Abstract
Introduction
Merkel cell carcinoma is a neuroendocrine malignancy. Suppressor of fused (SUFU) is a tumor suppressor oncogene that participates in the Hedgehog (Hh) signaling pathway. The aim of the study was to describe a patient whose Merkel cell carcinoma demonstrated a SUFU genomic alteration.
Case Study
The Hh signaling pathway is involved in the pathogenesis of several tumors, including nevoid basal cell carcinoma syndrome that is associated with an alteration of the patched-1 (PTCH1) gene. Targeted molecular therapy against smoothened (SMO) with vismodegib has been shown to be an effective therapeutic intervention for patients with PTCH-1 mutation. The reported patient was presented with metastatic Merkel cell carcinoma. Analysis of his tumor, using a next-generation sequencing-based assay, demonstrated a genomic aberration of SUFU protein, a component of the Hh signaling pathway that acts downstream to SMO and, therefore, is unlikely to be responsive to vismodegib. Of interest, arsenic trioxide or bromo and extra C-terminal inhibitors impact signals downstream to SUFU, making this aberration conceivably druggable. His tumor has initially been managed with chemotherapy (carboplatin and etoposide) and subsequent radiation therapy is planned.
Conclusion
The pathogenesis of Merkel cell carcinoma is multifactorial, and related to ultraviolet radiation exposure, immunosuppression, and Merkel cell polyomavirus. We report a patient with a mutation in SUFU, a potentially actionable component of the Hh signaling pathway.
Electronic supplementary material
The online version of this article (doi:10.1007/s13555-015-0074-5) contains supplementary material, which is available to authorized users.
Keywords: Carcinoma, Cell, Hedgehog, Merkel, Pathway, Signaling, Suppressor of fused (SUFU)
Introduction
Merkel cell carcinoma is an aggressive neuroendocrine cancer of the skin [1]. Suppressor of fused (SUFU), an integral component of the Hedgehog (Hh) signaling pathway, functions as a tumor suppressor gene [2]. A man with metastatic Merkel cell carcinoma is described whose tumor was associated with a SUFU genomic aberration. Potential therapeutic interventions based upon this unique abnormality of his tumor are postulated.
Case Report
A 66-year-old Caucasian man was referred for an evaluation of an asymptomatic left buttock mass that he had noticed 6 months earlier. His past medical history was significant for prostate cancer at age 64 years (stage T1c Gleason grade 3 + 3) that was successfully treated with intensity-modulated radiation therapy. His pretreatment and posttreatment nadir prostate-specific antigen were 4.5 and 0.88 nanograms per milliliter, respectively.
A month prior to his skin evaluation, he was being worked up for a possible left inguinal hernia repair. His computerized axial tomographic scan showed enlargement of the left inguinal, left pelvic sidewall and retroperitoneal lymph nodes and a soft tissue nodule in his left buttock. His surgeon performed a superficial inguinal lymph node biopsy that showed a high-grade neuroendocrine carcinoma; the tumor cells stained positive for CD56 (neural cell adhesion molecule), chromogranin, cytokeratin 20, cytokeratin AE1/AE3, and synaptophysin. The tumor cells were negative for CD45 (leukocyte common antigen), GATA3 (trans-acting T cell-specific transcription factor) and prostate-specific antigen. The morphologic and immunohistochemical features indicated a metastatic Merkel cell carcinoma.
He also had a history of multiple lipomas. He had considered the left buttock nodule, which he initially noticed 6 months earlier, to be another lipoma; however, it continued to increase in size. A positron emission tomography (PET)/computed tomography (CT) scan, which was performed after receiving the lymph node biopsy pathology report of metastatic Merkel cell carcinoma, demonstrated intense fluorodeoxyglucose uptake from the soft tissue density in the subcutaneous tissue of the left buttock. This was suspicious for a primary tumor.
Cutaneous examination (1 month after the lymph node biopsy and 6 months after the patient’s discovery of a left buttock mass) showed a painless flesh-colored 3 × 3 cm subcutaneous nodule with central erythema on his left buttock (Fig. 1). The nodule was firmly grasped and pushed toward the skin surface as a punch biopsy using a 4-mm circular blade was performed into its center. A cylindrical core specimen of tissue, extending from the epidermis to beyond the deep dermis, was obtained. The nodule was then firmly squeezed and additional aggregates of tumor (appearing yellowish-white and blood tinged) that extruded through the biopsy wound were collected (Fig. 2).
Fig. 1.

Distant (a) and closer (b) views with ink demarcating the borders of the primary Merkel cell carcinoma on the left buttock nodule of a 66-year-old man
Fig. 2.
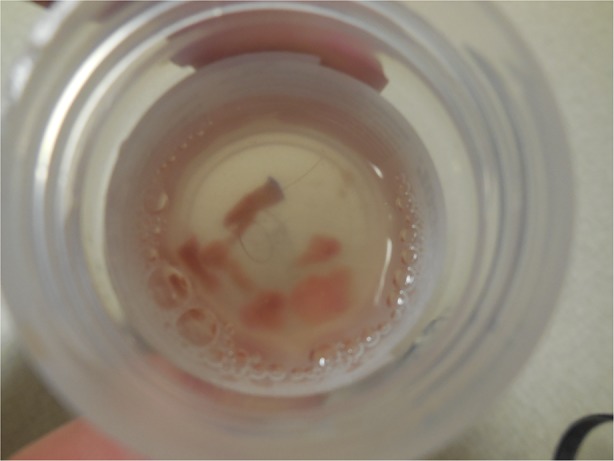
Gross examination of the punch biopsy specimen shows a cylindrical piece of tissue secured using the 4-mm circular blade and several blood-tinged yellowish-white lobules of tumor (morphologically mimicking adipose tissue). These were expressed through the hole created during the biopsy after applying firm pressure to the tumor
Microscopic examination of the skin biopsy specimen showed an infiltrate of small blue cells with minimal cytoplasm in the deep portion of the punch biopsy and all the pieces of subcutaneous tissue (Fig. 3). The basophilic cells were uniform in size with a vesicular nucleus and small nucleoli. Numerous mitoses were appreciated and there were large areas of necrosis. The tumor invaded fascia in the deeper specimens and focally grew in close approximation to multiple blood vessels; however, no intravascular or perineural invasion was identified (Fig. 4 ).
Fig. 3.
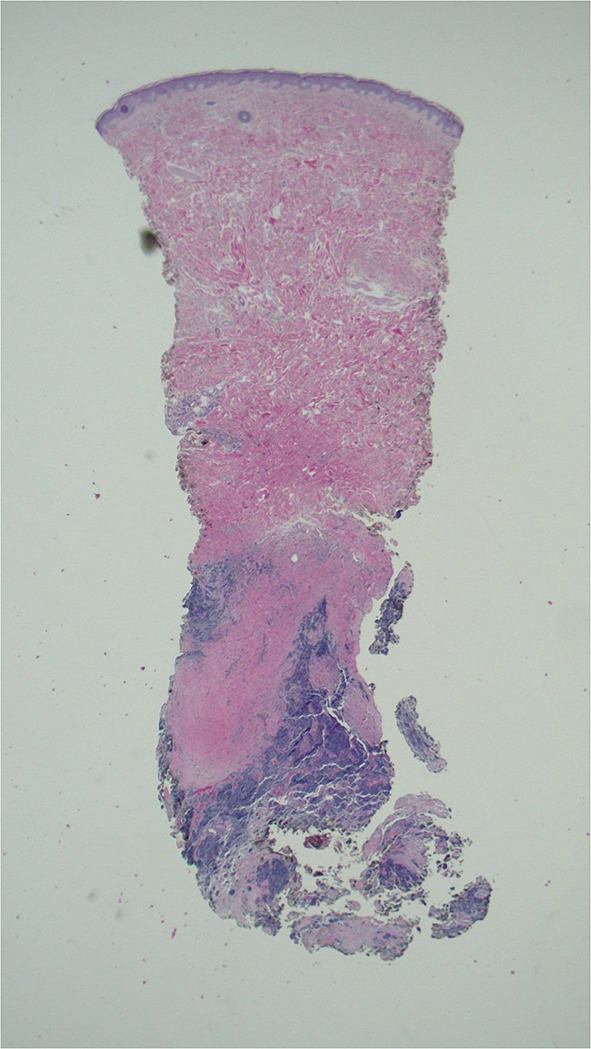
Tumor is present at the base of the punch biopsy specimen. It is in the deep dermis and extends into the subcutaneous tissue [hematoxylin and eosin, ×2]
Fig. 4.
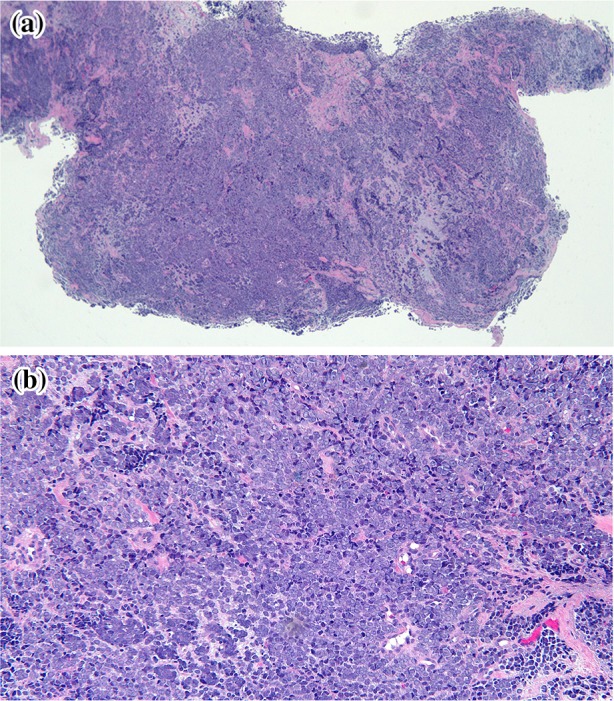
Distant (a) and closer (b) views of hematoxylin and eosin stained sections show the blue small cell tumor that extends from the deep dermis into the subcutaneous tissue [hematoxylin and eosin; a = ×4, b = ×20]
Immunohistochemical studies showed that the tumor cells stained positive for cytokeratin 20 (in a cytoplasmic and paranuclear dot-like pattern), CD56 (neural cell adhesion molecule), and synaptophysin (Fig. 5). The tumor cells were negative for prostate-specific antigen and thyroid transcription factor 1.
Fig. 5.
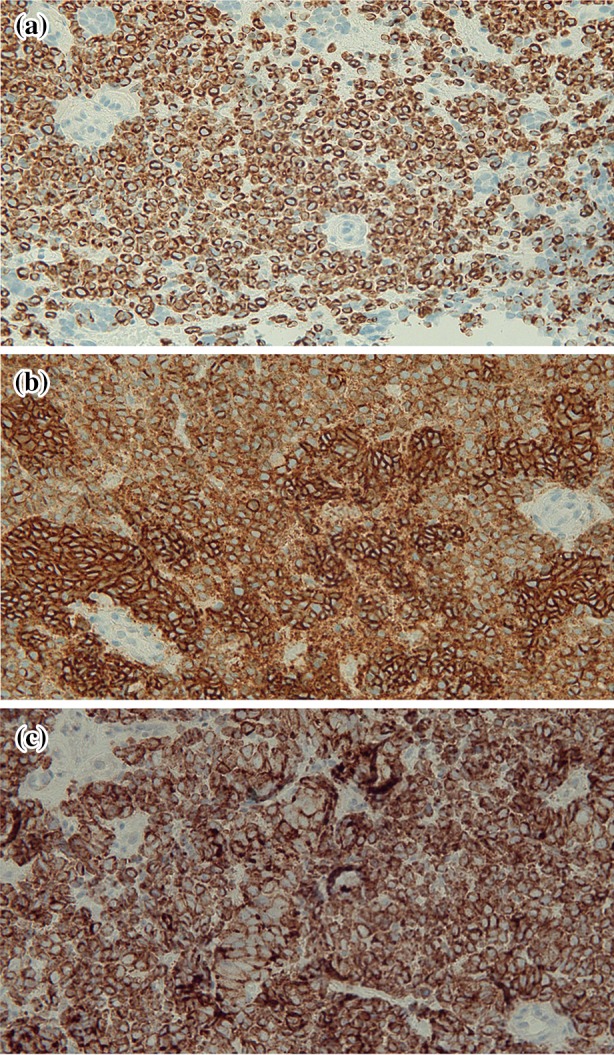
The tumor shows positive staining for cytokeratin 20 (a), CD56 (b) and synaptophysin (c) [immunoperoxidase: cytokeratin 20, a = ×40; CD56, b = ×40; synaptophysin, c = ×40]
Correlation of the history, clinical morphology, imaging studies, and pathology was diagnostic for metastatic Merkel cell carcinoma. The left buttock was the site of the primary tumor. Metastases had spread to the regional lymph nodes.
The biopsy tissue from the left buttock tumor was also sent for genomic evaluation. A next-generation sequencing-based assay was performed. Genomic alterations of our patient’s tumor, evaluated by next-generation sequencing included base substitutions, insertions, deletions, and copy number alterations [3].
DNA was extracted from 40 micrometer of formalin-fixed tissue (minimum 20% tumor cells) using the Maxwell® 16 FFPE Plus LEV DNA Purification kit (Promega, Wisconsin, USA) and quantified using a standardized PicoGreen fluorescence assay (Invitrogen). Library construction was performed using 50-200 nanogram of DNA sheared by sonication to approximately 100–400 base pairs before end-repair, dA addition and ligation of indexed Illumina sequencing adaptors (Illumina, Inc., San Diego, CA, USA). Enrichment of targeted sequences (including all coding exons of 315 cancer related genes) plus introns from 28 genes often rearranged or altered in cancer (Table 1) [4] was achieved by solution-based hybrid capture with custom biotinylated oligonucleotide bases. Enriched libraries were sequenced to an average median depth of >500× with 99% of bases covering >100× [Illumina HiSwq 2000 (Illumina, Inc.) platform using 49 × 49 paired-end reads] and mapped to the reference human genome (hg19) using the Burrows–Wheeler Aligner and the publicly available SAMtools, Picard, and Genome Analysis Toolkit. Point mutations were identified by a Bayesian algorithm; short insertions and deletions, determined by focal assembly; gene copy number alterations (amplification), by comparison to process matched normal controls; and gene fusions/rearrangements, by clustering chimeric reads mapped to targeted introns. Amplifications were called for ≥6 copies except for ErbB2 (≥5 copies). Six to seven copy numbers are called as equivocal and ≥8 are definitive; for ErbB2, equivocal amplification was 5–7 copies; all (equivocal or definitively amplified) were designated as positive for amplification in our patient. Aberrations, mutations or other alterations in kinases that were presumed to be inactivating based on wet lab experiments or structural modeling were not included [5].
Table 1.
Genes assayed by next-generation sequencing in Merkel cell carcinoma
| ABL1 | BRAF | CHEK1 | FANCC | GATA3 | JAK2 | MITF | PDCD1LG2 | RBM10 | STAT4 |
| ABL2 | BRCA1 | CHEK2 | FANCD2 | GATA4 | JAK3 | MLH1 | PDGFRA | RET | STK11 |
| ACVR1B | BRCA2 | CIC | FANCE | GATA6 | JUN | MPL | PDGFRB | RICTOR | SUFU |
| AKT1 | BRD4 | CREBBP | FANCF | GID4 (C17orf39) | KAT6A (MYST3) | MRE11A | PDK1 | RNF43 | SYK |
| AKT2 | BRIP1 | CRKL | FANCG | GLI1 | KDM5A | MSH2 | PIK3C2B | ROS1 | TAF1 |
| AKT3 | BTG1 | CRLF2 | FANCL | GNA11 | KDM5C | MSH6 | PIK3CA | RPTOR | TBX3 |
| ALK | BTK | CSF1R | FAS | GNA13 | KDM6A | MTOR | PIK3CB | RUNX1 | TERC |
| AMER1 (FAM123B) | C11orf30 (EMSY) | CTCF | FAT1 | GNAQ | KDR | MUTYH | PIK3CG | RUNX1T1 | TERT (promoter only) |
| APC | CARD11 | CTNNA1 | FBXW7 | GNAS | KEAP1 | MYC | PIK3R1 | SDHA | TET2 |
| AR | CBFB | CTNNB1 | FGF10 | GPR124 | KEL | MYCL (MYCL1) | PIK3R2 | SDHB | TGFBR2 |
| ARAF | CBL | CUL3 | FGF14 | GRIN2A | KIT | MYCN | PLCG2 | SDHC | TNFAIP3 |
| ARFRP1 | CCND1 | CYLD | FGF19 | GRM3 | KLHL6 | MYD88 | PMS2 | SDHD | TNFRSF14 |
| ARID1A | CCND2 | DAXX | FGF23 | GSK3B | KMT2A (MLL) | NF1 | POLD1 | SETD2 | TOP1 |
| ARID1B | CCND3 | DDR2 | FGF3 | H3F3A | KMT2C (MLL3) | NF2 | POLE | SF3B1 | TOP2A |
| ARID2 | CCNE1 | DICER1 | FGF4 | HGF | KMT2D (MML2) | NFE2L2 | PPP2R1A | SLIT2 | TP53 |
| ASXL1 | CD274 | DNMT3A | FGF6 | HNF1A | KRAS | NFKBIA | PRDM1 | SMAD2 | TSC1 |
| ATM | CD79A | DOT1L | FGFR1 | HRAS | LMO1 | NKX2-1 | PREX2 | SMAD3 | TSC2 |
| ATR | CD79B | EGFR | FGFR2 | HSD3B1 | LRP1B | NOTCH1 | PRKAR1A | SMAD4 | TSHR |
| ATRX | CDC73 | EP300 | FGFR3 | HSP90AA1 | LYN | NOTCH2 | PRKC1 | SMARCA4 | U2AF1 |
| AURKA | CDH1 | EPHA3 | FGFR4 | IDH1 | LZTR1 | NOTCH3 | PRKDC | SMARCB1 | VEGFA |
| AURKB | CDK12 | EPHA5 | FH | IDH2 | MAGI2 | NPM1 | PRSS8 | SMO | VHL |
| AXIN1 | CDK4 | EPHA7 | FLCN | IGF1R | MAP2K1 | NRAS | PTCH1 | SNCAIP | WISP3 |
| AXL | CDK6 | EPHB1 | FLT1 | IGF2 | MAP2K2 | NSD1 | PTEN | SOCS1 | WT1 |
| BAP1 | CDK8 | ERBB2 | FLT3 | IKBKE | MAP2K4 | NTRK1 | PTPN11 | SOX10 | XPO1 |
| BARD1 | CDKN1A | ERBB3 | FLT4 | IKZF1 | MAP3K1 | NTRK2 | QKI | SOX2 | ZBTB2 |
| BCL2 | CDKN1B | ERBB4 | FOXL2 | IL7R | MCL1 | NTRK3 | RAC1 | SOX9 | ZNF217 |
| BCL2L1 | CDKN2A | ERG | FOXP1 | INHBA | MDM2 | NUP93 | RAD50 | SPEN | ZNF703 |
| BCL2L2 | CDKN2B | ERRFI1 | FRS2 | INPP4B | MDM4 | PAK3 | RAD51 | SPOP | |
| BCL6 | CDKN2C | ESR1 | FUBP1 | IRF2 | MED12 | PALB2 | RAF1 | SPTA1 | |
| BCOR | CEBPA | EZH2 | GABRA6 | IRF4 | MEF2B | PARK2 | RANBP2 | SRC | |
| BCORL1 | CHD2 | FAM46C | GATA1 | IRS2 | MEN1 | PAX5 | RARA | STAG2 | |
| BLM | CHD4 | FANCA | GATA2 | JAK1 | MET | PBRM1 | RB1 | STAT3 | |
| Select rearrangements | |||||||||
| ALK | BRAF | BRD4 | ETV4 | FGFR1 | KIT | MYC | NTRK2 | RARA | TMPRSS2 |
| BCL2 | BRCA1 | EGFR | ETV5 | FGFR2 | MSH2 | NOTCH2 | PDGFRA | RET | |
| BCR | BRCA2 | ETV1 | ETV6 | FGFR3 | MYB | NTRK1 | RAF1 | ROS1 | |
All genes known to be somatically altered in human solid tumors that are validated targets for therapy, either approved or in clinical trials, and/or that are unambiguous drivers of oncogenesis were included. The assay interrogated 315 genes as well as introns of 28 genes involved in rearrangements
Alteration of the SUFU gene [SUFU R299Q (due to nucleotide mutation at position 299, arginine becomes glutamine in the SUFO protein)] was the only abnormality identified by the next-generation sequencing-based assay (315 gene panel; Foundation Medicine, Inc., Cambridge, MA, USA) [4]. Treatment with systemic chemotherapy was initiated with carboplatin and etoposide. He is receiving treatment every 3 weeks and adjuvant radiotherapy is planned.
Follow-up evaluation shows decrease in the size of the left buttock tumor and the previously enlarged lymph nodes. Consideration to use experimental agents that target the distal Hh pathway may be considered if his tumor progresses.
Informed consent was obtained from the patient for being included in the study and for publication of the images.
Discussion
Merkel Cell Carcinoma
Merkel cell carcinoma was initially described by Toker in 1972 as trabecular carcinoma of the skin [1]. It is a biologically aggressive tumor with a poor prognostic outcome resulting from local, regional and distant recurrences. Older men are more commonly affected and the primary tumor is usually located on sun-exposed skin [1, 6, 7].
Merkel cell carcinoma usually presents as an asymptomatic rapidly enlarging flesh-colored or blue-red nodule [6, 8]. In our patient, the tumor clinically appeared as a firm red nodule below the skin surface. Its gross appearance, yellow-white lobules, was similar in morphology to adipose tissue.
Histologically, Merkel cell carcinoma is a tumor composed of small monomorphous blue cells, often arranged in strands (trabeculae), which demonstrate numerous mitoses, apoptotic cells and occasional necrosis. The tumor shows a positive paranuclear dot-like pattern of staining for cytokeratin 20. In addition, the immunoperoxidase profile of the tumor often includes positive staining for CD56 (neural cell adhesion molecule), chromogranin A, gastrin, and somatostatin [6, 8, 9].
Non-invasive imaging methods (fluorine-18-fluorodeoxyglucose PET and CT) are typically used to assess the extent of disease in patients with Merkel cell carcinoma. They may be performed during the initial staging of the patient when distant metastases are suspected or when a cutaneous metastasis from a noncutaneous primary neuroendocrine carcinoma is considered [7, 10, 11]. Our patient had a history of multiple lipomas and presented with metastatic Merkel cell carcinoma to his left inguinal lymph node. His PET scan identified the primary tumor: a large and rapidly growing deep dermal nodule of his left buttock that had clinically been considered to be a lipoma.
The treatment of primary cutaneous Merkel cell carcinoma is a wide local surgical excision and sentinel lymph node biopsy. A positive sentinel lymph node may be followed by a complete lymph node dissection. Adjuvant radiotherapy is often included for patients with positive sentinel lymph nodes and considered in patients with tumors of the head and neck region that have a negative sentinel lymph node because of the higher risk of a false-negative result [1, 6–8].
The management of metastatic Merkel cell carcinoma includes a variety of treatments. Chemotherapy most often utilized platinum with or without etoposide. Although the tumor often initially responds to therapy, resistance frequently develops after two or three cycles of chemotherapy. Other treatments included radiation therapy and experimental novel approaches such as immunotherapy and targeted molecular therapy [1, 7, 12].
Exposure to ultraviolet radiation (such as psoriasis patients receiving psoralen and ultraviolet A therapy), immunosuppression (such as in AIDS patients, individuals with chronic lymphocytic leukemia, and organ transplant recipients) and Merkel cell polyomavirus infection are risk factors for the development of Merkel cell carcinoma [6, 12–15]. Merkel cell polyomavirus was discovered in 2008 [9, 16]. The virus is present in approximately 80 percent of Merkel cell carcinomas [17]. Indeed, there is improved survival for patients whose Merkel cell carcinoma is virus-positive as compared individuals with virus-negative tumors [14, 18]. However, the presence of Merkel cell polyomavirus is not specific for Merkel cell carcinoma. The virus has also been demonstrated in other malignancies, including cervical carcinoma [19], chronic lymphocytic leukemia [20], cutaneous squamous cell carcinoma [21], folliculotropic mycosis fungoides [22, 23], Langerhans cell sarcoma [24], and small cell carcinoma (extrapulmonary [25] and parotid [26]). In addition, although one group of researchers have found an association between Merkel cell polyomavirus infection and epidermal growth factor hotspot mutations in non-small-cell lung cancer [27], there is currently no evidence of an association between the virus and any specific genomic aberration—including a mutation in SUFU protein—in Merkel cell carcinoma [28].
Mutational analysis of Merkel cell carcinoma has revealed numerous abnormalities including mutations in telomerase activation, tumor suppressors, and tyrosine kinase signaling (Table 2) [9, 29]. Chromosomal analysis, utilizing comparative genomic hybridization to define copy number abnormalities (such as amplified or deleted regions) on chromosomes but not specific genes, has also found several chromosomal aberrations [9, 30]. A genomic alteration in SUFU protein—an integral component of the Hh signaling pathway—was identified in our patient’s Merkel cell carcinoma using a next-generation sequencing-based assay. Although one group of investigators detected over expression of Hh signaling pathway proteins in the Merkel cell carcinomas they studied [31], to the best of our knowledge, neither Merkel cell carcinoma associated with a SUFU protein mutation nor Merkel cell carcinoma related to a genomic aberration of the Hh signaling pathway has previously been described.
Table 2.
Mutations in telomerase activation, tumor suppressors and tyrosine kinase signaling in Merkel cell carcinoma
| Telomerase activation mutations and amplifications |
| TERT (telomerase reverse transcriptase) promoter |
| Tumor suppressor mutations |
| RB1 (human retinoblastoma gene) |
| SUFU (suppressor of fused) |
| TP53 (tumor protein p53) |
| Tyrosine kinase signaling mutations |
| AKT (protein kinase B) |
| KIT (also known as CD117 or mast/stem cell growth factor receptor) |
| PDGFRA (platelet-derived growth factor receptor, alpha) |
| PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha) |
| PTEN (phosphatase and tensin homolog) |
Hedgehog Signaling Pathway
The Hh signaling pathway has an essential role in not only embryonic development but also cell differentiation and proliferation. The term ‘hedgehog’ was introduced to describe the appearance (similar to that of the spiky hedgehog) of the mutated gene in the larva of the fruit fly Drosophila melanogaster. The Hh signaling pathway is suppressed in the majority of normal human cells [2, 32, 33].
In mammals, the primary cilium of the cell membrane contains a receptor and a signal transducer. The twelve-pass transmembrane receptor patched-1 (PTCH1) inhibits the seven-pass transmembrane G-coupled receptor-like protein smoothened (SMO). This allows for the intracellular inhibition of the associated oncogene glioma-1/2 transcription factor function by (SUFU) protein [2, 32, 34, 35].
The binding of the Hh ligand to the PTCH1 receptor activates the Hh signaling pathway. In humans, there are three Hh ligands: (1) sonic hedgehog (named after the Sega videogame and implicated in establishing left–right symmetry and development of the central nervous system, eyes and muscles), (2) desert hedgehog (named after a real hedgehog species and implicated in the development of male germ cells), and (3) Indian hedgehog (named after a real hedgehog species and implicated in the development of cartilage and regulation of bone growth). Binding of the Hh ligand to PTCH1 receptor (or the presence of a mutation in either the PTCH1 or SMO signal transducer) removes the suppression of SMO by PTCH1. This results in the inhibition of the regulation by SUFU protein and the subsequent release of glioma transcription activity in the nucleus [2, 32, 33, 36, 37].
The Hh signaling pathway plays a role in the development and persistence of human cancers. Constitutive activation of the Hh signaling pathway is involved in tumorigenesis of nevoid basal cell carcinoma syndrome (which is also known as Gorlin–Goltz syndrome or Gorlin syndrome) and associated with cancers that have a mutation in the PTCH1 gene: basal cell carcinoma, medulloblastoma, meningioma, and rhabdomyoblastoma. Hyperactivity of the Hh signaling pathway has also been linked to the development of other cancers including breast, colon, gastric, leukemias, lung, multiple myeloma, pancreas and prostate. In addition, aberrant activation of the Hh signaling pathway has been demonstrated to contribute not only to the maintenance and expression of leukemic stem cells, but also to chemotherapy resistance (in ovarian cancer, cervical cancer, and myeloid leukemia cells) and radiotherapy resistance (in pancreatic cancer and head and neck cancer) [2, 32, 38, 39].
Targeted therapy that affects the Hh signaling pathway—specifically with vismodegib that is an inhibitor of SMO—has shown efficacy in the treatment of locally advanced and metastatic basal cell carcinoma and other tumors with patched-1 mutations [34, 36, 40–45]. However, some patients’ basal cell carcinomas have acquired resistance to vismodegib [33, 46, 47]. Therefore, therapeutic agents that target downstream molecules in the Hh signaling pathway, distal to SMO, are being developed [38].
Suppressor of Fused Protein
SUFU is an intracellular negative regulator of the Hh signaling pathway. It is a tumor suppressor protein. SUFU functions by modulating the activity of glioma transcription factors [48–51]. SUFU mutation (R299Q), as seen in our patient, has previously been described in other cancers [52].
Mutations of SUFU have also been discerned in patients with either idiopathic or nevoid basal cell carcinoma syndrome-associated medulloblastomas [53–61]. Less than one percent (3/351) of cutaneous melanomas in The Cancer Genome Atlas (TCGA) dataset have a SUFU mutation [62]. To our knowledge, our patient represents the initial observation of this unique Hh signaling pathway aberration in Merkel cell carcinoma.
The SUFU protein acts downstream of SMO in the Hh signaling pathway. Therefore, it is expected that targeted therapeutic intervention using an inhibitor to SMO, such as vismodegib, would not be effective for individuals whose tumors were associated with a SUFU mutation. Indeed, SMO inhibitors failed to stop the growth of tumors in SUFU-mutant mouse cancer models and tumor-derived xenografts from a medulloblastoma patient that harbored a SUFU suppressor alteration [2, 56, 63].
Therapies that target the Hh signaling pathway downstream of SUFU—affecting glial transcription factors—may be appropriate for individuals whose cancer harbors a SUFU mutation, such as our patient’s Merkel cell carcinoma [2, 36, 64–66]. These may include agents such as arsenic trioxide that inhibits glial transcription factors [67–69] and bromo and extra C-terminal (BET) inhibitors that result in downregulation of glial transcriptional activity by inhibiting the BET bromodomain-containing protein 4 [70–72]. Treatment with BET inhibitors—such as JQ1 and OTX015—have not only resulted in the inhibition SUFU-mutant medulloblastoma growth but also have shown clinical activity in the treatment of patients with hematologic malignancies (such as acute myelogenous leukemia and lymphoma), respectively [70–72].
Conclusion
Merkel cell carcinoma is an aggressive neuroendocrine tumor typically presenting on sun-exposed skin of older men. In addition to ultraviolet radiation, immunosuppression and Merkel cell polyomavirus have been associated with the development of Merkel cell carcinoma. Chromosomal and mutational analysis of Merkel cell carcinoma has demonstrated several aberrations; however, neither a unifying abnormality nor an alteration in the Hh signaling pathway has been discovered. Our patient’s metastatic Merkel cell carcinoma has a unique, previously undescribed, genomic aberration: a SUFU abnormality was the sole alteration detected using a next-generation sequencing-based assay. We hypothesize that his tumor will not respond to inhibitors of SMO, such as vismodegib, since these agents act on the Hh signaling pathway upstream of the patient’s SUFU abnormality. However, we speculate that his Merkel cell carcinoma may be successfully treated with therapy that targets the Hh signaling pathway downstream to SUFU protein—such as arsenic trioxide or BET inhibitors—that result in downregulation of glial transcription activity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Dr. Philip R. Cohen is the guarantor for this article, and takes responsibility for the integrity of the work as a whole. All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work as a whole, and have given final approval for the version to be published. No editorial assistance was used in the preparation of the manuscript. No funding or sponsorship was received for this study or publication of this article.
Conflict of interest
Dr. Philip R. Cohen and Dr. Razelle Kurzrock declare no conflict of interest. As a member of the Editorial Board (Philip R. Cohen, MD), the funding of the article publication charges is waived for 2 manuscripts per year.
Compliance with ethics guidelines
Informed consent was obtained from the patient for being included in the study and for the publication of photographs.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Contributor Information
Philip R. Cohen, Email: mitehead@gmail.com
Razelle Kurzrock, Email: rkurzrock@ucsd.edu.
References
- 1.Hughes MP, Hardee ME, Cornelius LA, Hutchins LF, Becker JC, Gao L. Merkel cell carcinoma: epidemiology, target, and therapy. Curr Dermatol Rep. 2014;3:46–53. doi: 10.1007/s13671-014-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 3.Park JY, Kricka LJ, Fortina P. Next-generation sequencing in the clinic. Nat Biotechnol. 2013;31:990–992. doi: 10.1038/nbt.2743. [DOI] [PubMed] [Google Scholar]
- 4.Foundation Medicine, Inc., Cambridge. http://www.foundationmedicine.com. Accessed Mar 25, 2015.
- 5.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, Sun J, Juhn F, Brennan K, Iwanik K, MAillet A, Buell J, White E, Zhao M, Balasubramanian S, Terzic S, Richards T, Banning V, Garcia L, Mahoney K, Zwirko Z, Donahue A, Beltran H, Mosquera JM, Rubin MA, Dogan S, Hedvat CV, Berger MF, Pusztai L, Lechner M, Boshoff C, Jarosz M, Vietz C, Parker A, Miller VA, Ross JS, Curran J, Cronin MT, Stephens PJ, Lipson D, Yelensky R. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. (PMID = 24142049). [DOI] [PMC free article] [PubMed]
- 6.Marek L, Grzanka A, Chmielowska E, Jankowski M, Schwartz RA, Czajkowski R. Merkel cell carcinoma: an illustrative case and review. Postepy Dermatol Alergol. 2014;31:325–328. doi: 10.5114/pdia.2014.40797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz JL, Wong SL, McLean SA, Hayman JA, Lao CD, Kozlow JH, Malloy KM, Bradford CR, Frohm ML, Fullen DR, Lowe L, Bichakjian CK. NCCN guidelines implementation in the multidisciplinary Merkel cell carcinoma program at the university of Michigan. J Natl Compr Cancer Netw. 2014;12:434–441. doi: 10.6004/jnccn.2014.0043. [DOI] [PubMed] [Google Scholar]
- 8.Raju S, Vazirnia A, Totri C, Hata TR. Treatment of Merkel cell carcinoma of the head and neck: a systematic review. Dermatol Surg. 2014;40:1273–1283. doi: 10.1097/DSS.0000000000000131. [DOI] [PubMed] [Google Scholar]
- 9.Erstad DJ, Cusack JC., Jr Mutational analysis of Merkel cell carcinoma. Cancers (Basal) 2014;6:2116–2136. doi: 10.3390/cancers6042116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Treglia G, Kakhki VR, Giovanella L, Sadeghi R. Diagnostic performance of fluorine-18-fluorodeoxyglucose positron emission tomography in patients with Merkel cell carcinoma: a systematic review and meta-analysis. Am J Clin Dermatol. 2013;14:437–447. doi: 10.1007/s40257-013-0040-x. [DOI] [PubMed] [Google Scholar]
- 11.Ibrahim SF, Ahronowitz I, McCalmont TH, Hernandez Pampaloni M, Ryan JL, Yu SS. 18F-flurodeoxyglucose positron emission tomography-computed tomography imaging in the management of Merkel cell carcinoma: a single-institution retrospective study. Dermatol Surg. 2013;39:1323–1333. doi: 10.1111/dsu.12246. [DOI] [PubMed] [Google Scholar]
- 12.Aldabagh B, Joo J, Yu SS. Merkel cell carcinoma: current status of targeted and future potential for immunotherapies. Semin Cutan Med Surg. 2014;33:76–82. doi: 10.12788/j.sder.0084. [DOI] [PubMed] [Google Scholar]
- 13.Patel RV, Frankel A, Goldenberg G. An update on nonmelanoma skin cancer. J Clin Aesthet Dermatol. 2011;4(2):20–27. [PMC free article] [PubMed] [Google Scholar]
- 14.Czapiewski P, Biernat W. Merkel cell carcinoma—recent advances in the biology, diagnositics and treatment. Int J Biochem Cell Biol. 2014;53:536–546. doi: 10.1016/j.biocel.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 15.Ma JE, Brewer JD. Merkel cell carcinoma in immunosuppressed patients. Cancers (Basel) 2014;6:1328–1350. doi: 10.3390/cancers6031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sastre-Garau X, Peter M, Avril M-F, Laude H, Couturier J, Rozenberg F, Almeida A, Boitier F, Carlotti A, Courturaud B, Dupin N. Merkel cell carcinoma of the skin: pathological and molecular evidence for a causative role of MCV in oncogenesis. J Pathol. 2009;218:48–56. doi: 10.1002/path.2532. [DOI] [PubMed] [Google Scholar]
- 17.Samimi M, Touze A. Merkel cell carcinoma: the first human cancer shown to be associated with a polyomavirus. Presse Med. 2014;43:e405–e411. doi: 10.1016/j.lpm.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Leroux-Kozal V, Leveque N, Brodard V, Lesage C, Dudez O, Makeieff M, Kanagaratnam L, Diebold M-D. Merkel cell carcinoma: histopathologic and prognostic features according to the immunohistochemical expression of Merkel cell polyomavirus Large T antigen correlated with viral load. Hum Pathol. 2015;46:443–453. doi: 10.1016/j.humpath.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Salehi-Vaziri M, Sadeghi F, Alamsi-Hashiani A, Haeri H, Monavari SH, Keyvani H. Merkel cell polyomavirus and human papillomavirus infections in cervical disease in Iranian women. Arch Virol. 2015. [Epub ahead of print]. (PMID = 25721299). [DOI] [PubMed]
- 20.Trizuljak J, Srovnal J, Plevova K, Brychtova Y, Semerad L, Bakesova D, Letalova E, Benedikova A, Mayer J, Hajduch M, Posposilova S, Doubek M. Analysis of prognostic significance of Merkel cell polyomavirus in chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2015. doi:10.1016/j.clml.2015.02.003 [Epub ahead of print]. (PMID = 25758600). [DOI] [PubMed]
- 21.Falchook GS, Rady P, Hymes S, Nguyen HP, Trying SK, Prieto VG, Hong DS, Kurzrock R. Merkel cell polyomavirus and HPV-17 associated with cutaneous squamous cell carcinoma arising in a patient with melanoma treated with the BRAF inhibitor dabrafenib. JAMA Dermatol. 2013;149:322–326. doi: 10.1001/jamadermatol.2013.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gormley RH, Kim EJ, Rook AH, Kovarik CL, Sun A, Rady P, Tyring SK. Merkel cell polyomavirus in low levels in folliculotropic mycosis fungoides represents a passenger, not a driver. Int J Dermatol. 2015. doi:10.1111/ijd.12754 [Epub ahead of print]. (PMID = 25661198). [DOI] [PubMed]
- 23.Du-Thanh A, Dereure O, Guillot B, Foulongne V. Merkel cell polyomavirus: its putative involvement in a particular subset of cutaneous lymphoma with possibly unfavorable outcome. J Clin Virol. 2014;61:161–165. doi: 10.1016/j.jcv.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Murakami I, Matsushita M, Iwasaki T, Kuwamoto S, Kato M, Horie Y, Hayashi K, Gogusev J, Jaubert F, Nakamoto S, Yamakawa H, Takata K, Oka T, Yoshino T. High viral load of Merkel cell polyomavirus DNA sequences in Langerhans cell sarcoma tissues. Infect Agent Cancer. 2014;9:15. doi:10.1186/1750-9378-9-15 (eCollections 2014). (PMID = 24834110). [DOI] [PMC free article] [PubMed]
- 25.Hourdequin KC, Lefferts JA, Brennick JB, Ernstoff MS, Tsongalis GJ, Pipas JM. Merkel cell polyomavirus and extrapulmonary small cell carcinoma. Oncol Lett. 2013;6:1049–1052. doi: 10.3892/ol.2013.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher CA, Harms PW, McHugh JB, Edwards PC, Siddiqui J, Palanisamy N, Bichakjian CK, Benavides E, Danciu TE. Small cell carcinoma in the parotid harboring Merkel cell polyomavirus. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;118:703–712. doi: 10.1016/j.oooo.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Xu S, Jiang J, Yu X, Sheng D, Zhu T, Jin M. Association of Merkel cell polyomavirus infection with EGFR mutation status in Chinese non-small cell lung cancer patients. Lung Cancer. 2014;83:341–346. doi: 10.1016/j.lungcan.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Iwasaki T, Matsushita M, Nonaka D, Kuwamoto S, Kato M, Murakami I, Nagata K, Nakajima H, Sano S, Hayashi K. Comparison of Akt/mTOR/4E-BP1 pathway signaling activation and mutations of PIK3CA in Merkel cell polyomavirus-negative carcinomas. Hum Pathol. 2015;46:210–216. doi: 10.1016/j.humpath.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Xie H, Liu T, Wang N, Bjornhagen V, Hoog A, Larsson C, Lui WO, Xu D. TERT promoter mutations and gene amplification: promoting TERT expression in Merkel cell carcinoma. Oncotarget. 2014;5:10048–10057. doi: 10.18632/oncotarget.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garneski KM, Lemos BD, Feng B, Jaimes N, Penas PF, Bi X, Maher E, Cohen L, Leonard JH, Granter SR, Chin L, Nghiem P. Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-Myc. J Invest Dermatol. 2009;129:1547–1555. doi: 10.1038/jid.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunner M, Thurnher D, Pammer J, Heiduschka G, Petzelbauer P, Schmid C, Schneider S, Erovic BM. Expression of hedgehog signaling molecules in Merkel cell carcinoma. Head Neck. 2010;32:333–340. doi: 10.1002/hed.21191. [DOI] [PubMed] [Google Scholar]
- 32.Jai Y, Wang Y, Xie J. The Hedgehog pathway: role in cell differentiation, polarity and proliferation. Arch Toxicol. 2015 [Epub ahead of print]. (PMID = 25559776). [DOI] [PMC free article] [PubMed]
- 33.Cowey CL. Targeted therapy for advanced basal-cell carcinoma: vismodegib and beyond. Dermatol Ther (Heidelb) 2013;3:17–31. doi: 10.1007/s13555-013-0019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyons TG, O’Kane GM, Kelly CM. Efficacy and safety of vismodegib: a new therapeutic agent in the treatment of basal cell carcinoma. Expert Opin Drug Saf. 2014;13:1125–1132. doi: 10.1517/14740338.2014.939952. [DOI] [PubMed] [Google Scholar]
- 35.Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Abidi A. Hedgehog signaling pathway: a novel target for cancer therapy: vismodegib, a promising therapeutic option in treatment of basal cell carcinomas. Indian J Pharmacol. 2014;46:3–12. doi: 10.4103/0253-7613.124884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Salas V, Alegre M, Lopez-Ferrer A, Garces JR. Vismodegib: a review. Actas Dermosifiliogr. 2014;105:744–751. doi: 10.1016/j.ad.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 38.McMillan R, Matsui W. Molecular pathways: the hedgehog signaling pathway in cancer. Clin Cancer Res. 2012;18:4883–4888. doi: 10.1158/1078-0432.CCR-11-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dlugosz AA, Talaz M. Following the hedgehog to new cancer therapies. N Engl J Med. 2008;361:1202–1205. doi: 10.1056/NEJMe0906092. [DOI] [PubMed] [Google Scholar]
- 40.Meiss F, Zeiser R. Vismodigib. Recent Results Cancer Res. 2014;201:405–417. doi: 10.1007/978-3-642-54490-3_25. [DOI] [PubMed] [Google Scholar]
- 41.Aditya S, Rattan A. Vismodegib: a smoothened inhibitor for the treatment of advanced basal cell carcinoma. Indian Dermatol Online J. 2013;4:365–368. doi: 10.4103/2229-5178.120685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harms KL, Dlugosz AA. Harnessing hedgehog for the treatment of basal cell carcinoma. JAMA Dermatol. 2013;149:607–608. doi: 10.1001/jamadermatol.2013.448. [DOI] [PubMed] [Google Scholar]
- 43.Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander PA, Marmur E, Rudin CM, Chang ALS, Low JA, Mackey HM, Yauch RL, Graham RA, Reddy JC, Hauschild A. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366:2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, LoRusso PM, Von Hoff DD, de Sauvage FJ, Low JA. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Von Hoff DD, LoRusso PM, Rudin CM, Redy JC, Yauch RL, Tibes R, Weiss GJ, Borad MJ, Hann CL, Brahmer JR, Mackey HM, Lum BL, Darbonne WC, Marsters JC, Jr, de Sauvage FJ, Low JA. N Engl J Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 46.Brinkhuizen T, Reinders MG, van Geel M, Hendriksen AJ, Paulussen AD, Winnepenninckx VJ, Keymeulen KB, Soetekouw PM, van Steensel MA, Mosterd K. Acquired resistance to the Hedgehog pathway inhibitor vismodegib due to smoothened mutations in treatment of locally advanced basal cell carcinoma. J Am Acad Dermatol. 2014;71:1005–1008. doi: 10.1016/j.jaad.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Atwood SX, Chang AL, Oro AE. Hedgehog pathway inhibition and the race against tumor evolution. J Cell Biol. 2012;199:193–197. doi: 10.1083/jcb.201207140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith MJ, Beetz C, Williams SG, Bhaskar SS, O’Sullivan J, Anderson B, Baly SB Urquhart JE, Bholah Z, Oudit D, Cheesman E, Kelsey A, McCabe MG, Newman WG, Evans DG. Germline mutations in SUFU cause Gorlin syndrome-associated childhood medulloblastoma and redefine the risk associated with PTCH1 mutations. J Clin Oncol. 2014;32:4155–4161. (PMID = 25403219). [DOI] [PubMed]
- 49.Lin C, Yao E, Wang K, Nozawa Y, Shimizu H, Johnson JR, Chen JN, Krogan NJ, Chuang PT. Regulation of Sufu activity by p66B and Mycbp provides new insight into vertebrate Hedgehog signaling. Genes Dev. 2014;28:2547–2563. doi: 10.1101/gad.249425.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cherry AL, Finta C, Karlstrom M, Jin Q, Schwend T, Astorga-Wells J, Zubarev RA, Del Campo M, Criswell AR, de Sanctis D, Jovine L, Toftgard R. Structural basis of SUFU-GLI interaction in human Hedgehog signaling regulation. Acta Crystallogr D Biol Crystallogr. 2013;69:2563–2579. doi: 10.1107/S0907444913028473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toster U, Finta C, Rahman MF, Shimokawa T, Zaphiropoulos PG. Novel mechanism of action on Hedgehog signaling by a suppressor of fused carboxy terminal variant. PLoS One. 2012;7:e37761. doi: 10.1371/journal.pone.0037761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VE, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 53.Taylor MD, Liu L, Raffel C, Hui C-C, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, Hao A, Goldstein AM, Theodora S, Scherer SW, Dura WT, Wainwright B, Squire JA, Rutka JT, Hogg D. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 54.Aaikko M, Li SP, Saarinen S, Alhopuro P, Kaasinen E, Morgunova E, Li Y, Vesanen K, Smith MJ, Evans DG, Poyhonen M, Kiuru A, Auvinen A, Aaltonen LA, Taipale J, Vahteristo P. Loss of SUFU function in familial multiple meningioma. Am J Hum Genet. 2012;91:520–526. doi: 10.1016/j.ajhg.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brugieres L, Remenieras A, Pierron G, Varlet P, Forget S, Byrde B, Bombled J, Puget S, Caron O, Dufour C, Delatte O, Bressac-de-Paillerets B, Grill J. High frequency of germline SUFU mutations in children with desmoplastic/nodular medulloblastoma younger than 3 years of age. J Clin Oncol. 2012;30:2087–2093. doi: 10.1200/JCO.2011.38.7258. [DOI] [PubMed] [Google Scholar]
- 56.Lee Y, Kawagoe R, Sasai K, Li Y, Russell HR, Curran T, McKinnon PJ. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26:6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 57.Heby-Henricson K, Bergstrom A, Rozell B, Toftgard R, Teglund S. Loss of Trp53 promotes medulloblastoma development but not skin tumorigenesis in Sufu heterozygous mutant mice. Mol Carcinog. 2012;51:754–760. doi: 10.1002/mc.20852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, Pomeroy SL, Korshunov A, Lichter P, Taylor MD, Pfister SM. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12:818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Cameiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, Sogoloff B, Stojanov P, Tamayo P, Mesirov JP, Amani V, Teider N, Sengupta S, Francois JP, Northcott PA, Taylor MD, Yu F, Crabtree GR, Kautzman AG, Gabriel SB, Getz G, Jager N, Jones DT, Lichter P, Pfister SM, Roberts TM, Meyerson M, Pomeroy SL, Cho YJ. Medulloblastomas exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. (PMID = 22820256). [DOI] [PMC free article] [PubMed]
- 60.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, Chalhoub N, Baker SJ, Huether R, Kriwacki R, Curley N, Thiruvenkatam R, Wang J, Wu G, Rusch M, Hong X, Becksfort J, Gupta P, Ma J, Easton J, Vadodaria B, Onar-Thomas A, Lin T, Li S, Pounds S, Paugh S, Zhao D, Kawauchi D, Roussel MF, Finkelstein D, Ellison DW, Lau CC, Bouffet E, Hassall T, Gurarangan S, Cohn R, Fulton RS, Fulton LL, Dooling DJ, Ochoa K, Gajjar A, Mardis ER, Wilson RK, Downing JR, Zhang J, Gilbertson RJ. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. (PMID = 22722829). [DOI] [PMC free article] [PubMed]
- 61.Jones DT, Jager n, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Hovestadt V, Stutz AM, Rausch T, Warnatz HJ, Ryzhova M, Bender S Strum D, Pleier S, Cin H, Pfaff E, Seiber L, Wittmann A, Remke M, Witt H, Hutter S, Tzaridis T, Weischenfeldt J, Raeder B, Avci M, Amstislavskiy V, Zapatka M, Weber UD, Wang Q, Lasitschka B, Bartholomae CC, Schmidt M, von Kalle C, Ast V, Lawrenz C, Eils J, Kabbe R, Benes V, van Sluis P, Koster J, Volckmann R, Shih D, Betts MJ, Russell RB, Coco S, Tonini GP, Schuller U, Hans V, Graf N, Kim YJ, Monoranu C, Roggendorf W, Unterberg A, Herold-Mende C, Milde T, Kuozik AE, von Deimling A, Witt O, Maass E, Rossler J, Ebinger M, Schuhmann MU, Fruhwald MC, Hasselblatt M, Jabado N, Rutkowski S, von Bueren AO, Williamson D, Clifford SC, MCabe MG, Collins VP, Wolf S, Wiemann S, Lehrach H, Brors B, Scheurlen W, Felsberg J, Reifenberger G, Northcott PA, Taylor MD, Meyerson M, Pomeroy SL, Yaspo ML, Korbel JO, Korshunov A, Eils R, Pfister SM, Lichter P, ICGC PedBrain Tumor Project. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. (PMID = 22832583). [DOI] [PMC free article] [PubMed]
- 62.The Cancer Genome Atlas Data Portal. http://tcga-data.nci.nih.gov/tcga/. Accessed Dec 21, 2014.
- 63.Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, Milde T, Bourdeaut F, Ryzhova M, Strum D, Pfaff E, Stark S, Hutter S, Seker-Cin H, Johann P, Bender S, Schmidt C, Rausch T, Shih D, Reimand J, Sieber L, Wittmann A, Linke L, Witt H, Weber UD, Zapatka M, Konig R, Beroukim R, Bergthold G, van Sluis P, Volckmann R, Koster J, Versteeg R, Schmidt S, Wolf S, Lawerenz C, Bartholomae CC, von Kalle C, Unterberg A, Herold-Mende C, Hofer S, Kulozik AE, von Deimlig A, Scheurten W, Felsberg J, Reifenberger G, Hasselblatt M, Crawford JR, Grant GA, Jabado N, Perry A, Cowdrey C, Croul S, Zadeh G, Korbel JO, Doz F, Delattre O, Bader GD, McCabe MG, Collins VP, Kieran MW, Cho YJ, Pomeroy SL, Witt O, Brors B, Taylor MD, Schuller U, Korshunov A, Eils R, Wechsler-Reya RJ, Lichter P, Pfister SM, ICGC PedBrain Tumor Project. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25:393–405. (PMID = 24651015). [DOI] [PMC free article] [PubMed]
- 64.Ruch JM, Kim EJ. Hedgehog signaling pathway and cancer therapeutics: progress to date. Drugs. 2013;73:613–623. doi: 10.1007/s40265-013-0045-z. [DOI] [PubMed] [Google Scholar]
- 65.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, Sun M, Rack PG, Sinha S, Wu JJ, Solow-Cordero DE, Jiang J, Rowitch DH, Chen JK. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci. 2009;106:14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J, Aftab BT, Tang JY, Kim D, Lee AH, Rezaee M, Kim J, Chen B, King EM, Borodovsky A, Riggins GJ, Epstein EH, Jr, Beachy PA, Rudin CM. Itraconazole an arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer Cell. 2013;23:23–34. doi: 10.1016/j.ccr.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beauchamp EM, Ringer L, Bulut G, Saiwan KP, Hall MD, Lee YC, Peaceman D, Ozdemirli M, Rodriguez O, Macdonald TJ, Albanese C, Toretsky JA, Uren A. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J Clin Invest. 2011;121:148–160. doi: 10.1172/JCI42874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J, Lee JJ, Kim J, Gardner D, Beachy PA. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc Natl Acad Sci. 2010;107:13432–13437. doi: 10.1073/pnas.1006822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang Y, Gholamin S, Schubert S, Willardson M, Lee A, Bandopadhayay P, Bergthold G, Masoud S, Nguyen B, Vue N, Balansay B, Yu F, Oh S, Woo P, Chen S, Ponnuswami A, Monje M, Atwood SX, Whitson RJ, Mitra S, Cheshier SH, Qi J, Beroukhim R, Tang JY, Wechsler-Reva R, Oro AE, Link BA, Bradner JE, Cho YJ. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20:732–740. doi: 10.1038/nm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dombret H, Preudhomme C, Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roco C, Ethell M, Yee K, Bourdel F, Herait P Michallet M, Recher C, Roumier C, Quesnel B. A phase 1 study of the BET-bromodomain inhibitor OTX015 in patients with advanced acute leukemia [Abstract 117]. In: 56th ASH Annual Meeting and Exposition. San Francisco. 2014. https://ash.confex.com/ash/2014/webprogram/Paper70684.html. Accessed Jan 24, 2015.
- 72.Thieblemont C, Stathis A, Inghirami G, Korlin L, Morschhauser F, Gleeson M, Broussais F, Amorim S, Salles G, Facon T, Cunningham D, Vey N, Bourdel F, Herait P, Zucca E. A phase 1 study of the BET-bromodomain inhibitor OTX015 in patients with non-leukemic hematologic malignancies [Abstract 4417]. In: 56th ASH Annual Meeting and Exposition. San Francisco. 2014. https://ash.confex.com/ash/2014/webprogram/Paper70510.html. Accessed Jan 24, 2015.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


