Abstract
Tropomyosins are coiled-coil proteins that bind actin filaments and regulate multiple cytoskeletal functions, including actin network dynamics near the leading edge of motile cells. Previous work demonstrated that tropomyosins inhibit actin nucleation by the Arp2/3 complex and prevent filament disassembly by cofilin. We find that the Arp2/3 complex and cofilin, in turn, regulate the binding of tropomyosin to actin filaments. Using fluorescence microscopy we show that tropomyosin (non-muscle Drosophila Tm1A) polymerizes along actin filaments, starting from “nuclei” that appear preferentially on ADP-bound regions of the filament, near the pointed end. Tropomyosin fails to bind dendritic actin networks created in vitro by the Arp2/3 complex, in part because the Arp2/3 complex blocks pointed ends. Cofilin promotes phosphate dissociation and severs filaments, generating new pointed ends and rendering Arp2/3-generated networks competent to bind tropomyosin. Tropomyosin’s attraction to pointed ends reflects a strong preference for conformations localized to that region of the filament and reveals a basic molecular mechanism by which lamellipodial actin networks are insulated from the effects of tropomyosin.
Introduction
Amoeboid cell movement relies on multiple, dynamic networks of actin filaments to generate force and move leading edge membranes. These networks are defined by several properties, including: (i) their three-dimensional architecture; (ii) the spectrum of actin regulatory proteins associated with them; and (iii) the rates at which filaments within them assemble and disassemble [1, 2, 3, 4]. Beneath leading edge membranes, in a compartment called the lamellipod, short-lived and highly branched actin filaments are nucleated by the Arp2/3 complex and disassembled by cofilin [3, 4]. Adjacent to the lamellipod, in a slower-moving network called the lamellum, longer actin filaments are stabilized by the coiled-coil protein tropomyosin.
Previous work focused on the role of tropomyosin in establishing the boundary between lamellipodial and lamellar actin networks. In vitro, tropomyosin blocks binding of the Arp2/3 complex and cofilin to actin filaments, inhibiting both nucleation [5, 6] and disassembly of filaments [7]. When added to branched, Arp2/3-generated actin networks reconstituted in vitro, skeletal muscle tropomyosin mimics the transition from lamellipodial to lamellar networks by rescuing some of the filaments from depolymerization by cofilin [8]. In vivo, injection of skeletal muscle tropomyosin into crawling or ruffling cells expands the lamellar network at the expense of the lamellipod [9], while RNAi-mediated depletion of tropomyosin has the opposite effect, expanding the lamellipod while shrinking the lamellum [4].
Tropomyosins are alpha-helical, coiled-coil dimers that self-associate, head-to-tail to form polarized polymers. On its own, the self-association of tropomyosin is weak but it contributes significantly to the cooperative nature of tropomyosin’s interaction with actin filaments [10]. When tropomyosin coats a filament, it alters the spectrum of accessory factors that interact with that filament, and different tropomyosins are thought to alter this spectrum of interactions in different ways [11, 12]. This is significant because many eukaryotic genomes contain multiple tropomyosin genes, each of which can produce multiple splice variants, leading to the expression of multiple tropomyosin isoforms in a single cell. A mammalian cell, for example, can express 10 or more tropomyosins [10], associated with different actin networks and different cellular functions [13, 14].
What molecular mechanisms determine the different subcellular locations of tropomyosin isoforms? In fission yeast, S. pombe, competition with proteins, such as fimbrin [15] and formins [16] helps determine the localization of tropomyosin (Cdc8) which, in turn, regulates the localization and activity of other actin-binding proteins, including various myosins [17, 18] and the actin severing protein, Adf1 [15, 16]. We do not, however, fully understand how binding of tropomyosins to actin filaments is regulated, especially near the leading edge of migrating cells. Why, for example, are filaments in the lamellum coated with tropomyosin while filaments in the adjacent lamellipod are not? To address this question we created a fluorescent derivative of Tm1A, a non-muscle tropomyosin isoform from Drosophila that localizes to lamellar and cortical actin networks in S2 cells (Goins and Mullins, in press).
We used Total Internal Reflection Fluorescence (TIRF) microscopy to follow the binding of Tm1A to single actin filaments and reconstituted dendritic networks, and found that Tm1A binds preferentially near the pointed ends of actin filaments. This interaction is blocked when pointed ends are capped by the Arp2/3 complex, explaining why Arp2/3-generated actin networks fail to bind tropomyosin. The dynamics of tropomyosin spreading on actin filaments and the effects of nucleotide analogs indicate that tropomyosin does not bind to the pointed end per se, but preferentially binds older, ADP-bound actin filaments and appears to recognize filament conformations found near the pointed end. Severing of actin filaments by cofilin generates free pointed ends capable of recruiting tropomyosin, even in the presence of the Arp2/3 complex. Our work reveals how tropomyosin is excluded from lamellipodial actin networks and suggests a cofilin-dependent mechanism for conversion of branched lamellipodial actin filaments into tropomyosin-coated lamellar filaments.
Results
Interaction of Tm1A with actin filaments
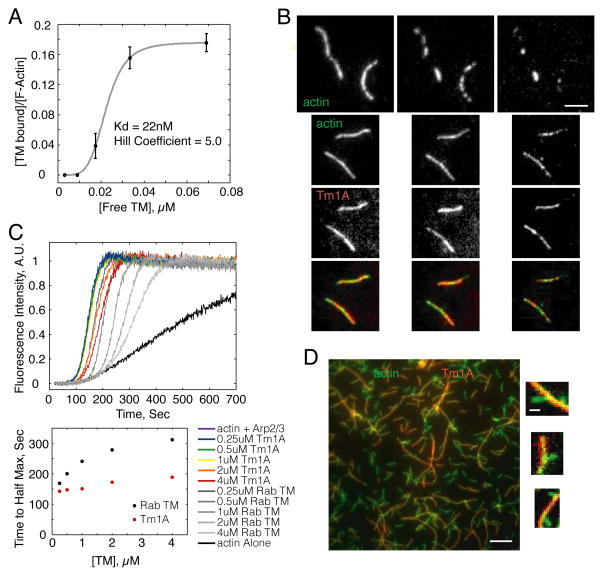
Using total internal reflection fluorescence (TIRF) microscopy, we imaged the interaction between Alexa-488-labeled actin filaments and Cy5-Tm1A for over 2 hours, and found that our labeled tropomyosin binds actin (Figure 1A) with an affinity (Kd: 22 nM) and cooperativity (Hill coefficient: 5.0) similar to other, previously studied tropomyosins [19, 20, 21]. These values agree with the affinity and cooperativity of unlabeled Tm1A measured by high-speed copelleting with actin filaments (Kd: 46 nM, Figure S1B). These results confirm that: (1) recombinant Tm1A binds actin, and (2) fluorescent labeling of our engineered protein has little or no effect on its actin binding. Single filament fluorescence microscopy also verified that —similar to other tropomyosins— Cy5-Tm1A blocks filament severing and depolymerization by cofilin (Figure 1B and [7, 22, 6]).
Figure 1. Drosophila non-muscle tropomyosin isoform, Tm1A, protects actin filaments from cofilin severing, and only weakly inhibits nucleation by Arp2/3 complex.
(A) Binding affinity of Tm1A with actin filaments using single filament TIRF. (B) Actin filaments pre-bound by Tm1A (bottom three rows) are protected from cofilin’s severing activity, as seen in single filament TIRF. Top row: Actin filaments without Tm1A are severed within 16min. Rows 1–2: Alexa488-actin. Row 3: Cy5-Tm1A. Row 4: merge of rows 2–3. Prebinding conditions: 20% A488 600nM actin +/− 450nM Tm1A-Cy5. Severing conditions flowed into chamber 10min after the start of the reaction: 150nM cofilin, +/− 450nM Tm1A-Cy5. Scale bar: 3μm. (C) Tm1A binding to actin filaments has a weak effect on nucleation by the Arp2/3 complex. Time-resolved pyrene fluorescence of polymerizing actin filaments nucleated by the Arp2/3 complex in the absence and presence of Tm1A and RSK muscle TM. Below: Plot of time to half max vs. TM concentration for Tm1A and RSK TM. (D) TIRF microscopy of actin filaments nucleated from the sides of pre-existing, Tm1A-coated, mother filaments. Large scale bar: 5μm. Small scale bar: 1μm. (See also Figure S1).
Tm1A has less effect on actin nucleation by the Arp2/3 complex than previously studied tropomyosins. Blanchoin et al. (2001) [5] characterized three tropomyosin isoforms and found that they inhibit Arp2/3 complex activity with different efficiencies, likely because they occupy different positions on the filament that occlude the Arp2/3 binding site to different extents. These authors found mammalian skeletal muscle tropomyosin is the least effective Arp2/3 inhibitor, so we compared the relative abilities of rabbit skeletal muscle tropomyosin and Tm1A to inhibit the Arp2/3 complex. Both tropomyosins bind actin with similar affinity, but Tm1A turns out to be much less effective at inhibiting Arp2/3 activity in pyrene actin assembly assays (Figures 1C & S1D). To see whether inefficient inhibition was due to incomplete coating of filaments by Tm1A, we used TIRF microscopy to observe dendritic nucleation in the presence of Cy5-Tm1A. When we mixed actin filaments that had been pre-bound to Cy5-Tm1A with the Arp2/3 complex —plus monomeric actin and an Arp2/3 activator from L. monocytogenes, ActA— we observed new daughter filaments branching directly from the sides of Cy5-Tm1A-coated mothers (Figure 1D). This result argues that the footprint of Tm1A on the actin filament overlaps minimally with that of the Arp2/3 complex.
Cofilin promotes binding of Tm1A to Arp2/3-generated dendritic actin networks
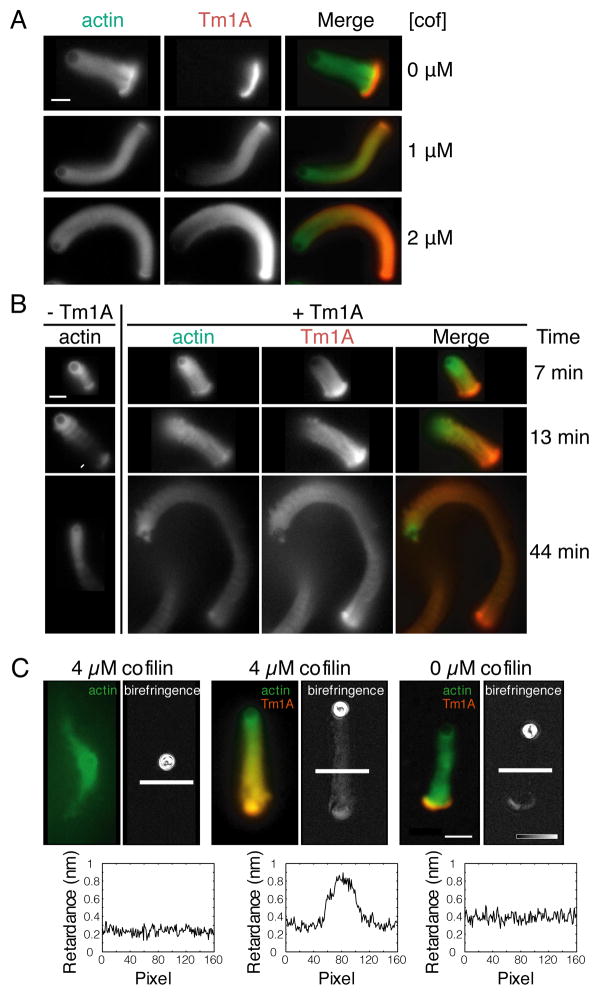
To study the binding of Tm1A to dendritic actin networks we used the Arp2/3 complex, capping protein, and monomeric actin to create motile actin ‘comet tails’ [23] on 5 μm polystyrene microspheres coated with ActA. At optimal concentrations of all the components, a symmetrical actin “shell” forms around the microsphere and eventually ruptures, generating a motile “comet tail,” structurally and functionally similar to a lamellipodial actin network [24]. Remarkably, Cy3-Tm1A binds only to the original actin shell, formed before the network ‘broke symmetry’ and began to move. The rest of the actin network excludes Cy3-Tm1A (Figure 2A, top row) and this exclusion continues for at least two hours after initiation of the reaction (Figure S2B, Movie S1).
Figure 2. Cofilin promotes the binding of tropomyosin to motile actin networks generated by the Arp2/3 complex and capping protein.
(A) Actin networks generated from ActA-coated polystyrene microspheres by the Arp2/3 complex and capping protein exclude tropomyosin in the absence of cofilin. Cofilin renders dendritic actin networks competent to bind tropomyosin. Left: Alexa488-actin. Middle: 1μM Cy3- Tm1A. Right: merge of actin and Tm1A fluorescence. Each row has the indicated concentration of added cofilin. Scale bar: 10μm. (B) Tropomyosin decreases the depolymerizing activity of cofilin. First Column: Motile actin networks generated by the Arp2/3 complex in the presence of capping protein and cofilin (arrow points to severing event). Second-Fourth Columns: Addition of 1μM Tm1A. Second Column: Alexa488-actin. Third Column: Cy3-Tm1A. Fourth Column: Merge. Scale bar: 10μm. (C) Polarized light microscopy of dendritic actin networks show that filaments align only when both cofilin and tropomyosin are present. In each pair of images: the left is Alexa488-actin and Cy3-labeled Tm1A, if present; the right is optical birefringence of the same comet tail. Left pair: 4μM cofilin, 0μM Tm1A. Middle pair: 4μM cofilin, 1μM Tm1A. Right pair: 0μM cofilin, 1μM Tm1A. Line scans of the birefringence (converted to retardance) are displayed below each birefringence image. Scale bar: 10μm. (See also Figure S2).
Strikingly, addition of the actin severing and depolymerizing factor, cofilin, to the ‘comet tail’ reaction causes Cy3-Tm1A to bind robustly throughout the actin network (Figure 2A, middle and bottom rows; Figure S2A, Movie S2). Increasing the concentration of cofilin increases the level of Cy3-Tm1A incorporation, but the distribution of tropomyosin along the tail is not uniform: more Cy3-Tm1A associates with the oldest regions of the network, most distal from the ActA-coated bead (we observed the same results with rabbit skeletal muscle tropomyosin: Figure S2D). Based on line scans, the distribution of labeled Tm1A is similar to our previous measurements of cofilin distribution along actin comet tails (data not shown). Significantly, in single-filament TIRF experiments, at concentrations of cofilin sufficient to coat actin filaments, Tm1A binding is blocked (Figure S1E). This result argues that cofilin does not directly recruit Tm1A to actin filaments but facilitates tropomyosin binding by severing or de-branching older filaments. The non-uniform association of Cy3-Tm1A with motile actin networks fits a previous suggestion by Bugyi et al. (2010) [8] that tropomyosin prefers to bind unbranched (or de-branched) actin filaments.
Although cofilin renders dendritic actin networks competent to bind Tm1A, the binding of Tm1A inhibits further network disassembly by cofilin (Figure 2B, S2C). In comet tail reactions without Tm1A, the actin network attached to the microsphere detaches from the original, broken actin shell in 13 ± 5 minutes (n = 7). Without tropomyosin we never observed an actin shell remain attached to a growing tail for longer than 25 minutes. In the presence of Tm1A, however, none of the shells completely detached from the rest of the network, even by 45 minutes (n = 11).
Using polarized light microscopy [25] we found that tropomyosin and cofilin collaborate to change the architecture of the actin comet tail. The optical birefringence of polymer networks increases with increasing parallel alignment of the constituent filaments. Therefore, measuring the difference in optical path length (retardance) of light polarized in different directions provides a measure of filament alignment [26]. In the absence of cofilin, the birefringence of dendritic actin networks formed from ActA-coated microspheres is very low, both in the presence and absence of Tm1A (retardance measured along the ‘slow’ optical axis is < 0.05 nm, Figure 2C: left and right). This reflects the tendency of Arp2/3-dependent branching to oppose parallel alignment of filaments [27]. The only exception to this rule is found in the ruptured actin shell attached to the base of the growing tail. Filaments in this shell are much more aligned than in the rest of the network, probably due to the tensile forces that initially break the shell and polarize the network. Addition of cofilin increases the birefringence of growing dendritic networks constructed in the presence of Tm1A (measured retardance is > 0.4 nm along the ‘slow’ optical axis, Figure 2C: center), especially in older regions of the network distal to the microsphere. In the absence of Cy3-Tm1A, however, high concentrations of cofilin disassemble the older regions of the actin comet tail without detectably increasing network birefringence (Figure 2C: left). These results argue that Cy3-Tm1A binds and stabilizes filaments after de-branching: once they are no longer linked to the dendritic network by the Arp2/3 complex and are free to re-align. Note that comet tails produced in the presence of cofilin but without Tm1A (Figure 2C) look different from those produced under other conditions. This difference reflects the fact that we optimized concentrations of Arp2/3 complex and capping protein to produce the most robust comet tails in the absence of cofilin. Optimizing the concentrations of Arp2/3 complex and capping protein to produce single comet tails in the presence of cofilin (without tropomyosin) alters the dynamics of network assembly such that, when cofilin is removed, actin comet tails often fail to form at all. To maintain consistency, we chose one set of Arp2/3 and capping protein concentrations to use in all of our comet tail assays.
The Arp2/3 complex excludes tropomyosin by blocking the pointed end of actin filaments
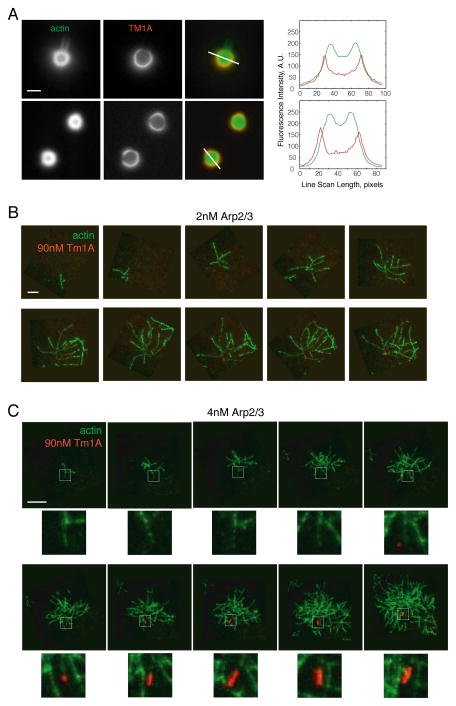
Bugyi et al. (2010) [8] suggested that the apparent preference of tropomyosin for unbranched actin filaments was caused by daughter filaments interfering with tropomyosin binding to the mother filaments from which they sprouted. To test this idea we first changed the average distance between branch sites in Arp2/3- and ActA-generated dendritic networks by changing the concentration of capping protein. Decreasing capping protein concentration produces networks with longer filaments and sparser Arp2/3-generated branches [24], but even when we dropped capping protein concentration to very low levels, below those that support symmetry breaking and directed motion, we observed no increase in the binding of Cy3-Tm1A (Figure 3A). Once again, we detected Cy3-Tm1A only on the oldest regions of the network, at the extreme outer edge of the original actin shell.
Figure 3. Tm1A binding is blocked by the Arp2/3 complex.
(A) At low capping protein concentrations Tm1A is still excluded from dendritic actin networks. Top row: 10nM CP. Bottom row: 50nM CP. Scale bar: 5μm. Right: fluorescence intensity line scans. (B–C) Frames from a time-lapse TIRF movie of Cy5-Tm1A binding to Arp2/3 branched actin filaments. B: 5% 600nM A488-actin, 2nM Arp2/3, 40nM ActA, 90nM Cy5-Tm1A. C: 5% 600nM A488-actin, 4nM Arp2/3, 40nM ActA, 90nM Cy5-Tm1A. An arrow indicates the pointed end of the original mother filament in each frame. Stars indicate frames where Tm1A is bound. Areas outlined by white dashed box are enlarged beneath each frame. Scale bar: 10μm. (See also Figure S3).
To further increase spacing between daughter filaments, we formed small, branched actin arbors using the Arp2/3 complex and soluble nucleation promoting factors. We added Cy5-Tm1A and imaged network formation by TIRF microscopy, starting from the initial branching events. To avoid potential artifacts we did not attach filaments to the coverslip. Interestingly, the initial interaction of Cy5-Tm1A with these growing filament arbors was quite slow (e.g. t1/2 = 726 ± 73 seconds under conditions shown in Figure 3C, n=3) but, once formed, foci of Cy5-Tm1A spread rapidly along a filament. Remarkably, in these experiments Cy5-Tm1A binds only one filament in each arbor: the initial mother —or “Eve” filament— that seeds formation of the rest of the network (Figure 3B–C, Movie S4). Under these conditions no other filament in the network binds detectable amounts of tropomyosin. The presence of daughter filaments clearly does not block binding of tropomyosin since the Eve filament is neither longer nor less branched than other filaments in the arbor. The major difference between the Eve filament and her descendants is the presence of the Arp2/3 complex at the pointed end. Although, the Arp2/3 complex can bind free pointed ends [28, 29], it dissociates from them much more rapidly (>100-fold) than from stable branches [30]. The Eve filament is, therefore, the only filament in the arbor that could possibly have a free pointed end. The slow binding of tropomyosin in these experiments might reflect a combination of factors: concomitant polymerization of the actin network and partial occupancy of the pointed end of the Eve filament by the Arp2/3 complex.
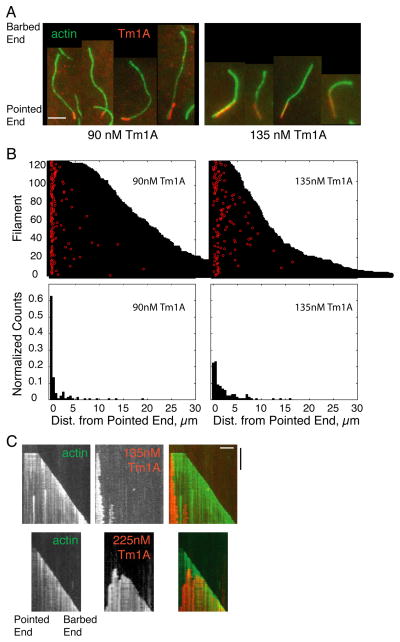
To test whether tropomyosin binds actin filaments preferentially at or near pointed ends, we followed binding of Cy5-Tm1A to individual filaments by TIRF microscopy in the absence of the Arp2/3 complex. We identified barbed and pointed ends based on growth rate and photo-bleaching of the labeled filaments. At all concentrations of Cy5-Tm1A tested we observed a strong preference for the initial binding event to occur near the pointed end (Figure 4A–B, Figures S4D & S5A, Movie S3). This preference was strongest at the lowest Cy5-Tm1A concentrations and was less pronounced at higher concentrations (compare data in the two top panels of Figure 4B and in Figure 5A). At low concentrations (~90 nM) Cy5-Tm1A coated only regions of the filaments immediately adjacent to the pointed end (Figure 4A).
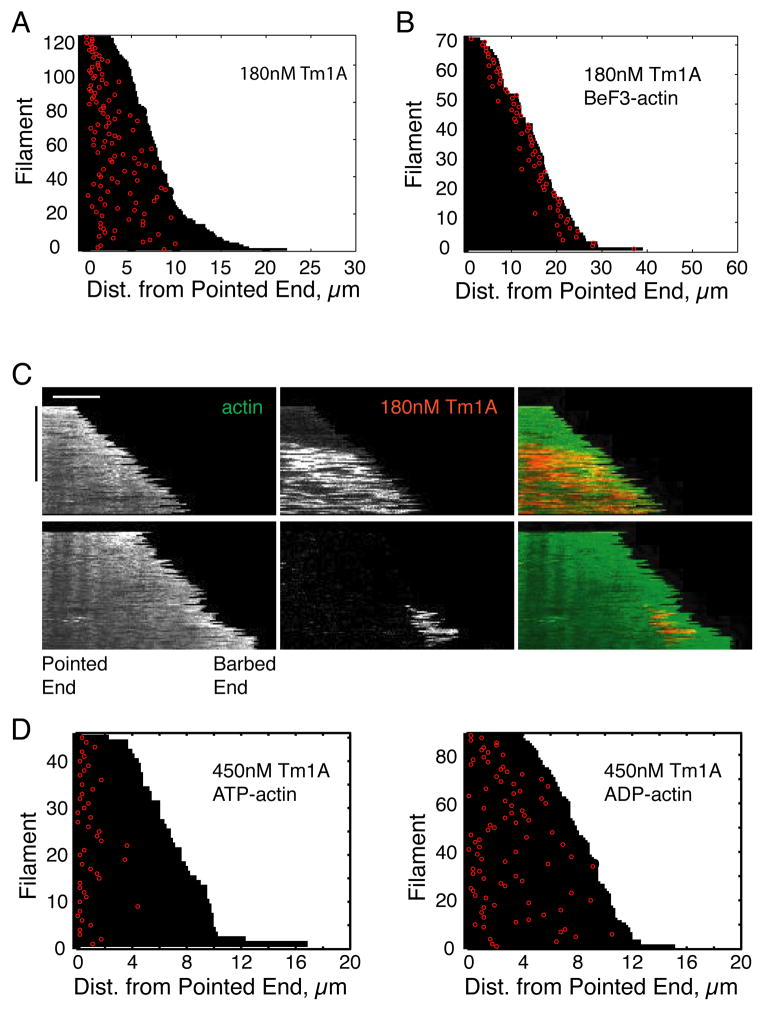
Figure 4. Tm1A binds near the pointed end.
(A) Tm1A binds near the pointed end of actin filaments in single filament TIRF. Red: Cy5-Tm1A. Green: 600nM A488-actin. Left: 90nM Tm1A. Right: 135nM Tm1A. (B) Top panel: Visual representation of each binding event of Cy5-Tm1A to actin filaments, as shown in A. Each filament’s length is plotted as a black bar, and the distance from the pointed end of each binding event is plotted in red. Bottom panel: Normalized histograms of Tm1A binding events as a function of the distance from the pointed end of actin filaments. 90nM: 126 binding events, 135nM: 121 binding events. (C) Kymographs of Tm1A binding and spreading along actin filaments. Top panel: 135nM Cy5-Tm1A. Bottom panel: 225nM Cy5-Tm1A. White scale bar: 5μm. Black time bar: 500 sec. (See also Figure S4).
Figure 5. In the presence of the phosphate mimic, BeF3, Tm1A binding shifts to the barbed end, and becomes transient.
(A–B) Visual representation of each binding event of 180nM Cy5-Tm1A to actin filaments formed in ATP (A), or in BeF3 (B). Each filament’s length is plotted as a black bar, and the distance from the pointed end of each binding event is plotted in red. (C) Binding of Tm1A to BeF3-bound actin filaments is transient. Representative hand-drawn kymographs of A488-actin and Cy5-Tm1A, with and without BeF3. White scale bar: 5μm. Black time bar: 500 sec. (D) Visual representation of each binding event of 450nM Cy5-Tm1A to ATP-actin filaments (left) or ADP-actin filaments (right). (See also Figure S5).
We observed the same pointed-end preference of Tm1A binding when filaments were tacked to coverslips (Figure 4C) by suface-immobilized phalloidin as when they remained unattached (Figure S4B). However, with attached filaments, binding required higher concentrations of Tm1A (450nM). At low concentrations of Tm1A (135 nM) on unattached filaments, initial binding occurred almost exclusively near the pointed end (Figure 4C, top panels). From this initial site of interaction, Tm1A spread toward the barbed end of the growing filament, but stopped growing at a more-or-less stable, steady-state length. We interpret the slow and relatively rare initial binding events as the assembly of tropomyosin “nuclei”: clusters of tropomyosin large enough to remain stably associated with the filament and support further tropomyosin binding. We interpret the rapid spreading of labeled Tm1A as recruitment of soluble Tm1A dimers into stable polymers on the actin filament (Figure 6A). Interestingly, the steady-state length of Tm1A decoration was not constant but fluctuated detectably, suggesting the existence of phases of cooperative assembly and disassembly of Tm1A polymers. At higher concentrations (Figure 4C, bottom panels) tropomyosin spreading did not stop at a stable length but continued until it coated the entire filament. At these higher concentrations we also observed multiple tropomyosin nucleation events on the same filament. Spreading of Tm1A was often saltatory —even at the highest concentrations tested— with periods of rapid spreading separated by occasional pauses.
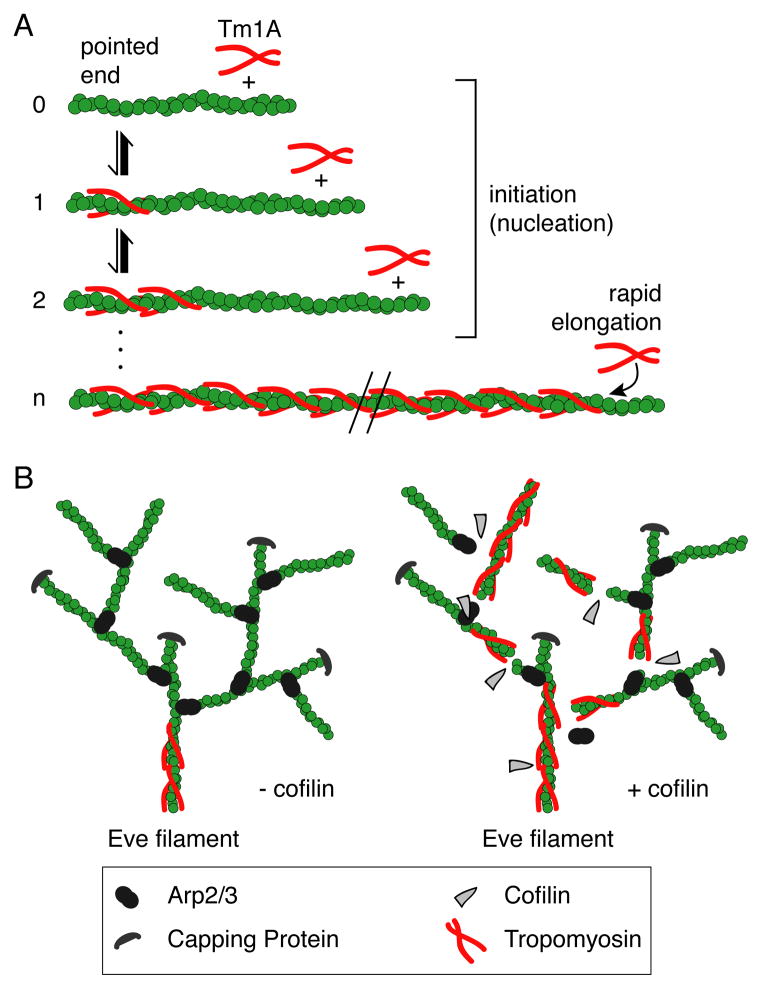
Figure 6. Model: Cooperation of the severing activity of cofilin and tropomyosin binding help establish the border between the lamellipod and lamellum.
(A) Initiation of tropomyosin binding to actin filaments is slow, but once a “nucleus” of tropomyosin is bound, elongation is rapid. (B) Left branched network: tropomyosin binding is blocked by Arp2/3 branches in the absence of cofilin. Right branched network: in the presence of cofilin, new pointed ends are created, which allows tropomyosin to bind. Once tropomyosin is bound, it protects the actin filaments from further cofilin severing, possibly resulting in the transition from the lamellipod to the lamellum.
Although Tm1A binds near the pointed end, binding does not inhibit pointed end polymerization, as judged by assembly of pyrene-labeled actin from capping protein-generated filament seeds (Figure S4C).
Tropomyosin binding is sensitive to actin filament conformation
The preference of Tm1A for the pointed end of actin filaments could reflect either: (1) contact with surfaces exposed only at pointed ends, or (2) interaction with filament conformations found predominantly near the pointed end. Although initial tropomyosin binding events are biased toward the pointed end of the filament not all of them occur at the extreme pointed end (Figure 4B–C & S4D), arguing against the idea that Tm1A binds directly to exposed ends. To test the role of filament conformation, we polymerized ATP-actin in the presence of beryllium fluoride (BeF3), a phosphomimic that stabilizes filaments in the ATP or ADP-Pi conformation [31]. We observed two striking effects of BeF3: (1) Binding of Cy5-Tm1A was destabilized. Low concentrations of Cy5-Tm1A (~90 nM) that partially coated filaments in the absence of BeF3 (Figure 4A) failed to bind at all in the presence of 1 mM BeF3 (plus 200 μM ATP), and higher concentrations of Cy5-Tm1A (180 nM) produced only transient interactions that failed to spread along the filament (Figure 5C). (2) More remarkably, all of these transient Cy5-Tm1A binding events were biased toward the barbed end (<2.5 μm) rather than the pointed end (Figure 5B).
To understand this result it helps to recognize that, in the presence of BeF3, growing actin filaments can be divided into four zones: (1) ATP-bound protomers at the growing barbed end; (2) slightly older, ADP-Pi-bound protomers near the barbed end; (3) ADP-bound protomers that begin to appear in regions slightly more distal to the barbed end; and (4) ADP-BeF3 subunits created by the binding of BeF3 to ADP-bound protomers. We used previously determined rate constants [32, 33] to model the spatial distribution of these different nucleotide states in our filaments (Figure S5C), and obtained results consistent with transient binding of Cy5-Tm1A to ADP-actin subunits that that appear transiently between the ADP-Pi-bound and ADP-BeF3-bound protomers.
We next asked whether Tm1A binding is biased toward the pointed ends of filaments formed from ADP-actin monomers. For this experiment, we added Cy5-Tm1A to polarity-marked actin filaments grown using either ATP- or ADP-actin monomers imaged binding by TIRF microscopy. As in the previous experiments, we found that Tm1A binds preferentially near the pointed ends of filaments generated from ATP-actin but binds more-or-less uniformly along ADP-actin filaments (Figure 5D). These results reveal that filament conformations that promote interaction with Tm1A appear only after hydrolysis of ATP and dissociation of inorganic phosphate (Figure S4A).
Discussion
Spreading of tropomyosin from a single initiation site at one end of an actin filament solves two potential problems: (i) gaps in tropomyosin’s coverage of the filament and (ii) mixing of multiple tropomyosin isoforms on the same filament. Gaps in tropomyosin coverage arise when two tropomyosin polymers spread from different initiation sites on the same filament. Each tropomyosin dimer spans 6 or 7 protomers and so two adjacent polymers have only a 1-in-6 or 1-in-7 chance of merging seamlessly, without a gap. Such gaps may have important consequences for filament stability and can be eliminated only by very slow processes [34]. Similarly, binding of multiple tropomyosin isoforms or post-translationally modified variants to the same filament can be avoided by having a single site of initiation, access to which could be controlled by regulatory molecules. For example, Johnson et al. (2014) [16] found that the two fission yeast formins promote binding of two different tropomyosin variants (acetylated versus unacetylated) to the filaments they create. Formins create actin filaments with free pointed ends and, if they interact even weakly with tropomyosins, they could specify from the outset which tropomyosin variant will assemble on a newly formed filament.
Dendritic actin networks: strengths and limitations of biomimetic assays
Tropomyosin splits reconstituted dendritic actin networks into two sets of filaments [8]: a tropomyosin-coated set that does not bind cofilin or the Arp2/3 complex, and a tropomyosin-free set that is competent to promote nucleation by the Arp2/3 complex. These two filament populations are stably maintained because they are insulated from each other by the Arp2/3 complex, which blocks binding of tropomyosin to the new filaments it creates. Cofilin initiates a transition from Arp2/3-crosslinked networks to tropomyosin-bound filaments by severing and debranching the dendritic network. On the surface, this appears to differ from the results of Bugyi et al. (2010) [8] who claimed that tropomyosin binding to reconstituted dendritic networks is insensitive to the concentration of cofilin. These authors, however, included the potent actin severing protein gelsolin in all of their network assembly assays.
Although the Arp2/3-branched and the tropomyosin-bound filaments overlap spatially, they have different network geometries: the tropomyosin-stabilized filaments reorganize into a more parallel orientation (overall order parameter approximately 0.025) with higher optical birefringence than the parent dendritic network. These composite actin networks —with one subpopulation branched by the Arp2/3 complex and another bound to tropomyosin— resemble the actin networks that underlie lamellipodia and lamella at the leading edge of migrating cells. Despite biochemical and architectural similarities between the in vivo and in vitro networks, however, it remains unclear whether lamellar filaments in motile cells begin life as lamellipodial filaments or whether they are nucleated de novo by factors other than the Arp2/3 complex (e.g. formin-family proteins).
Non-uniform association of tropomyosin with actin filaments
Binding of cytoskeletal tropomyosin to actin filaments turns out to be more complicated than previously appreciated (Figure 6). Both nucleation and spreading of tropomyosin are strongly influenced by the conformation of the actin filament and the presence of other regulatory proteins (Table 1). The pointed end bias of tropomyosin binding disappears in filaments assembled from ADP-actin monomers, but it cannot be explained solely by ATP hydrolysis since we observe strong pointed-end association even on filaments that have hydrolyzed most of their ATP and released most of their inorganic phosphate. Using previously measured rate constants for ATP hydrolysis [32] and phosphate dissociation [35] we estimate that many of the initial tropomyosin binding events we observed by TIRF on actin filaments assembled in ATP occurred at times when >95% of the protomers were actually bound to ADP (Figure S4A). In addition, both tropomyosin and cofilin bind preferentially to ADP-actin filaments, but the two proteins stabilize different filament conformations [36, 37], arguing that the nucleotide state of the filament is not sufficient to explain tropomyosin’s pointed end preference.
Table 1. Pointed-end binding is dependent on nucleotide state of actin as well as Tm1A concentration.
Quantification of pointed-end bias (percentage of initial Tm1A binding events that occur on the pointed end half of the filament at the time of binding).
| Actin Nucleotide State | Filaments Attached/Detached | [Tm1A] | Pointed-end Bias | N events |
|---|---|---|---|---|
| ATP | Detached | 90nM | 98.4% | 127 |
| ATP | Detached | 135nM | 87.6% | 121 |
| ATP | Detached | 180nM | 74.8% | 115 |
| ATP | Attached | 450nM | 97.8% | 45 |
| ADP | Attached | 450nM | 46.3% | 134 |
| BeF3 | Detached | 135nM | 3.7% | 82 |
| BeF3 | Detached | 180nM | 1.4% | 72 |
At high concentrations, Tm1A exhibits a preference for binding near the pointed end, but also begins to bind distal sites on uncapped/unbranched actin filaments. Motile dendritic networks however, fail to bind Tm1A even at high concentrations, suggesting that the influence of Arp2/3 on filament architecture is not limited to the extreme pointed end (Figure 6B). It is unclear how Arp2/3 bound at the pointed end could influence TM binding further from the pointed end, but a similar ‘action at a distance’ has been proposed to underlie the ability of cofilin to promote dissociation of nearby Arp2/3 complexes from a filament [38]. We suggest that the Arp2/3 complex could change the conformation of the filament near the pointed end, possibly by delaying ATP hydrolysis and/or phosphate release.
Finally, our results may explain previous observations, such as Michele et al. (1999) [39], who found that tropomyosin loads onto thin filaments of cardiac myocytes preferentially at their pointed ends, near the middle of the sarcomere. Part of this bias may be due to the effects of pointed end-capping proteins such as tropomodulin [40], but our results indicate that tropomyosin also has an intrinsic preference for pointed ends.
Materials & Methods
Design of labeled tropomyosin
We constructed a recombinant version of the Tm1A isoform of Drosophila tropomyosin. We labeled our engineered protein with Cy3- or Cy5-maleimide to produce Cy3-Tm1A and Cy5-Tm1A. We designed our labeled tropomyosin so that the label would be least disruptive to the protein’s structure and function. We replaced the only endogenous cysteine with an alanine, C32A, and chose a serine to replace with a cysteine, S82C. Since Tm1A is acetylated in vivo (Goins and Mullins, in press), we also genetically encoded an acetylation mimic to the N terminus of the protein: AS-Tm1A. The first amino acids in our Tm1A construct are MASMTTS.
Bead Motility Assay
Carboxylated, polystyrene, 5μm diameter beads (Bangs Labs) were coated with ActA30-612-KCK-6XHis using EDC-SulfoNHS chemistry, as described in Akin and Mullins (2008) [24]. For motility assays without recycling agents (cofilin and profilin), we mixed the ActA-coated beads with 7.4μM 3% Alexa488 labeled actin, 210nM capping protein, 125nM Arp2/3, and varying amounts of labeled Tm1A. For motility assays with recycling agents, we included the indicated concentration of hCofilin1, and added an equal concentration of hProfilin1.
Single Filament TIRF
Images were acquired at RT on a software-controlled (NIS Elements 4.1) Nikon Ti-E inverted TIRF microscope equipped with an EM CCD camera (Andor DU897), and a Nikon 100X oil immersion objective. For assays where the filaments were not attached to the coverslip, we used a protein mix of labeled actin, tropomyosin, with or without Arp2/3 and hcofilin1. For assays where the filaments were attached, the PEG-biotin glass chamber was first treated with 1μM Streptavidin and 1μM biotin-phalloidin. For assays with BeF3, 1mM BeCl2 and 5mM KF were added to the protein mix before starting the reaction. Images were taken at 10 second intervals.
Polarized Light Microscopy
Birefringence and fluorescence images were acquired at RT on a software-controlled (Micromanager 1.4.15) Nikon Microphot-FXA polarized light microscope equipped with a CRI Abrio camera, and a Nikon 60X (N.A. 1.40) oil immersion objective.
For complete materials and methods, please see the Supplemental Materials.
Supplementary Material
Actin networks generated from ActA-coated polystyrene microspheres by the Arp2/3 complex and capping protein exclude Tm1A in the absence of cofilin. Green: Alexa488-actin. Red: Cy3-Tm1A. 7.4μM 3% A488 actin, 125nM Arp2/3, 210nM CP, 450nM Cy3-Tm1A. 1min intervals.
Actin networks generated from ActA-coated polystyrene microspheres. Cofilin renders dendritic actin networks competent to bind Tm1A. Green: Alexa488-actin. Red: Cy3-Tm1A. 7.4μM 3% A488 actin, 125nM Arp2/3, 210nM CP, 450nM Cy3-Tm1A, 1μM cofilin, 1μM profilin. 1min intervals.
Tm1A binds near pointed ends in single filament TIRF. Green: Alexa488-actin. Red: Cy5-Tm1A. 135nM Cy5-Tm1A, 600nM 5% A488-actin. 10sec intervals.
Tm1A binds only free pointed ends in single filament TIRF. Green: Alexa488-actin. Red: Cy5-Tm1A. 90nM Cy5-Tm1A, 600nM 5% A488-actin, 4nM Arp2/3, 40nM ActA.10sec intervals.
Highlights.
Tropomyosin (TM) binding is biased towards ADP-bound actin, near the pointed end.
Arp2/3 blocks TM binding to branched actin networks by blocking the pointed end.
Cofilin severs actin filaments, generating free pointed ends competent to bind TM.
Acknowledgments
This work was supported by grants to RDM from the National Institutes of Health (3R01GM061010) and the Howard Hughes Medical Institute. RDM acknowledges J.R. L., F.R. Nada, and C.R. Amit for streamlining the operation. The authors would also like to thank Orkun Akin for generously giving us his ActA and capping protein constructs, and for teaching JYH how to do bead motility assays. We would like to thank Peter Bieling for giving us his hCofilin1 and hProfilin1 constructs. We are grateful to Elena Ingerman for lending us her NWASP WWCA-Sepharose column, and helping JYH with the Arp2/3 prep, as well as teaching JYH how to do single filament TIRF assays. We are also grateful to Chris Rivera, who provided many intellectually stimulating conversations. We would like to thank Kurt Thorn and DeLaine Larsen from the UCSF Nikon Imaging Center, who helped us operate and maintain the TIRF scope used in these assays. JYH would like to dedicate this work to her parents, Tony and Sue Hsiao.
Footnotes
Contributions
JYH did the cloning, protein purifications, microscopy assays, image analysis, modeling, figure making and writing. LMG made the WT AS-Tm1A construct from which the AS-Tm1A C32A/S82C construct was made. LMG also optimized the Tm1A purification protocol, and gave useful scientific guidance throughout the project. NP collected the electron micrographs of Cy5-Tm1A. RDM collected the birefringence data, determined the concentration and labeling percentage of Cy5-Tm1A, gave useful scientific guidance throughout the project, and contributed to the writing of this paper.
Conflict of Interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–6. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 2.Vallotton P, Gupton SL, Waterman-Storer CM, Danuser G. Simultaneous mapping of filamentous actin flow and turnover in migrating cells by quantitative fluorescent speckle microscopy. Proc Natl Acad Sci USA. 2004;101:9660–5. doi: 10.1073/pnas.0300552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salmon WC, Adams MC, Waterman-Storer CM. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J Cell Biol. 2002;158:31–7. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 6.DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. Spatial regulation of actin dynamics: a tropomyosin- free, actin-rich compartment at the leading edge. J Cell Science. 2002;115:4649–4660. doi: 10.1242/jcs.00147. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- 8.Bugyi B, Didry D, Carlier MF. How tropomyosin regulates lamellipodial actin-based motility: a combined biochemical and reconstituted motility approach. EMBO Journal. 2010;29:14–26. doi: 10.1038/emboj.2009.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, et al. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J Cell Biol. 2005;168:619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunning P, O’Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 11.Lehrer SS, Morris EP. Comparison of the effects of smooth and skeletal tropomyosin on skeletal actomyosin subfragment 1 ATPase. J Biol Chem. 1984;259:2070–2072. [PubMed] [Google Scholar]
- 12.Barua B, Nagy A, Sellers JR, Hitchcock-DeGregori SE. Regulation of nonmuscle myosin II by tropomyosin. Biochem. 2014;53:4015–4024. doi: 10.1021/bi500162z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creed SJ, Bryce N, Naumanen P, Weinberger R, Lappalainen P, Stehn J, Gunning P. Tropomyosin isoforms define distinct microfilament populations with different drug susceptibility. Eur J Cell Biol. 2008;87:709–20. doi: 10.1016/j.ejcb.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. A molecular pathway for myosin II recruitment to stress fibers. Curr Biol. 2011;21:539–50. doi: 10.1016/j.cub.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Skau CT, Kovar DR. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr Biol. 2010;20:1415–1422. doi: 10.1016/j.cub.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson M, East DA, Mulvihill DP. Formins determine the functional properties of actin filaments in yeast. Curr Biol. 2014;24:1525–1530. doi: 10.1016/j.cub.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 17.Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol. 2010;20:1423–1431. doi: 10.1016/j.cub.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 18.Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Bio Cell. 2010;21:989–1000. doi: 10.1091/mbc.E09-10-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Korn ED, Eisenberg E. Cooperative binding of tropomyosin to muscle and Acanthamoeba actin. J Biol Chem. 1979;254:7137–40. [PubMed] [Google Scholar]
- 20.Dabrowska R, Nowak E, Drabikowski W. Some functional properties of non-polymerizable and polymerizable tropomyosin. J Muscle Res Cell Motil. 1983;4:143–161. doi: 10.1007/BF00712027. [DOI] [PubMed] [Google Scholar]
- 21.Pittenger MF, Helfman DM. In vitro and in vivo characterization of four fibro-blast tropomyosins produced in bacteria: TM-2, TM-3, TM-5a, and TM-5b are co-localized in interphase fibroblasts. J Cell Biol. 1992;118:841–58. doi: 10.1083/jcb.118.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–76. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–6. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 24.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oldenbourg R, Mei G. New polarized light microscope with precision universal compensator. J Microsc. 1995;180:140–7. doi: 10.1111/j.1365-2818.1995.tb03669.x. [DOI] [PubMed] [Google Scholar]
- 26.de Gennes PG. The physics of liquid crystals. Oxford, UK: Clarendon Press; 1974. [Google Scholar]
- 27.Cameron LA, Svitkina TM, Vignjevic D, Theriot JA, Borisy GG. Dendritic organization of actin comet tails. Curr Biol. 2001;11:130–135. doi: 10.1016/s0960-9822(01)00022-7. [DOI] [PubMed] [Google Scholar]
- 28.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volkmann N, Page C, Li R, Hanein D. Three-dimensional reconstructions of actin filaments capped by Arp2/3 complex. Eur J Cell Biol. 2014;S0171-9335:00004–1. doi: 10.1016/j.ejcb.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchoin L, Pollard TD, Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000;10:1273–1282. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- 31.Combeau C, Carlier MF. Probing the mechanism of ATP hydrolysis on F-actin using vanadate and the structural analogs of phosphate BeF-3 and A1F-4. J Biol Chem. 1988;263:17429–36. [PubMed] [Google Scholar]
- 32.Blanchoin L, Pollard TD. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41:597–602. doi: 10.1021/bi011214b. [DOI] [PubMed] [Google Scholar]
- 33.Jégou A, Niedermayer T, Orbán J, Didry D, Lipowsky R, Carlier MF, Romet-Lemonne G. Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 2011;9:e1001161. doi: 10.1371/journal.pbio.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilfan A. The binding dynamics of tropomyosin on actin. Biophys J. 2001;81:3146–3155. doi: 10.1016/S0006-3495(01)75951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlier MF. Measurement of Pi dissociation from actin filaments following ATP hydrolysis using a linked enzyme assay. Biochemical and Biophysical Research Communications. 1987;143:1069–1075. doi: 10.1016/0006-291x(87)90361-5. [DOI] [PubMed] [Google Scholar]
- 36.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li XE, Tobacman LS, Mun JY, Craig R, Fischer S, Lehman W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys J. 2011;100:1005–13. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michele DE, Albayya FP, Metzger JM. Thin filament protein dynamics in fully differentiated adult cardiac myocytes: toward a model of sarcomere maintenance. J Cell Biol. 1999;145:1483–95. doi: 10.1083/jcb.145.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–51. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Actin networks generated from ActA-coated polystyrene microspheres by the Arp2/3 complex and capping protein exclude Tm1A in the absence of cofilin. Green: Alexa488-actin. Red: Cy3-Tm1A. 7.4μM 3% A488 actin, 125nM Arp2/3, 210nM CP, 450nM Cy3-Tm1A. 1min intervals.
Actin networks generated from ActA-coated polystyrene microspheres. Cofilin renders dendritic actin networks competent to bind Tm1A. Green: Alexa488-actin. Red: Cy3-Tm1A. 7.4μM 3% A488 actin, 125nM Arp2/3, 210nM CP, 450nM Cy3-Tm1A, 1μM cofilin, 1μM profilin. 1min intervals.
Tm1A binds near pointed ends in single filament TIRF. Green: Alexa488-actin. Red: Cy5-Tm1A. 135nM Cy5-Tm1A, 600nM 5% A488-actin. 10sec intervals.
Tm1A binds only free pointed ends in single filament TIRF. Green: Alexa488-actin. Red: Cy5-Tm1A. 90nM Cy5-Tm1A, 600nM 5% A488-actin, 4nM Arp2/3, 40nM ActA.10sec intervals.