table 2.
Clinical and pathological description of OPTN and TBK1 mutation carriers
| Case ID |
Mutation(s) | Gender | Clinical Diagnosis |
Onset Age |
Pathological Diagnosis |
TDP-43 Subtype |
Age at death |
Family History |
|---|---|---|---|---|---|---|---|---|
| Case A | OPTN p.[Gln235*];[Ala481Val] | M | AD | 64 | FTLD | A | 70 | Yes (paternal uncle) |
| Case B | TBK1 p.Arg117*; OPTN p.Gly538Glufs*27 | M | agPPA | 68 | FTLD | A | 72 | No |
| Case C | TBK1 p.Glu696Lys | F | AD | 78 | FTLD-MND | B | 84 | Unknown |
| Case D | TBK1 p.Leu306Ile | F | FTD-ALS | 70 | FTLD-MND | B | 72 | No |
| Case E | TBK1 p.Lys401Glu | F | AD | 80 | FTLD | A | 90 | Unknown |
AD: Alzheimer’s disease, agPPA: progressive non fluent/agrammatic variant of primary progressive aphasia, FTD-ALS: frontotemporal dementia-amyotrophic lateral sclerosis, FTLD: frontotemporal lobar degeneration, MND: motor neuron disease.
Case B (OPTN p.Gly538Glufs27; TBK1 p.Arg117*) was a Caucasian male who started having significant cognitive difficulties at the age of 68. He would repeat the same questions, had a flat affect, and would substitute words. Positron emission tomography scan found hyperperfusion in the left temporal lobe, frontal lobe, and perisylvian area. Magnetic resonance imaging revealed a similar pattern of cortical atrophy with ventricular enlargement (mildly asymmetric, left > right). By the age of 69, he was almost completely mute, mainly communicating through gesturing. He also had trouble swallowing and frequent choking. He could not write and his memory was also worse. He developed jerky movements of his hands and he was moving slower. He was prescribed carbidopa/levodopa, but it was not helpful. On neurological examination he was attentive, but unable to speak. His understanding was much better than his speech. He had difficulty naming and copying a design. The remainder of his neurologic exam was unremarkable with normal strength, tone, coordination and gait. His Mini-Mental State Exam (MMSE) scores were 26/30 at age 68, 19/30 at age 69, and 3/30 at age 70. Formal neuropsychologic assessment was consistent with a nonfluent aphasia with mild comprehension difficulties. He also had features consistent with frontal dementia including divided attention, impaired organization, perseverations, echolalia and echopraxia. A clinical diagnosis of agPPA was made. His family history was negative for neurologic disease in his parents and 2 siblings. He died at the age of 72.
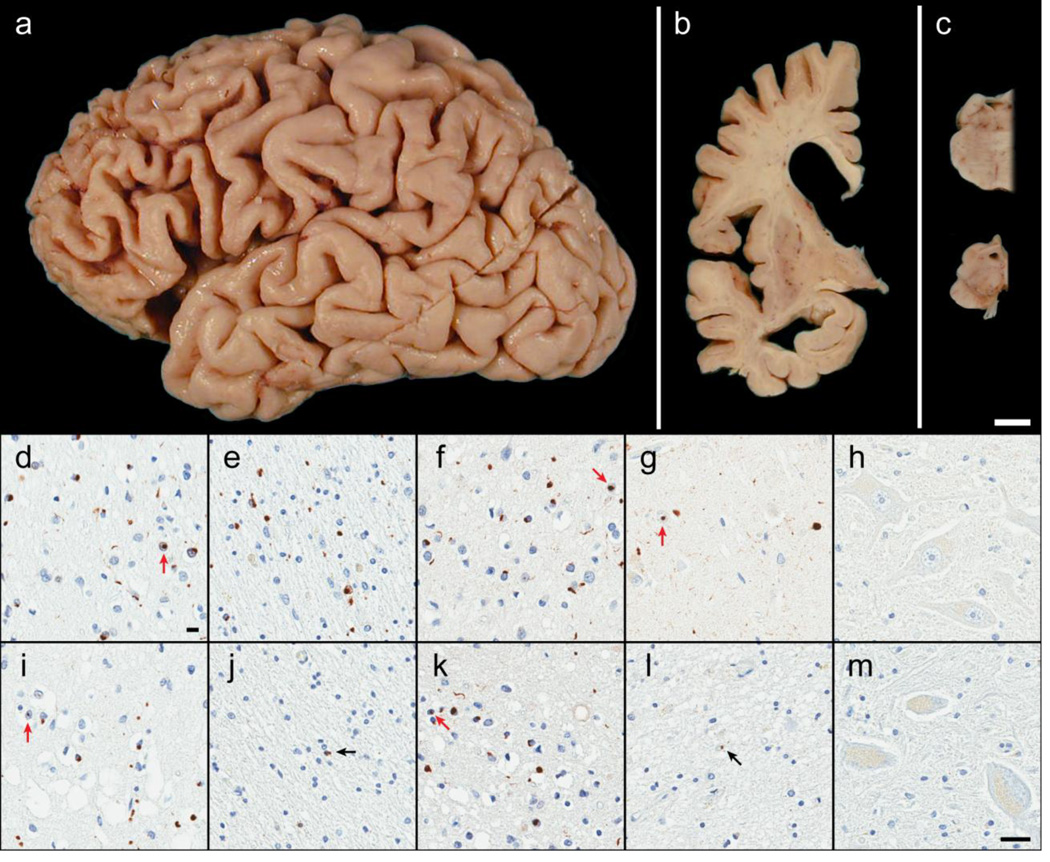
Post-mortem examination of case B revealed severe focal cortical atrophy of the frontal lobe (Figure 4a). Coronal sections showed enlargement of the lateral ventricles, atrophy of the amygdala and hippocampus, and atrophy of the caudate nucleus of the basal ganglia with white matter attenuation in the anterior limb of the internal capsule (Figure 4b). Sections of the brainstem demonstrated normal pigment in the locus ceruleus, but loss of pigment in the substantia nigra with midbrain atrophy (Figure 4c). Microscopic evaluation (Figure 4d–h) showed thinning of the cortical ribbon and superficial spongiosis, most prominently in the frontal cortex. Immunohistochemistry with TDP-43 and p62 uncovered many neuronal and glial cytoplasmic inclusions, neuronal intranuclear inclusions and dystrophic neurites, consistent with FTLD-TDP Type A. P62 immunoreactivity was more pronounced in the white matter than in the gray matter of the frontal cortex, and was at least 3.7-fold higher in case B than any of the other 19 FTLD-TDP cases analyzed. No pathology was noted with TBK1 and OPTN immunohistochemistry. In the hippocampus, neuronal loss in the CA1 sector and subiculum in addition to TDP-43 fine neurites and lesions was consistent with hippocampal sclerosis. There was no neuronal loss in the motor cortex or brainstem with preservation of Betz cells and brainstem motor neurons.
Figure 4. Neuropathology of OPTN and TBK1 double mutant carriers.
Case B exhibited focal cortical atrophy of the frontal lobe (a), ventricular enlargement of the frontal and temporal horns (b), atrophy or ‘flattening’ of the caudate (b), and loss of pigmentation in the substantia nigra (c) consistent with frontotemporal lobar degeneration. Microscopically, case B had abundant neuronal cytoplasmic inclusions, neuronal intranuclear inclusions (red arrows), glial cytoplasmic inclusions, and dystrophic neurites immunoreactive for p62/sequestosome-1 (d and e) as well as TDP-43 (f and g). These lesions, along with neuronal loss and superficial cortical spongiosis, were found in the gray matter of the frontal cortex (layer II; d and f) and, surprisingly, the underlying white matter of the frontal cortex (e). Neuronal loss and fine TDP-43-immunoreactive neurites in the CA1 of the hippocampus was indicative of hippocampal sclerosis (g). No pathology was found in motor neurons (hypoglossal nucleus) including with immunohistochemistry for optineurin protein (h). Case A had similar pathology in the gray matter of the frontal cortex (i and k) with p62 (i) and TDP-43 (k) immunohistochemistry in addition to pronounced superficial cortical spongiosis. Only rare immunoreactive glial cytoplasmic inclusions (black arrows) were observed the cortical white matter (p62; j) and in the hippocampal CA1 (TDP-43; l). Despite the paucity of hippocampal TDP-43 pathology, case A was also consistent with hippocampal sclerosis due to the vacuolation of the neuropil and severe neuronal loss. Again, no pathology was observed in the motor neurons (optineurin immunohistochemistry of the trigeminal motor nucleus; m). [white bar = 1 cm (a–c), black bar = 25 µm (d–m)]
Case A (OPTN p.[Gln235*];[Ala481Val]) was a Caucasian male who had received his GED diploma and served in the Army for 20 years before working as a heavy equipment operator. He had a medical history of cardiac issues including arrhythmia and 2 heart attacks. He had previously smoked tobacco and marijuana. Carotid angiodynography revealed minimal bilateral intraluminal thickening with 40% stenosis in the right proximal internal carotid artery, no stenosis in the left internal carotid artery, and patent vertebral arteries. He began experiencing progressive cognitive decline at the age of 64. By age 66, he had significant loss of executive function including agitation, restlessness, hyperactivity, and inappropriate behaviors. On neuropsychological testing at age 68, he scored 3/30 on the MMSE, demonstrating significant impairment. He was diagnosed as early-onset AD with delusions and hypomanic state. He had a positive family history of early-onset dementia in a paternal uncle. He died at age 70.
Post-mortem examination of case A revealed mild cortical atrophy with extensive softening and fragmentation of the temporal lobe. Coronal sections showed mild ventricular enlargement and mild atrophy in the amygdala and hippocampus, but no visible atrophy of the basal ganglia or thalamus. The substantia nigra had normal pigmentation. Microscopic evaluation (Figure 4 i–m) also demonstrated thinning of the cortical ribbon and significant superficial spongiosis, p62 and TDP-43 pathology in the cortical gray matter was morphologically similar to case B and also characteristic of FTLD-TDP Type A. P62 immunoreactivity in the white matter was minimal. There was a paucity of TDP-43 pathology in the hippocampal CA1 and subiculum, however, marked neuronal loss and vacuolation of the neuropil was also consistent with hippocampal sclerosis. No pathology was noted with TBK1 and OPTN immunohistochemistry. Similar to case B, no neuronal loss in the motor cortex or brainstem was noted.
DISCUSSION
In this study, we queried whole-genome sequence data to study the contribution of rare SNVs and CNVs in 21 known neurodegenerative genes to FTLD-TDP using in a large cohort of 104 pathologically confirmed patients from the Mayo Clinic brain bank negative for mutations in C9ORF72 and GRN as well as 3 GRN mutation carriers included as positive controls. Unexpectedly, we found 5 patients (4.8%) with variants in OPTN and TBK1 that are predicted to be highly pathogenic, including two double mutants. Based on all FTLD-TDP patients in our Mayo Clinic brain bank (including GRN and C9ORF72 mutation carriers), OPTN and TBK1 therefore represent ~3% of all FTLD-TDP. Subsequent study of mRNA and protein levels in the cerebellum showed a striking reduction of OPTN and/or TBK1 expression in 4 out of 5 patients, supporting pathogenicity in these specific cases and suggesting a loss-of-function disease mechanism. Pathological examination showed typical FTLD pathology without clinical motor neuron disease in 3 of the 4 cases carrying likely pathogenic mutations, extending the spectrum of OPTN and TBK1 mutations to pure FTLD. Importantly, our brain bank series shows TBK1 mutations as the third most common FTLD-TDP gene after C9ORF72 repeat expansions and mutations in GRN.
TBK1 and OPTN are functionally related proteins which recently gained increased attention from the ALS research community. TBK1 was identified as a novel risk factor for ALS through two independent exome sequencing studies [18,11]. Dominant TBK1 variants which were predicted to be damaging were identified in ~1% of nearly 3000 sporadic ALS cases (5-fold more frequent than in controls) [11]; while an even higher frequency of dominant TBK1 mutations (especially of loss-of-function mutations; 1.8%), was reported in a smaller study focused on familial ALS patients [18]. In contrast, OPTN mutations were first described as the cause of autosomal recessive ALS in Japanese ALS families [35]. Since then, heterozygous mutations have also been reported in familial ALS cases; however, no segregation or very limited segregation analyses of these potential dominant mutations with disease has been performed [38,15,52,5,50,58,13]. Interestingly, the recent exome-based sequencing study in sporadic ALS patients, which identified TBK1 as a novel ALS risk gene, also showed an increase in rare damaging variants in OPTN in patients compared to controls using a dominant model [11]. This led the authors to suggest that dominant acting variants in OPTN appeared to make a substantial contribution to sporadic ALS. An alternative hypothesis based on our present findings however supports a more complex mode of inheritance and suggests that sporadic individuals with heterozygous OPTN mutations who develop disease symptoms may do so because of compound heterozygote mutations (with the second hit not “qualifying” as a pathogenic variant) or because they also carry a mutation(s) in another gene.
Our study is the first to report OPTN mutations in FTD. Case A was compound heterozygote carrying the nonsense mutation p.Gln235*, previously reported in a sporadic ALS patient [8] together with the p.Ala481Val missense variant reported by Belzil et al. 2011 [5] in a familial ALS case. Our initial filtering had excluded p.Ala481Val because of its presence in EVS, however, only a single individual was reported and the frequency in the data from the Exome Aggregation Consortium (ExAC), Cambridge, MA (URL: http://exac.broadinstitute.org) [March, 2015] confirms that it is very rare (observed only once in 66,736 alleles in non-Finnish Europeans). cDNA sequencing further showed that these mutations were inherited in trans supporting a ‘double’ hit to this patient. The use of whole-genome sequencing also allowed us to identify an OPTN mutation in case B. This case carried a partial deletion of OPTN encompassing exons 13–15 which had not previously been reported. In contrast to case A with the compound heterozygous variants in OPTN, case B did not have any other rare variants in OPTN. However, case B also harbors the nonsense mutation p.Arg117* in TBK1. Interestingly, cases A and B both presented with pure dementia phenotypes in the absence of any obvious clinical or pathological features of motor neuron disease. We cannot however rule out the presence of very mild subclinical motor neuron degeneration .Pathologic examination showed FTLD-TDP pathology type A, which differs from the type B that was reported in one case with a TBK1 mutation with autopsy available in the recent study by Freischmidt et al.[18]. Additionally, p62 staining in case B carrying the OPTN/TBK1 double mutation revealed an unusual high burden of pathology in the white matter. The mechanism underlying this unique pathology remains to be elucidated, but it may be related to the oligogenic nature of the disease in this patient.
Our study also identified 3 other rare missense variants in TBK1. Based on the combined analysis of the frequency of these variants in ExAC and the recently published ALS exome studies [11,18,9], their observed effect on TBK1 expression in cerebellar tissue in this study and the evolutionary conservation and location within a known functional protein domain, we speculate that 2 out of the 3 observed mutations (p.Glu696Lys and p.Lys401Glu) are likely involved in disease pathogenesis. Mutation p.Glu696Lys observed in case C is located in the coiled-coil domain (CCD2) required for OPTN binding and was recently reported in two unrelated ALS patients [18]. It was shown that this mutation impaired the binding of TBK1 to OPTN without affecting phosphorylation and activation of IRF3, which binds the more N-terminal ubiquitin-like domain. Mutation p.Lys401Glu (case E) is a very rare variant, only observed once in ExAC in an African American individual, associated with low expression of TBK1 protein levels in the patient’s brain. Functional characterization of this specific mutation has not been performed; however, the p.Lys401Arg mutation affecting the same amino acid results in a loss of TBK1 activation in vitro, suggesting that this amino acid is important for TBK1 function [51]. In contrast, the pathogenicity of mutation p.Leu306Ile (case D) is questionable. TBK1 protein levels in cerebellar brain tissue of case D seemed normal and this variant has already been reported in 2 cases with glaucoma among a cohort of 254 patients [17]. It is also more frequent in the European population compared to the other observed missense mutations (observed 36 times in ExAC or roughly in 1 out of 1000 individuals).
It is striking that despite the fact that we started with a candidate list of 21 genes, we observed likely pathogenic mutations only in OPTN and TBK1, two genes encoding highly functionally connected proteins. Together with SQSTM1 (p62), OPTN and TBK1 play important roles in the autophagy pathway in regulating protein aggregates [60,19]. Remarkably, TBK1 directly interacts and phosphorylates OPTN, thereby increasing OPTN binding to LC3, an essential autophagosome protein [59,39,21]. It is therefore evident that mutations leading to a loss of OPTN and TBK1expression (as identified in this study) could affect the protective role of the OPTN/TBK1 pathway in neurodegenerative disorders such as FTLD and ALS associated with intracellular protein aggregation. In parallel to their function in the autophagy pathway, p62, TBK1 and OPTN also play regulatory roles in neuroinflammation, in part through their involvement in the NF-κB pathway [39,24].
The availability of whole-genome sequence data in this study sheds new light on the involvement of the OPTN/TBK1 pathways in the ALS-FTD disease spectrum. It allowed us to study both SNVs and CNVs and resulted in the detection of double mutants, either as compound heterozygote (case A) or with mutations in two separate genes (case B). While the functional consequence of each individual variant reported in our study may not be fully understood, the idea of an oligogenic basis of ALS and FTD, in particular with respect to the OPTN/TBK1 pathway, is of interest and could explain the somewhat contradictory earlier reports on the inheritance patterns associated with these genes. Interestingly, an oligogenic mechanism was also suggested by Freischmidt et al. 2015 [18] with the co-occurrence of a TBK1 loss-of-function mutation and the p.Arg524Gly mutation in FUS in a German ALS family. Of note, the term oligogenic was used by those authors and in our current study in a broad sense. It is used to refer to multiple genes contributing to a disease phenotype, where either both genes are necessary to cause disease or where the presence of multiple gene mutations may affect the disease penetrance or presentation. The observation of additional mutations in patients carrying a loss-of-function mutation in TBK1 or OPTN, is reminiscent of the emerging data from patients carrying C9ORF72 repeat expansions [54]. Similar to TBK1, repeat expansions in C9ORF72 clearly segregate with disease in some extended families, but expansions have also been reported in 6% of sporadic FTD and ALS patients and in a few elderly controls [34]. While in the case of C9ORF72, variable repeat sizes or specific methylation states of promoter CpG islands may influence disease penetrance [36,30,46], it has become clear that a striking number of patients carrying a C9ORF72 repeat expansion also carry a mutation in another known neurodegenerative disease gene [55]. The frequency of these ‘double mutations’ is remarkable and is tempting to speculate that the presence of these additional mutations may influence disease penetrance and presentation. A similar scenario may be at play with the OPTN/TBK1 mutations in FTD and ALS. In the case of OPTN, a second mutation may truly be necessary to cause disease, while loss-of-function mutations in TBK1 are likely to lead to disease in a dominant fashion in the majority of cases, though likely not in all. Fortunately, the strong advances in sequencing technologies will soon allow the study of oligogenic mechanisms at a larger scale to further clarify the role of oligogenic mechanisms in FTD, ALS and related neurodegenerative diseases.
In conclusion, we report the first genetic characterization of a large cohort of pathologically confirmed FTLD-TDP patients using whole-genome sequencing, focusing on the known neurodegenerative disease genes. These are the first exciting results using our FTLD-TDP whole-genome sequence on which more in-depth analysis is required. Interestingly, we demonstrated that loss of function variants in OPTN and TBK1 are associated with clinical and pathological FTD without motor neuron disease and we identify TBK1 mutations as a common cause of FTLD-TDP. Importantly, our data suggest a potential oligogenic basis for at least a subset of FTLD-TDP. Finally, our results re-emphasize that ALS and FTD are part of a disease spectrum with an overlapping genetic basis.
Supplementary Material
ACKNOWLEDGEMENTS
This project was supported by the State of Florida Alzheimer’s Disease Initiative, the Cure PSP brain bank, NIH grants R01 AG037491 (KAJ), R01 NS076471 (RR), R01 NS080882 (RR), R01 AG026251 (RR), P50 AG016574 (RP), UO1 AG006786 (RP). Whole genome sequencing in the Mayo Clinic FTLD-TDP cohort and the Wellderly dataset was performed by Complete Genomics.
Footnotes
Compliance with Ethical Standards
Research involving Human Participants and/or Animals:
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Disclosure of potential conflicts of interest:
The authors declare that they have no conflict of interest.
Informed consent:
Informed consent was obtained from all individual participants included in the study.
References
- 1.Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 2.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 3.Arai T, Nonaka T, Hasegawa M, Akiyama H, Yoshida M, Hashizume Y, Tsuchiya K, Oda T, Ikeda K. Neuronal and glial inclusions in frontotemporal dementia with or without motor neuron disease are immunopositive for p62. Neurosci Lett. 2003;342:41–44. doi: 10.1016/s0304-3940(03)00216-7. [DOI] [PubMed] [Google Scholar]
- 4.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 5.Belzil VV, Daoud H, Desjarlais A, Bouchard JP, Dupre N, Camu W, Dion PA, Rouleau GA. Analysis of OPTN as a causative gene for amyotrophic lateral sclerosis. Neurobiology of aging. 2011;32:555, e513–e554. doi: 10.1016/j.neurobiolaging.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Bennion Callister J, Pickering-Brown SM. Pathogenesis/genetics of frontotemporal dementia and how it relates to ALS. Exp Neurol. 2014;262(Pt B):84–90. doi: 10.1016/j.expneurol.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boomsma DI, Wijmenga C, Slagboom EP, Swertz MA, Karssen LC, Abdellaoui A, Ye K, Guryev V, Vermaat M, van Dijk F, Francioli LC, Hottenga JJ, Laros JF, Li Q, Li Y, Cao H, Chen R, Du Y, Li N, Cao S, van Setten J, Menelaou A, Pulit SL, Hehir-Kwa JY, Beekman M, Elbers CC, Byelas H, de Craen AJ, Deelen P, Dijkstra M, den Dunnen JT, de Knijff P, Houwing-Duistermaat J, Koval V, Estrada K, Hofman A, Kanterakis A, Enckevort D, Mai H, Kattenberg M, van Leeuwen EM, Neerincx PB, Oostra B, Rivadeneira F, Suchiman EH, Uitterlinden AG, Willemsen G, Wolffenbuttel BH, Wang J, de Bakker PI, van Ommen GJ, van Duijn CM. The Genome of the Netherlands: design, and project goals. Eur J Hum Genet. 2014;22:221–227. doi: 10.1038/ejhg.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cady J, Allred P, Bali T, Pestronk A, Goate A, Miller TM, Mitra RD, Ravits J, Harms MB, Baloh RH. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 2015;77:100–113. doi: 10.1002/ana.24306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chio A, Calvo A, Mazzini L, Cantello R, Mora G, Moglia C, Corrado L, D'Alfonso S, Majounie E, Renton A, Pisano F, Ossola I, Brunetti M, Traynor BJ, Restagno G. Extensive genetics of ALS: a population-based study in Italy. Neurology. 2012;79:1983–1989. doi: 10.1212/WNL.0b013e3182735d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chio A, Restagno G, Brunetti M, Ossola I, Calvo A, Canosa A, Moglia C, Floris G, Tacconi P, Marrosu F, Marrosu MG, Murru MR, Majounie E, Renton AE, Abramzon Y, Pugliatti M, Sotgiu MA, Traynor BJ, Borghero G. ALS/FTD phenotype in two Sardinian families carrying both C9ORF72 and TARDBP mutations. Journal of neurology, neurosurgery, and psychiatry. 2012;83:730–733. doi: 10.1136/jnnp-2012-302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, Ren Z, Keebler J, Han Y, Levy SE, Boone BE, Wimbish JR, Waite LL, Jones AL, Carulli JP, Day-Williams AG, Staropoli JF, Xin WW, Chesi A, Raphael AR, McKenna-Yasek D, Cady J, Vianney de Jong JM, Kenna KP, Smith BN, Topp S, Miller J, Gkazi A, Al-Chalabi A, van den Berg LH, Veldink J, Silani V, Ticozzi N, Shaw CE, Baloh RH, Appel S, Simpson E, Lagier-Tourenne C, Pulst SM, Gibson S, Trojanowski JQ, Elman L, McCluskey L, Grossman M, Shneider NA, Chung WK, Ravits JM, Glass JD, Sims KB, Van Deerlin VM, Maniatis T, Hayes SD, Ordureau A, Swarup S, Landers J, Baas F, Allen AS, Bedlack RS, Harper JW, Gitler AD, Rouleau GA, Brown R, Harms MB, Cooper GM, Harris T, Myers RM, Goldstein DB. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, van Duijn C, Peeters K, Sciot R, Santens P, De Pooter T, Mattheijssens M, Van den Broeck M, Cuijt I, Vennekens K, De Deyn PP, Kumar-Singh S, Van Broeckhoven C. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 13.Czell D, Andersen PM, Morita M, Neuwirth C, Perren F, Weber M. Phenotypes in Swiss patients with familial ALS carrying TARDBP mutations. Neurodegener Dis. 2013;12:150–155. doi: 10.1159/000345835. [DOI] [PubMed] [Google Scholar]
- 14.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Bo R, Tiloca C, Pensato V, Corrado L, Ratti A, Ticozzi N, Corti S, Castellotti B, Mazzini L, Soraru G, Cereda C, D'Alfonso S, Gellera C, Comi GP, Silani V. Novel optineurin mutations in patients with familial and sporadic amyotrophic lateral sclerosis. Journal of neurology, neurosurgery, and psychiatry. 2011;82:1239–1243. doi: 10.1136/jnnp.2011.242313. [DOI] [PubMed] [Google Scholar]
- 16.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, Dahl F, Fernandez A, Staker B, Pant KP, Baccash J, Borcherding AP, Brownley A, Cedeno R, Chen L, Chernikoff D, Cheung A, Chirita R, Curson B, Ebert JC, Hacker CR, Hartlage R, Hauser B, Huang S, Jiang Y, Karpinchyk V, Koenig M, Kong C, Landers T, Le C, Liu J, McBride CE, Morenzoni M, Morey RE, Mutch K, Perazich H, Perry K, Peters BA, Peterson J, Pethiyagoda CL, Pothuraju K, Richter C, Rosenbaum AM, Roy S, Shafto J, Sharanhovich U, Shannon KW, Sheppy CG, Sun M, Thakuria JV, Tran A, Vu D, Zaranek AW, Wu X, Drmanac S, Oliphant AR, Banyai WC, Martin B, Ballinger DG, Church GM, Reid CA. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 17.Fingert JH, Robin AL, Stone JL, Roos BR, Davis LK, Scheetz TE, Bennett SR, Wassink TH, Kwon YH, Alward WL, Mullins RF, Sheffield VC, Stone EM. Copy number variations on chromosome 12q14 in patients with normal tension glaucoma. Human molecular genetics. 2011;20:2482–2494. doi: 10.1093/hmg/ddr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Muller K, Marroquin N, Nordin F, Hubers A, Weydt P, Pinto S, Press R, Millecamps S, Molko N, Bernard E, Desnuelle C, Soriani MH, Dorst J, Graf E, Nordstrom U, Feiler MS, Putz S, Boeckers TM, Meyer T, Winkler AS, Winkelman J, de Carvalho M, Thal DR, Otto M, Brannstrom T, Volk AE, Kursula P, Danzer KM, Lichtner P, Dikic I, Meitinger T, Ludolph AC, Strom TM, Andersen PM, Weishaupt JH. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015 doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 19.Galluzzi L, Kepp O, Kroemer G. Autophagy and innate immunity ally against bacterial invasion. Embo J. 2011;30:3213–3214. doi: 10.1038/emboj.2011.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, Leach R, Klein R, Tearle R, Bo T, Pfundt R, Yntema HG, de Vries BB, Kleefstra T, Brunner HG, Vissers LE, Veltman JA. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 21.Gleason CE, Ordureau A, Gourlay R, Arthur JS, Cohen P. Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon beta. The Journal of biological chemistry. 2011;286:35663–35674. doi: 10.1074/jbc.M111.267567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graff-Radford NR, Woodruff BK. Frontotemporal dementia. Semin Neurol. 2007;27:48–57. doi: 10.1055/s-2006-956755. [DOI] [PubMed] [Google Scholar]
- 23.Hardy J, Rogaeva E. Motor neuron disease and frontotemporal dementia: sometimes related, sometimes not. Exp Neurol. 2014;262(Pt B):75–83. doi: 10.1016/j.expneurol.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Hiscott J. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 2007;18:483–490. doi: 10.1016/j.cytogfr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Huey ED, Ferrari R, Moreno JH, Jensen C, Morris CM, Potocnik F, Kalaria RN, Tierney M, Wassermann EM, Hardy J, Grafman J, Momeni P. FUS and TDP43 genetic variability in FTD and CBS. Neurobiology of aging. 2012;33:1016, e1019–e1017. doi: 10.1016/j.neurobiolaging.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5'-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 27.Ingram EM, Spillantini MG. Tau gene mutations: dissecting the pathogenesis of FTDP-17. Trends Mol Med. 2002;8:555–562. doi: 10.1016/s1471-4914(02)02440-1. [DOI] [PubMed] [Google Scholar]
- 28.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 29.Leverenz JB, Yu CE, Montine TJ, Steinbart E, Bekris LM, Zabetian C, Kwong LK, Lee VM, Schellenberg GD, Bird TD. A novel progranulin mutation associated with variable clinical presentation and tau, TDP43 and alpha-synuclein pathology. Brain. 2007;130:1360–1374. doi: 10.1093/brain/awm069. [DOI] [PubMed] [Google Scholar]
- 30.Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, Irwin DJ, Van Deerlin VM, Lee EB. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014;128:525–541. doi: 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 32.Luigetti M, Lattante S, Zollino M, Conte A, Marangi G, Del Grande A, Sabatelli M. SOD1 G93D sporadic amyotrophic lateral sclerosis (SALS) patient with rapid progression and concomitant novel ANG variant. Neurobiology of aging. 2011;32:1924, e1915–e1928. doi: 10.1016/j.neurobiolaging.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Mackenzie IR, Baborie A, Pickering-Brown S, Du Plessis D, Jaros E, Perry RH, Neary D, Snowden JS, Mann DM. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 2006;112:539–549. doi: 10.1007/s00401-006-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 36.McMillan CT, Russ J, Wood EM, Irwin DJ, Grossman M, McCluskey L, Elman L, Van Deerlin V, Lee EB. C9orf72 promoter hypermethylation is neuroprotective: Neuroimaging and neuropathologic evidence. Neurology. 2015 doi: 10.1212/WNL.0000000000001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A, Wu W, Corominas R, Peoples A, Koren A, Gore A, Kang S, Lin GN, Estabillo J, Gadomski T, Singh B, Zhang K, Akshoomoff N, Corsello C, McCarroll S, Iakoucheva LM, Li Y, Wang J, Sebat J. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millecamps S, Boillee S, Chabrol E, Camu W, Cazeneuve C, Salachas F, Pradat PF, Danel-Brunaud V, Vandenberghe N, Corcia P, Le Forestier N, Lacomblez L, Bruneteau G, Seilhean D, Brice A, Feingold J, Meininger V, LeGuern E. Screening of OPTN in French familial amyotrophic lateral sclerosis. Neurobiology of aging. 2011;32:557, e511–e553. doi: 10.1016/j.neurobiolaging.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 2008;582:997–1002. doi: 10.1016/j.febslet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee O, Pastor P, Cairns NJ, Chakraverty S, Kauwe JS, Shears S, Behrens MI, Budde J, Hinrichs AL, Norton J, Levitch D, Taylor-Reinwald L, Gitcho M, Tu PH, Tenenholz Grinberg L, Liscic RM, Armendariz J, Morris JC, Goate AM. HDDD2 is a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions caused by a missense mutation in the signal peptide of progranulin. Ann Neurol. 2006;60:314–322. doi: 10.1002/ana.20963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 43.Rademakers R, Baker M, Nicholson AM, Rutherford NJ, Finch N, Soto-Ortolaza A, Lash J, Wider C, Wojtas A, DeJesus-Hernandez M, Adamson J, Kouri N, Sundal C, Shuster EA, Aasly J, MacKenzie J, Roeber S, Kretzschmar HA, Boeve BF, Knopman DS, Petersen RC, Cairns NJ, Ghetti B, Spina S, Garbern J, Tselis AC, Uitti R, Das P, Van Gerpen JA, Meschia JF, Levy S, Broderick DF, Graff-Radford N, Ross OA, Miller BB, Swerdlow RH, Dickson DW, Wszolek ZK. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nature genetics. 2012;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rademakers R, Neumann M, Mackenzie IR. Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol. 2012;8:423–434. doi: 10.1038/nrneurol.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russ J, Liu EY, Wu K, Neal D, Suh E, Irwin DJ, McMillan CT, Harms MB, Cairns NJ, Wood EM, Xie SX, Elman L, McCluskey L, Grossman M, Van Deerlin VM, Lee EB. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol. 2015;129:39–52. doi: 10.1007/s00401-014-1365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampathu DM, Neumann M, Kwong LK, Chou TT, Micsenyi M, Truax A, Bruce J, Grossman M, Trojanowski JQ, Lee VM. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am J Pathol. 2006;169:1343–1352. doi: 10.2353/ajpath.2006.060438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sieben A, Van Langenhove T, Engelborghs S, Martin JJ, Boon P, Cras P, De Deyn PP, Santens P, Van Broeckhoven C, Cruts M. The genetics and neuropathology of frontotemporal lobar degeneration. Acta Neuropathol. 2012;124:353–372. doi: 10.1007/s00401-012-1029-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solski JA, Williams KL, Yang S, Nicholson GA, Blair IP. Mutation analysis of the optineurin gene in familial amyotrophic lateral sclerosis. Neurobiology of aging. 2012;33:210, e219–e210. doi: 10.1016/j.neurobiolaging.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 51.Tu D, Zhu Z, Zhou AY, Yun CH, Lee KE, Toms AV, Li Y, Dunn GP, Chan E, Thai T, Yang S, Ficarro SB, Marto JA, Jeon H, Hahn WC, Barbie DA, Eck MJ. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep. 2013;3:747–758. doi: 10.1016/j.celrep.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tumer Z, Bertelsen B, Gredal O, Magyari M, Nielsen KC, Lucamp, Gronskov K, Brondum-Nielsen K. Novel heterozygous nonsense mutation of the OPTN gene segregating in a Danish family with ALS. Neurobiology of aging. 2012;33:208, e201–e205. doi: 10.1016/j.neurobiolaging.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 53.van Blitterswijk M, Baker MC, Bieniek KF, Knopman DS, Josephs KA, Boeve B, Caselli R, Wszolek ZK, Petersen R, Graff-Radford NR, Boylan KB, Dickson DW, Rademakers R. Profilin-1 mutations are rare in patients with amyotrophic lateral sclerosis and frontotemporal dementia. Amyotroph Lateral Scler Frontotemporal Degener. 2013;14:463–469. doi: 10.3109/21678421.2013.787630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Blitterswijk M, Baker MC, DeJesus-Hernandez M, Ghidoni R, Benussi L, Finger E, Hsiung GY, Kelley BJ, Murray ME, Rutherford NJ, Brown PE, Ravenscroft T, Mullen B, Ash PE, Bieniek KF, Hatanpaa KJ, Karydas A, Wood EM, Coppola G, Bigio EH, Lippa C, Strong MJ, Beach TG, Knopman DS, Huey ED, Mesulam M, Bird T, White CL, 3rd, Kertesz A, Geschwind DH, Van Deerlin VM, Petersen RC, Binetti G, Miller BL, Petrucelli L, Wszolek ZK, Boylan KB, Graff-Radford NR, Mackenzie IR, Boeve BF, Dickson DW, Rademakers R. C9ORF72 repeat expansions in cases with previously identified pathogenic mutations. Neurology. 2013;81:1332–1341. doi: 10.1212/WNL.0b013e3182a8250c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Blitterswijk M, DeJesus-Hernandez M, Rademakers R. How do C9ORF72 repeat expansions cause amyotrophic lateral sclerosis and frontotemporal dementia: can we learn from other noncoding repeat expansion disorders? Curr Opin Neurol. 2012;25:689–700. doi: 10.1097/WCO.0b013e32835a3efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Blitterswijk M, van Es MA, Hennekam EA, Dooijes D, van Rheenen W, Medic J, Bourque PR, Schelhaas HJ, van der Kooi AJ, de Visser M, de Bakker PI, Veldink JH, van den Berg LH. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Human molecular genetics. 2012;21:3776–3784. doi: 10.1093/hmg/dds199. [DOI] [PubMed] [Google Scholar]
- 57.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weishaupt JH, Waibel S, Birve A, Volk AE, Mayer B, Meyer T, Ludolph AC, Andersen PM. A novel optineurin truncating mutation and three glaucoma-associated missense variants in patients with familial amyotrophic lateral sclerosis in Germany. Neurobiology of aging. 2013;34:1516, e1519–e1515. doi: 10.1016/j.neurobiolaging.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 59.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu CH, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, Lowe P, Koppers M, McKenna-Yasek D, Baron DM, Kost JE, Gonzalez-Perez P, Fox AD, Adams J, Taroni F, Tiloca C, Leclerc AL, Chafe SC, Mangroo D, Moore MJ, Zitzewitz JA, Xu ZS, van den Berg LH, Glass JD, Siciliano G, Cirulli ET, Goldstein DB, Salachas F, Meininger V, Rossoll W, Ratti A, Gellera C, Bosco DA, Bassell GJ, Silani V, Drory VE, Brown RH, Jr, Landers JE. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.