Abstract
Prospective analytic study was conducted in NICUs of three Egyptian Neonatal Network (EGNN) participants in Mansoura Hospitals in Egypt over a period of 18 months from March 2011 to August 2012. By using EGNN 28-day discharge form, all demographic, clinical, and laboratory data were recorded and studied. During the study period, 357 neonates were diagnosed as suspected sepsis with an incidence of 45.9% (357/778) among the admitted neonates at the three neonatal intensive care units. 344 neonates (sex ratio = 1.3:1) were enrolled in the study in which 152 (44.2%) were classified as early onset sepsis EOS (≤72 hr) and 192 (55.8%) as late onset sepsis LOS (>72 hr). Among the LOS cases, 33.9% (65/192) were caused by nosocomial infections. In 40.7% (140/344), sepsis was confirmed by positive blood culture. The total mortality rate for the proven neonatal sepsis was 51% (25/49) and 42.9% (39/91) for EOS and LOS, respectively. Coagulase negative staphylococci were predominant isolates in both EOS and LOS, followed by Klebsiella pneumoniae. Most of the bacterial isolates had low sensitivity to the commonly used empiric antibiotics. However, 70.1% (89/127) exhibited multidrug resistance. Best sensitivities among Gram-positive isolates were found against imipenem, ciprofloxacin, vancomycin, and amikacin.
1. Introduction
Globally, sepsis is still one of the major causes of morbidity and mortality in neonates, in spite of recent advances in health care units [1]. More than 40% of under-five deaths globally occur in the neonatal period, resulting in 3.1 million newborn deaths each year [2]. The majority of these deaths usually occur in low-income countries and almost 1 million of these deaths are attributed to infectious causes including neonatal sepsis, meningitis, and pneumonia [3]. On the other hand, the survivors of neonatal sepsis are vulnerable to short- and long-term neurodevelopmental morbidity [4–6].
Neonatal sepsis is defined as a clinical syndrome in an infant 28 days of life or younger, manifested by systemic signs of infection and isolation of a bacterial pathogen from the bloodstream [7]. Diagnosis and management of sepsis are a great challenge facing neonatologists in NICUs. Clinical diagnosis of presentation is difficult due to nonspecific signs and symptoms. In addition, laboratory diagnosis is time consuming. This matter necessitates the initiation of empirical antibiotic therapy till the suspected sepsis is ruled out. At the same time, increased multidrug resistant organisms make the treatment options fewer and the effective treatment is delayed [8].
Neonatal sepsis is caused by Gram-positive and Gram-negative bacteria and Candida [9]. The diversity of organisms causing sepsis varies from region to another and changes over time even in the same place [10, 11]. This is attributed to the changing pattern of antibiotic use and changes in lifestyle. Many factors contribute to the susceptibility of the neonate to sepsis, which can influence the incidence of neonatal sepsis. Incidence also varies from nursery to nursery depending on conditions predisposing infants to infection [9, 12].
The aim of the present study was to evaluate the incidence of neonatal sepsis and characterize the microbiological pattern of neonatal sepsis and the antibiotic susceptibility of the isolates to evaluate the empirical antibiotic used in neonatal units of three referral hospitals in Mansoura, Egypt.
2. Materials and Methods
2.1. Study Design and Population
This study was prospectively conducted over a period of 18 months between March 2011 and August 2012, at three NICUs in Mansoura City, Egypt, namely, Mansoura University Children Hospital (MUCH), Health Insurance Hospital (HIH), and Mansoura General Hospital (MGH). During the study period, all admitted neonates with clinical signs and symptoms of sepsis at the time of admission or who developed sepsis during their hospital stay were assessed using EGNN sepsis screening tool and included in the study.
2.2. Patient Data
Using EGNN guidelines, a standard structured data collection form was designed to obtain social demographic, clinical, and laboratory data that were recorded by qualified medical staff. All neonates were subjected to full clinical examination stressing on gestational age, birth weight, mode of delivery, and risk factors for sepsis: premature rupture of membranes (PROM), maternal fever, insertion of an umbilical catheter, and so forth.
According to the Egyptian Neonatal Network (EGNN), sepsis is defined as presence of at least 3 out of the following four criteria [13]:
presence of risk factors of sepsis (e.g., prematurity, chorioamnionitis),
presence of two or more clinical signs of sepsis (poor reflexes, lethargy, respiratory distress, bradycardia, apnea, convulsions, abdominal distension, and bleeding),
abnormal hemogram and positive CRP and positive culture.
Patient receives antibiotics and antifungal for at least five days (or <5 days if he is transferred or died before completion of these five days).
According to the infant age, at the onset of symptoms, neonates were classified into two groups: EoNS (≤72 hours of life) and LoNS (>72 hours of life) [14].
2.3. Nosocomial Infection
It was defined by Standard Center for Disease Control and Prevention [15] as an infection acquired during hospitalization (>48 hrs) and resulted from an organism inoculation that was not present in the patient at the time of admission [15, 16], excluding cases of early-onset sepsis.
2.4. Collection of Specimens
Blood samples were collected from the neonates with suspected sepsis for CRP, CBC, and blood cultures. Blood was collected from a peripheral vein. Approximately 1 mL of blood was inoculated directly into blood culture medium vials and sent to our clinical microbiology laboratory for cultivation and subsequent processing.
2.5. Processing of Specimens
The blood cultures were incubated aerobically at 37°C and observed daily for the first 3 days for the presence of visible microbial growth by one of the following: haemolysis, air bubbles (gas production), and coagulation of broth. At the same time, subcultures were made during 3 successive days on enriched and selective media including blood, chocolate, MacConkey, and mannitol salt agar plates and examined for growth after 24–48 hours of incubation. The same protocol was repeated until the 7th day before blood culture was considered to be free of microorganisms. Isolates obtained were identified by standard microbiological techniques, namely, Gram staining, colony characteristics, and biochemical properties including catalase, coagulase (free and bound), DNase production, growth on mannitol salt agar, and hemolytic activity on blood agar plates for Gram-positive isolates, and triple sugar iron (TSI), motility, indole, citrate utilization, urease, oxidase and hydrogen sulphide production, Voges-Proskauer (VP) test, and growth on cetrimide agar for Gram-negative bacilli [17]. API 20E identification kits (bioMe rieux) were also used to confirm the identification of Gram-negative isolates and the results were read using API 32 GN reader. Candida isolates were confirmed by growth on Sabouraud media.
2.6. Identification of Staphylococci Species
Staphylococcus species were identified by PCR-Restriction Fragment Length Polymorphism of gap Gene, using AluI as restriction enzyme, resulting in a distinctive RFLP pattern for every species as previously described [18].
2.7. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing of all bacterial isolates was performed by the Kirby-Bauer disc diffusion method on Mueller-Hinton agar (Oxoid) according to the recommendations of the CLSI (2010). The antibiotics tested were ampicillin (10 μg), oxacillin (1 μg), amoxicillin-clavulanic acid (30 μg), cefoxitin (30 μg), cefotaxime (30 μg), ceftriaxone (30 μg), ceftazidime (30 μg), imipenem (10 μg), vancomycin (30 μg), gentamicin (10 μg), amikacin (30 μg), erythromycin (15 μg), azithromycin (15 μg), ciprofloxacin (5 μg), and norfloxacin (10 μg).
Multidrug Resistant (MDR) Bacteria. They were defined by resistance to three or more antimicrobial classes [19].
2.8. Statistical Analysis
Summary of measures was reported as mean ± standard deviation (SD) for quantitative variables and percentages for categorical variables. The differences in distribution were evaluated using the chi-square test for categorical variables. P value ≤ 0.05 was considered statistically significant. All the statistical analyses were performed using GraphPad InStat version 3.05.
3. Results
3.1. Studied Population
During the study period, a total of 357 neonates with suspected cases of sepsis were enrolled. Thirteen cases were excluded: eleven blood cultures were lost and the other two cultures were contaminated. As a result, the final number subjected for the report was 344. The incidence of suspected neonatal sepsis among the admitted neonates at the neonatal intensive care units of the three included hospitals during the study period was 45.9% (357/778). Among the studied neonates, sepsis was recognized as EOS in 152 (44.2%) cases and as LOS in 192 (55.8%) cases according to infant age at the onset of symptoms. 33.9% (65/192) of LOS were due to nosocomial infection (Table 1).
Table 1.
Age and sex distribution among 344 neonates with suspected sepsis at Mansoura hospitals.
| Category | P value | |||
|---|---|---|---|---|
| Neonates with EOS (≤72 hr) number (%) | Neonates with LOS (>72 hr) number (%) | Total number (%) | ||
| Total | 152 (44.19) | 192 (55.81) | 344 | |
| Blood culture results | ||||
| Proven sepsis | 49 (32.2) | 91 (47.4) | 140 (40.7) | 0.0063 (<0.05) |
| Possible sepsis | 103 (67.8) | 101 (52.6) | 204 (59.3) | |
| Sex | ||||
| Male | 93 (61.2) | 102 (53.1) | 195 (56.7) | 0.1650 (>0.05) |
| Female | 59 (38.8) | 90 (46.9) | 149 (43.3) | |
EOS: early-onset sepsis, LOS: late-onset sepsis.
The sepsis was proved in 140 (40.7%) cases by positive blood culture: 49 from early-onset and 91 from late-onset sepsis. There was a significant difference in the positivity rate between EOS and LOS groups (P < 0.05).
Among the studied neonates, 195 (56.7%) were males and 149 (43.3%) were females resulting in an overall male to female ratio of 1.3 : 1. However, no significant difference was detected with regard to sex (P > 0.05). The total mortality rate for the confirmed neonatal sepsis was estimated as 51% for EOS and 42.9% for LOS.
3.2. Maternal and Neonatal Characteristics and Clinical Features
Maternal and demographic data and clinical information were available for 304 patients (88.4%) as shown in Tables 2 and 3. Of the 304 neonates, 179 (58.9%) were preterm. 296 (97.4%) were born in the health care facilities (hospitals/clinics) and 8 (2.6%) were born at home. Referring to the delivery mode, 212 (69.7%) were delivered by caesarean section whereas 92 (30.3%) were delivered vaginally. Approximately, 212 (69.7%) neonates with sepsis had low birth weight (<2500 g), and out of these 81 (38.2%) had very low birth weight (<1500 g). The mean gestational age and birth weight of the study population were 34.4 ± 3.8 weeks and 2124 ± 828 grams, respectively. The most prevalent clinical feature was respiratory distress (41.3%).
Table 2.
Maternal and neonatal data of the 304 neonates investigated for sepsis at Mansoura hospitals.
| Characteristics | Total (n = 304) number (%) |
|---|---|
| (I) Maternal data | |
| Gestational age | |
| ≤33 weeks (preterm) | 108 (35.5) |
| 34–36 weeks (late preterm) | 71 (23.4) |
| ≥37 weeks (term) | 125 (41.1) |
| Place of delivery | |
| Hospital | 230 (75.7) |
| Clinic | 66 (21.7) |
| Home | 8 (2.6) |
| Mode of delivery | |
| Vaginal | 92 (30.3) |
| Caesarean section | 212 (69.7) |
|
| |
| (II) Neonatal data | |
| Weight at birth | |
| ≤1000 g (VLBW) | 21 (6.9) |
| 1001–1500 g (VLBW) | 60 (19.7) |
| 1501–2500 g (LBW) | 131 (43.1) |
| >2500 g | 92 (30.3) |
VLBW: very low birth weight, LBW: low birth weight.
Table 3.
Clinical signs/accompanied diagnoses among neonates with suspected sepsis at Mansoura hospitals.
| Clinical signs/accompanied diagnoses | Total (n = 304) |
|---|---|
| Respiratory distress | 142 |
| Pneumonia | 24 |
| Temperature instability | 7 |
| Convulsions | 5 |
| Hypoglycemia | 4 |
| Fetal distress | 4 |
| Meningitis | 9 |
| Surgical problems | |
| Diaphragmatic hernia without obstruction or gangrene | 3 |
| Esophageal atresia/TEF | 11 |
| Choanal atresia | 1 |
| Gastroschisis | 1 |
| Obstruction of duodenum | 1 |
| Congenital heart disease | 11 |
| Diseases of genitourinary | 5 |
| Cardiovascular collapse (shock) | 3 |
| Hematological symptoms (purpura/DIC) | 2 |
| Hypotonia/poor activities | 4 |
| Neonatal jaundice | 15 |
| Septic arthritis | 1 |
The neonate could have more than one of the above clinical findings.
3.3. Other Investigations
3.3.1. C-Reactive Protein Results
Among the 344 neonates admitted with suspected cases of sepsis, the CRP level was measured in 326 cases where it was positive (>6 mg/L) in 278 (85.3%) cases.
3.3.2. Hematological Parameters (White Blood Cell and Platelet Counts)
CBC was determined in 319 cases. Abnormalities in the CBC were found in 213 (66.8%) neonates with 22 (6.9%) having leucopenia (WBC < 5,000/mm3), 71 (22.3%) leukocytosis (WBC > 20,000/mm3), 74 (23.2%) neutropenia, and 145 (45.5%) thrombocytopenia (platelets < 140,000/mm3).
3.3.3. Isolated Pathogens
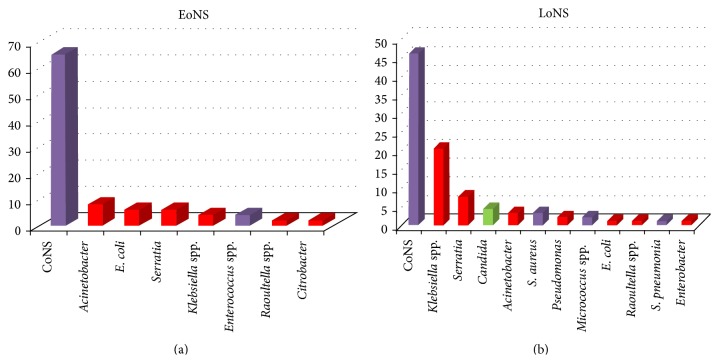
Out of the 344 blood cultures, only 140 (40.7%) showed growth of different bacteria and fungi. The type and frequency of isolated pathogen in relation to the type of sepsis were shown in Table 4 and Figure 1. Gram-positive bacteria were responsible for most cases of neonatal sepsis. Coagulase negative staphylococci (CoNS) were the most frequent isolated pathogens in EoNS and LoNS, followed by Klebsiella pneumoniae and Serratia marcescens.
Table 4.
Microbiological profile found in positive blood cultures from neonates with early- and late-onset sepsis.
| Isolated microorganism | Total (%) | EoNS number (%) | LoNS number (%) |
|---|---|---|---|
| Gram-positive bacteria | 82 (58.57) | 34 (69.39) | 48 (52.75) |
| Staphylococcus aureus | 3 (2.14) | — | 3 (3.30) |
| Coagulase negative staphylococci | 74 (52.86) | 32 (65.31) | 42 (46.15) |
| Streptococcus pneumoniae | 1 (0.71) | — | 1 (1.10) |
| Enterococcus faecalis | 2 (1.43) | 2 (4.08) | — |
| Micrococci spp. | 2 (1.43) | — | 2 (2.20) |
| Gram-negative bacteria | 54 (38.57) | 15 (30.61) | 39 (42.86) |
| Enterobacteriaceae | |||
| Escherichia coli ∗ | 4 (2.86) | 3 (6.12)∗ | 1 (1.10) |
| Klebsiella pneumoniae | 20 (14.29) | 2 (4.08) | 18 (19.78) |
| Klebsiella oxytoca | 1 (0.71) | — | 1 (1.10) |
| Raoultella spp. | 2 (1.43) | 1 (2.04) | 1 (1.10) |
| Enterobacter cloacae | 1 (0.71) | — | 1 (1.10) |
| Citrobacter freundii | 1 (0.71) | 1 (2.04) | — |
| Serratia marcescens | 10 (7.14) | 3 (6.12) | 7 (7.69) |
| Other Gram-negative bacilli | |||
| Acinetobacter baumannii | 7 (5.00) | 4 (8.16) | 3 (3.30) |
| Pseudomonas aeruginosa | 2 (1.43) | — | 2 (2.20) |
| Not identified | 6 | 1 | 5 |
| Fungi | |||
| Candida spp. | 4 (2.86) | — | 4 (4.40) |
| Total | 140 | 49 (34.75) | 91 (65.00) |
∗Two isolates of E. coli were metabolically inactive E. coli.
Figure 1.
Microbiological profile found in positive blood cultures from neonates with EoNS (a) and LoNS (b).
3.3.4. Antibiotic Susceptibility Pattern
The sensitivity patterns of the bacterial isolates to first- and second-line empiric antibiotics commonly used in neonatal infection were illustrated in Tables 5 and 6 and Figures 2, 3, and 4. Quinolones (ciprofloxacin) are not recommended for use in young children, but they may be used in culture-proven sepsis with bacteria resistant to other antibiotics.
Table 5.
Distribution of bacterial isolates according to the global sensitivities.
| Antibiotics | Global resistances (%) | Gram-positive cocci resistances (%) (n = 73) | Gram-negative resistances (%) (n = 54) |
|---|---|---|---|
| Ampicillin | 122 (96.06) | 70 (95.89) | 52 (96.30) |
| Oxacillin | — | 64 (87.67) | NT |
| Amoxicillin-clavulanic acid | 86 (67.72) | 37 (50.68) | 49 (90.74) |
| Cefoxitin | 106 (83.46) | 64 (87.67) | 42 (77.78) |
| Cefotaxime | 92 (72.44) | 43 (58.90) | 49 (90.74) |
| Ceftriaxone | 92 (72.44) | 43 (58.90) | 49 (90.74) |
| Ceftazidime | 115 (90.55) | 69 (94.52) | 46 (85.19) |
| Imipenem | 28 (22.05) | 12 (16.44) | 16 (29.63) |
|
| |||
| Vancomycin | — | 0 (0) | NT |
|
| |||
| Gentamicin | 78 (61.42) | 42 (57.53) | 36 (66.67) |
| Amikacin | 50 (39.37) | 13 (17.81) | 37 (68.52) |
|
| |||
| Erythromycin | — | 45 (61.64) | NT |
| Azithromycin | — | 45 (61.64) | NT |
|
| |||
| Ciprofloxacin | 43 (33.86) | 22 (30.14) | 21 (38.89) |
| Norfloxacin | 44 (34.65) | 22 (30.14) | 22 (40.74) |
NT: not tested.
Table 6.
Comparative resistance percentage of Gram-negative bacteria to different antimicrobial agents.
| Etiologic agents | Beta-lactams | Amino-glycosides | Quinolones | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillins | Cephalosporins | Carbapenem | ||||||||||
| AMP | AMC | FOX | CTX | CRO | CAZ | IPM | CN | AK | CIP | NOR | ||
| Klebsiella species | n = 21 | 100 | 95.24 | 66.67 | 95.24 | 95.24 | 95.24 | 23.81 | 61.90 | 66.67 | 38.10 | 42.86 |
| Serratia marcescens | n = 10 | 100 | 100 | 80 | 100 | 100 | 70 | 0 | 70 | 100 | 0 | 0 |
| Acinetobacter baumannii | n = 7 | 100 | 100 | 100 | 100 | 100 | 100 | 71.43 | 100 | 71.43 | 100 | 100 |
| Escherichia coli | n = 4 | 100 | 50 | 50 | 75 | 75 | 75 | 0 | 50 | 0 | 25 | 25 |
| Pseudomonas aeruginosa | n = 2 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Raoultella species | n = 2 | 100 | 100 | 100 | 100 | 100 | 100 | 50 | 100 | 100 | 100 | 100 |
| Enterobacter cloacae | n = 1 | 100 | 100 | 100 | 100 | 100 | 100 | —∗ | 0 | 100 | 0 | 0 |
| Citrobacter freundii | n = 1 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
∗Imipenem had an intermediate effect on this isolate.
Figure 2.
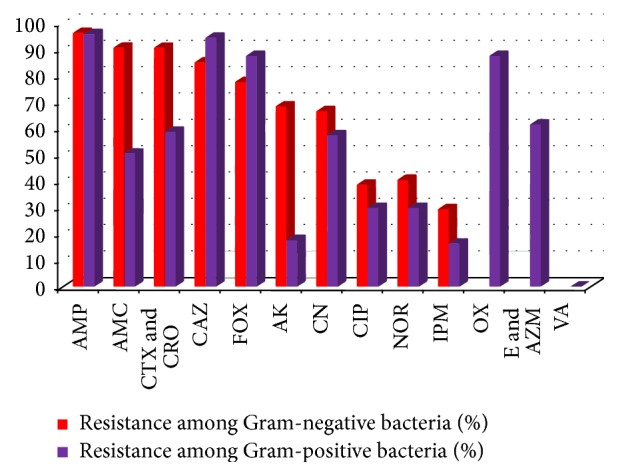
Comparative percentage of resistance to the tested antimicrobial agents among Gram-negative isolates and Gram-positive isolates.
Figure 3.
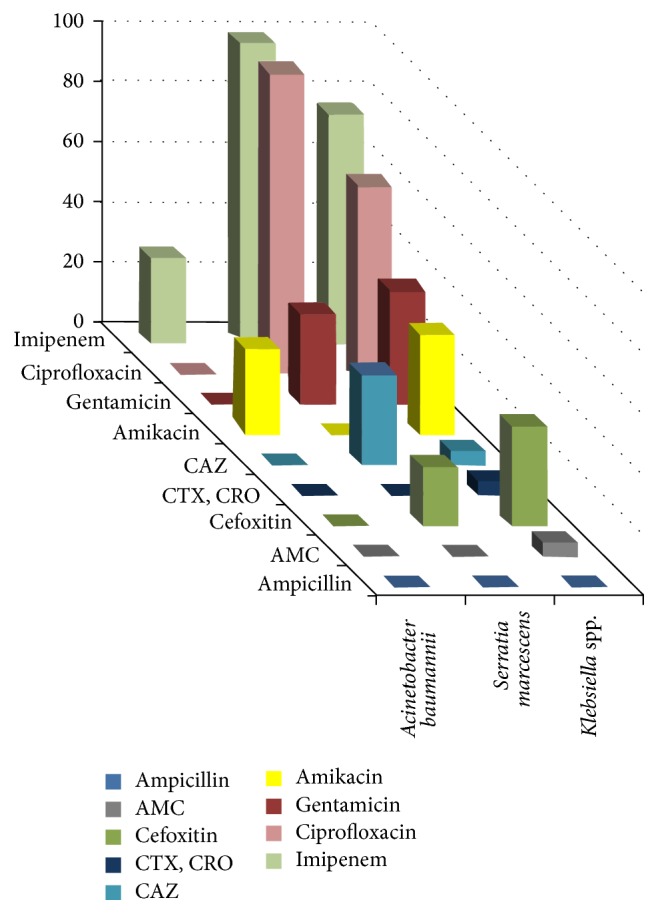
Comparative sensitivities of different Gram-negative bacteria to different antimicrobial agents.
Figure 4.
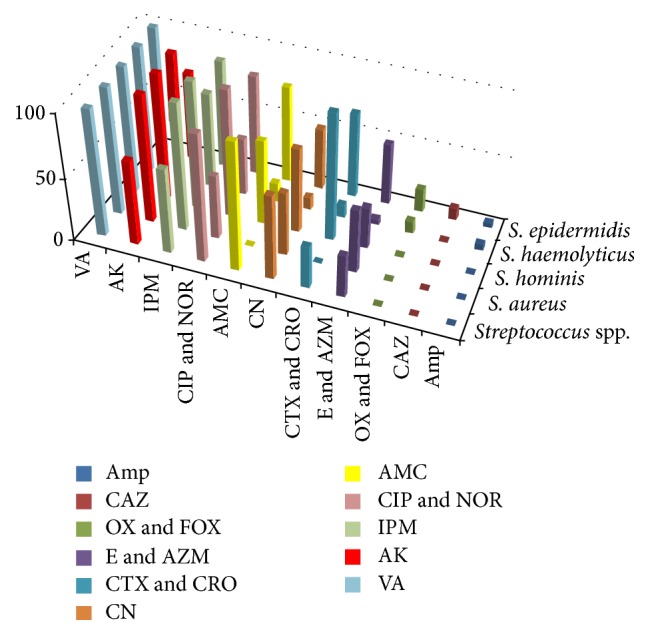
Comparative sensitivities of different Gram-positive bacteria to different antimicrobial agents.
Gram-Positive Bacteria. They showed high resistance to ampicillin (95.9%). The intermediate effect was observed (50.7%) with amoxicillin-clavulanic acid. In contrast to gentamicin, amikacin was highly effective on Gram-positive isolates. Best sensitivity was also observed to imipenem and ciprofloxacin. All isolates were sensitive to vancomycin. Among the Staphylococci spp., S. haemolyticus isolates were highly resistant.
Gram-Negative Bacteria. They were highly resistant to the first- and second-line empiric antibiotics: ampicillin (96.3%), amoxicillin-clavulanic acid (90.7%), gentamicin (66.7%) and amikacin (68.5%), and 3rd generation cephalosporins (>85%). Best sensitivity was observed to imipenem and ciprofloxacin. The effect of the tested antimicrobial agents was variable according to the genus as illustrated in Table 6 and Figure 3. It was found that imipenem and ciprofloxacin had a strong effect on Serratia isolates followed by Klebsiella isolates, whereas Acinetobacter isolates were resistant to all antimicrobial agents except imipenem and amikacin on a small number.
3.3.5. Multidrug Resistance (MDR)
MDR was observed in 89 isolates (70.1%). Among the Gram-positive isolates, 53.4% (39/73) were multidrug resistant while, among Gram-negative isolates, MDR was detected in 92.6% (50/54).
4. Discussion
The clinical signs and symptoms of neonatal sepsis are subtle and nonspecific, making its early diagnosis difficult, and it can interfere with other life-threatening diseases, such as necrotizing enterocolitis and perinatal asphyxia [20, 21]. Blood culture is still the gold standard for definitive diagnosis of neonatal sepsis, in spite of some drawbacks of blood cultures as being time consuming, low sensitivity, and possible contamination especially with commensal CoNS that could be produced.
In our study, the incidence of suspected neonatal sepsis during the study period was 45.9% with a mortality rate of 51% for proven EOS and 42.9% for proven LOS. Similar high rates were previously reported in Egypt [22] and other developing countries such as Tanzania 39% [23] and Cameroon 34.7% [24]. In contrast, very low rates were reported in the developed countries [25], which can be explained by the high quality of life and high standard measures of health care and hospital services in these countries.
During the study period, 344 neonates with suspected neonatal sepsis (using clinical criteria) were enrolled. Only 140 (40.7%) were confirmed to have bloodstream infection by using blood culture. This rate is comparable to rates reported in other developing African and Asian countries as Bangladesh (34.88%) [26], Uganda (37%) [27], Ethiopia (44.7%) [28], and Nigeria (45.9%) [29]. However, negative blood culture does not exclude sepsis as about 26% of all neonatal sepsis could be due to anaerobes [30]. Furthermore, the etiological agent may not be isolated by media used in our study such as viral (e.g., rubella, cytomegalovirus), protozoal (e.g., Toxoplasma gondii), and treponemal (e.g., Treponema pallidum) pathogens.
Among the studied neonates, LOS (55.8%) was more common than EOS (44.2%), which is in agreement with reports from other African and Asian countries [23, 31, 32]. However, the opposite was documented in some previous reports [33–35].
In our study, the incidence of sepsis was higher in neonates born via CS than in those born via VD. This finding is similar to other previous studies [33, 36, 37]. For example, in the study of Utomo et al. (2010), in Indonesia (Surabaya), it was reported that infants delivered via CS have 1.89 times higher risk to develop sepsis than noncaesarean.
In our study, the incidence of neonatal sepsis in both EOS and LOS was predominantly associated with Gram-positive cocci, specifically CoNS compared to Gram-negative and Candida spp. Similar findings were obtained in other studies in Egypt [38] and other different countries (including China, Mexico, South Africa, and Kenya) [25, 31, 39–41]. High rates of CoNS infections were reported in the Middle East, Southeast Asia, and Latin America [42]. In some studies, CoNS were more common to cause LOS. However, true EOS caused by CoNS was proved in other studies [43, 44]. On the contrary, Gram-negative neonatal sepsis was predominant in other studies [24, 28, 33–35, 45–48].
The extensive use of invasive devices for caring for the immunologically immature neonates especially preterm and LBW is the main cause of CoNS bacteremia in NICU. This finding is supported by the study of Kerur et al., as, in more than 50% of the cases of CoNS bacteremia in NICU, the infection could be correlated with the use of venous catheters.
Despite the importance and role of CoNS as etiological agents of neonatal sepsis as proved in many studies, determination of the identity of CoNS isolates whether being true pathogens or contaminants is still problematic. In our study, S. epidermidis was the most frequently recovered CoNS isolate in blood cultures, followed by S. haemolyticus. These two species were present at 55% and 33.3% in blood cultures, respectively. Similar findings were reported in other previous studies [25, 49, 50].
Gram-negative bacteria were the 2nd cause of neonatal sepsis especially LOS following CoNS, with increased mortality rate. Klebsiella spp. (15%), mainly K. pneumoniae, were the most predominant Gram-negative pathogen, followed by Serratia marcescens (7.14%) and Acinetobacter baumannii (5%). In our study, Klebsiella isolates were responsible for 4.08% and 20.88% of EOS and LOS, respectively. Other Gram-negative bacilli were recovered but in a few numbers. The predominance of Klebsiella among the causative Gram-negative pathogens was also reported in other studies in Egypt [45, 48] and other different countries [24, 25, 39, 40, 46, 51]. On the contrary, other Gram-negative bacteria such as E. coli [35, 47, 52], P. aeruginosa [34, 53], and Enterobacter spp. [33] could be identified as the most common Gram-negative isolates associated with neonatal sepsis.
Serratia marcescens was the 2nd higher Gram-negative isolate. This was in consistency with a study in Bangladesh in which S. marcescens was the 2nd Gram-negative isolate following K. pneumoniae with a rate of 18.27% [54]. Moreover, in another study in Europe, S. marcescens caused 5% of neonatal bloodstream infections in NICU [55]. However, in other studies S. marcescens could not be detected among pathogens isolated from cases of neonatal sepsis [25, 31, 39]. In the last decade, several outbreaks in NICU were documented [56–58], causing potentially fatal sepsis, meningitis, or pneumonitis in very premature or low birth weight neonates with high mortality rate.
Acinetobacter baumannii was isolated in our study from 5% of positive blood cultures of septic neonates accounting for 8.16% and 3.30% in EOS and LOS, respectively. Similarly, septic neonates infection ranging from 3.5% up to 7.7% has been previously recognized [24, 35, 36, 42, 46].
Candida spp. were isolated only in 4 cases (2.86%) causing LOS; two were born preterm, a known risk factor for candidemia [59]. Similar findings were found in other studies in Kenya (2.41%) [40] and India (2.63%) [36].
Ampicillin and aminoglycosides (mainly gentamicin) are the first-line empirical antibiotics used in our NICUs. Quinolones (ciprofloxacin) are not recommended for use in young children. However, they may be used in culture-proven sepsis with bacteria resistant to other antibiotics. For this reason such sensitivities were tested.
Among the Gram-negative isolates, all Klebsiella pneumoniae, Serratia marcescens, and Acinetobacter baumannii isolates were resistant to ampicillin, amoxicillin-clavulanic acid, cefotaxime, and ceftriaxone. Ceftazidime was only effective against 30% of all Serratia isolates.
Aminoglycosides, gentamicin and amikacin, had an intermediate effect on Klebsiella isolates. Similar results were observed for gentamicin against Serratia isolates. In contrast, amikacin had no effect against Serratia isolates.
The best sensitivity was observed with imipenem and quinolones, which varied from complete sensitivity by all Serratia isolates to lower level by Klebsiella isolates. Despite the high resistance of Acinetobacter isolates to quinolones including all strains, 29.57% of these isolates were sensitive to imipenem.
The resistance of all Klebsiella isolates to ampicillin was previously reported [60]. In addition, in another study in Iran, all Klebsiella isolates from neonates were resistant to ampicillin, while 31%, 46%, and 27% were resistant to ceftriaxone, amikacin, and gentamicin, respectively [53].
In our study, only two isolates of Pseudomonas aeruginosa were recovered from blood cultures exhibiting resistance to all antibiotics tested in this study.
In our study, all CoNS isolates showed high resistance to ampicillin and oxacillin. S. haemolyticus isolates had the highest level of resistance (≥85%) among the other CoNS isolates to amoxicillin-clavulanic acid, cefotaxime, ceftriaxone, erythromycin, azithromycin, and gentamicin. These results are in agreement with previous studies [61, 62]. Amikacin was effective against 69.70% of S. epidermidis isolates and all of S. haemolyticus and S. hominis isolates. The sensitivity of different CoNS spp. to amikacin, imipenem, and quinolones was variable. S. epidermidis was highly sensitive to imipenem, followed by quinolones, then amikacin. S. haemolyticus was 100% sensitive to amikacin, followed by imipenem. Concerning S. hominis isolates, these strains were all sensitive to amikacin and imipenem, while exhibiting lower activity to quinolones.
Interestingly, all staphylococcal isolates were sensitive to vancomycin as previously found in other reports [35, 46, 63], but its overprescription may result in the development of vancomycin-resistant strains such as enterococci.
According to our finding, best sensitivity among Gram-negative isolates was observed with imipenem followed by quinolones, while among Gram-positive isolates, vancomycin is followed by imipenem, amikacin, and finally quinolones.
5. Conclusion
Appropriate identification of the sepsis source, prompt antibiotic prescription, and aggressive management can effectively prevent adverse events following neonatal sepsis. Determination of the neonatal sepsis incidence, causative pathogens, and the patterns and rates of antibiotic resistance among all the neonate and infant populations are necessary to prevent complications.
Acknowledgments
All authors thank and express appreciation to Department of Pediatrics, Faculty of Medicine, Mansoura University, Mansoura University Children Hospital (MUCH), Health Insurance Hospital (HIH), and Mansoura General Hospital (MGH) for providing clinical isolates. The authors also thank and express appreciation to Dr. Maysaa El Sayed Zaki (Professor of clinical pathology).
Disclosure
This work was performed at Microbiology Department, Faculty of Pharmacy, Mansoura University, Egypt.
Conflict of Interests
The authors declare that they have no conflict of interests.
References
- 1.Wu J. H., Chen C. Y., Tsao P. N., Hsieh W. S., Chou H. C. Neonatal sepsis: a 6-year analysis in a neonatal care unit in Taiwan. Pediatrics and Neonatology. 2009;50(3):88–95. doi: 10.1016/s1875-9572(09)60042-5. [DOI] [PubMed] [Google Scholar]
- 2.UNICEF. Levels and Trends in Child Mortality. New York, NY, USA: UNICEF; 2011. [Google Scholar]
- 3.Black R. E., Cousens S., Johnson H. L., et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. The Lancet. 2010;375(9730):1969–1987. doi: 10.1016/s0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 4.Stoll B. J., Hansen N., Fanaroff A. A., et al. Changes in pathogens causing early-onset sepsis in very-low-birth-weight infants. The New England Journal of Medicine. 2002;347(4):240–247. doi: 10.1056/nejmoa012657. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira R. C., Mello R. R., Silva K. S. Neonatal sepsis as a risk factor for neurodevelopmental changes in preterm infants with very low birth weight. Jornal de Pediatria. 2014;90(3):293–299. doi: 10.1016/j.jped.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Dammann O., Kuban K. C. K., Leviton A. Perinatal infection, fetal inflammatory response, white matter damage, and cognitive limitations in children born preterm. Mental Retardation and Developmental Disabilities Research Reviews. 2002;8(1):46–50. doi: 10.1002/mrdd.10005. [DOI] [PubMed] [Google Scholar]
- 7.Edwards M. S., Baker C. J. Sepsis in the newborn. In: Gershon A. A., Hotez P. J., Katz S. L., editors. Krugman's Infectious Diseases of Children. Philadelphia, Pa, USA: Mosby; 2004. p. p. 545. [Google Scholar]
- 8.Patel S. J., Saiman L. Antibiotic resistance in neonatal intensive care unit pathogens: mechanisms, clinical impact, and prevention including antibiotic stewardship. Clinics in Perinatology. 2010;37(3):547–563. doi: 10.1016/j.clp.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jumah D. S., Hassan M. K. Predictor of mortality outcome in neonatal sepsis. Medical Journal of Basrah University. 2007;25:11–18. [Google Scholar]
- 10.Shrestha S., Adhikari N., Rai B. K., Shreepaili A. Antibiotic resistance pattern of bacterial isolates in neonatal care unit. Journal of the Nepal Medical Association. 2010;50(4):277–281. [PubMed] [Google Scholar]
- 11.Ghotaslou R., Ghorashi Z., Nahaei M.-R. Klebsiella pneumoniae in neonatal sepsis: a 3-year-study in the pediatric hospital of Tabriz, Iran. Japanese Journal of Infectious Diseases. 2007;60(2-3):126–128. [PubMed] [Google Scholar]
- 12.Klein J. O., Remington J. S. Current concepts of infection of the fetus and newborn infant. In: Remington J., Klein J., editors. Infectious Diseases of the Fetus and Newborn. Philadelphia, Pa, USA: WB Saunders; 2000. pp. 1–24. [Google Scholar]
- 13. Egyptian Neonatal Network (EGNN), 2010.
- 14.Bizzarro M. J., Dembry L.-M., Baltimore R. S., Gallagher P. G. Changing patterns in neonatal Escherichia coli sepsis and ampicillin resistance in the era of intrapartum antibiotic prophylaxis. Pediatrics. 2008;121(4):689–696. doi: 10.1542/peds.2007-2171. [DOI] [PubMed] [Google Scholar]
- 15.Garner J. S., Jarvis W. R., Emori T. G., Horan T. C., Hughes J. M. CDC Definitions for nosocomial infections, 1988. American Journal of Infection Control. 1988;16(3):128–140. doi: 10.1016/0196-6553(88)90053-3. [DOI] [PubMed] [Google Scholar]
- 16.Clark R., Powers R., White R., Bloom B., Sanchez P., Benjamin D. K., Jr. Nosocomial infection in the NICU: a medical complication or unavoidable problem? Journal of Perinatology. 2004;24(6):382–388. doi: 10.1038/sj.jp.7211120. [DOI] [PubMed] [Google Scholar]
- 17.Murray B., Pfaller T. Manual of Clinical Microbiology. 6th. Washington, DC, USA: American Society for Microbiology Press; 1999. [Google Scholar]
- 18.Yugueros J., Temprano A., Berzal B., et al. Glyceraldehyde-3-phosphate dehydrogenase-encoding gene as a useful taxonomic tool for Staphylococcus spp. Journal of Clinical Microbiology. 2000;38(12):4351–4355. doi: 10.1128/jcm.38.12.4351-4355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magiorakos A.-P., Srinivasan A., Carey R. B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 20.English M., Ngama M., Mwalekwa L., Peshu N. Signs of illness in Kenyan infants aged less than 60 days. Bulletin of the World Health Organization. 2004;82(5):323–329. [PMC free article] [PubMed] [Google Scholar]
- 21.The Young Infant Clinical Study Group. Clinical signs that predict severe illness in children under age 2 months: a multicentre study. The Lancet. 2008;371(9607):135–142. doi: 10.1016/s0140-6736(08)60106-3. [DOI] [PubMed] [Google Scholar]
- 22.Moore K. L., Kainer M. A., Badrawi N., et al. Neonatal sepsis in Egypt associated with bacterial contamination of glucose-containing intravenous fluids. Pediatric Infectious Disease Journal. 2005;24(7):590–594. doi: 10.1097/01.inf.0000168804.09875.95. [DOI] [PubMed] [Google Scholar]
- 23.Kayange N., Kamugisha E., Mwizamholya D. L., Jeremiah S., Mshana S. E. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatrics. 2010;10, article 39 doi: 10.1186/1471-2431-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiabi A., Djoupomb M., Mah E., et al. The clinical and bacteriogical spectrum of neonatal sepsis in a tertiary hospital in Yaounde, Cameroon. Iranian Journal of Pediatrics. 2011;21(4):441–448. [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z., Xiao Z., Zhong Q., Zhang Y., Xu F. 116 cases of neonatal early-onset or late-onset sepsis: a single center retrospective analysis on pathogenic bacteria species distribution and antimicrobial susceptibility. International Journal of Clinical and Experimental Medicine. 2013;6(8):693–699. [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed A. S., Chowdhury M. A., Hoque M., Darmstadt G. L. Clinical and bacteriological profile of neonatal septicemia in a tertiary level pediatric hospital in Bangladesh. Indian Pediatrics. 2002;39(11):1034–1039. [PubMed] [Google Scholar]
- 27.Mugalu J., Nakakeeto M. K., Kiguli S., Kaddu-Mulindwa D. H. Aetiology, risk factors and immediate outcome of bacteriologically confirmed neonatal septicaemia in Mulago hospital, Uganda. African Health Sciences. 2006;6(2):120–126. doi: 10.5555/afhs.2006.6.2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shitaye D. Neonatal sepsis: bacterial etiologic agents and their antibiotic susceptibility pattern in Tikur Anbessa University Hospital [M.S. thesis] Addis Ababa, Ethiopia: Addis Ababa University; 2008. [Google Scholar]
- 29.Meremikwu M. M., Nwachukwu C. E., Asuquo A. E., Okebe J. U., Utsalo S. J. Bacterial isolates from blood cultures of children with suspected septicaemia in Calabar, Nigeria. BMC Infectious Diseases. 2005;5, article 110 doi: 10.1186/1471-2334-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shrestha P., Das B. K., Bhatta N. K., et al. Clinical and bacteriological profiles of blood culture positive sepsis in newborns. Journal of Nepal Paediatric Society. 2008;27:64–67. [Google Scholar]
- 31.Ballot D. E., Nana T., Sriruttan C., Cooper P. A. Bacterial bloodstream infections in neonates in a developing country. ISRN Pediatrics. 2012;2012:6. doi: 10.5402/2012/508512.508512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw C. K., Shaw P., Thapalial A. Neonatal sepsis bacterial isolates and antibiotic susceptibility patterns at a NICU in a tertiary care hospital in western Nepal: a retrospective analysis. Kathmandu University Medical Journal. 2007;5(18):153–160. [PubMed] [Google Scholar]
- 33.Afsharpaiman S., Torkaman M., Saburi A., Farzaampur A., Amirsalari S., Kavehmanesh Z. Trends in incidence of neonatal sepsis and antibiotic susceptibility of causative agents in two neonatal intensive care units in Tehran, I.R Iran. Journal of Clinical Neonatology. 2012;1(3):124–130. doi: 10.4103/2249-4847.101692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cecilia C. M., Mary Ann C. B., Elizabeth E. G., Jonathan G. L., Joanne J. L., Cecille Y. A. Etiology of neonatal sepsis in five urban hospitals in the Philippines. PIDSP Journal. 2011;12:75–85. [Google Scholar]
- 35.Shah A. J., Mulla S. A., Revdiwala S. B. Neonatal sepsis: high antibiotic resistance of the bacterial pathogens in a neonatal intensive care unit of a tertiary care hospital. Journal of Clinical Neonatology. 2012;1(2):72–75. doi: 10.4103/2249-4847.96753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gandhi S., Ranjan K., Ranjan N., Sapre N., Masani M. Incidence of neonatal sepsis in tertiary care hospital: an overview. International Journal of Medical Science and Public Health. 2013;2(3):548–552. doi: 10.5455/ijmsph.2013.090320131. [DOI] [Google Scholar]
- 37.Utomo M. T. Risk factors of neonatal sepsis: a preliminary study in Dr. Soetomo hospital. Indonesian Journal of Tropical and Infectious Diseases. 2010;1:23–26. [Google Scholar]
- 38.Shokry M., Bassyouni M. I., Abu-El-Moon S., Maoz M., Tamer S. Evaluation of 16s rDNA amplification by PCR and some immunological mediators assessment compared with blood culture in diagnosis of neonatal sepsis. El-Minia Medical Bulletin. 2007;18:1–17. [Google Scholar]
- 39.Leal Y. A., Álvarez-Nemegyei J., Velázquez J. R., et al. Risk factors and prognosis for neonatal sepsis in southeastern Mexico: analysis of a four-year historic cohort follow-up. BMC Pregnancy and Childbirth. 2012;12, article 48 doi: 10.1186/1471-2393-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohli-Kochhar R., Omuse G., Revathi G. A ten-year review of neonatal bloodstream infections in a tertiary private hospital in Kenya. Journal of Infection in Developing Countries. 2011;5(11):799–803. doi: 10.3855/jidc.1674. [DOI] [PubMed] [Google Scholar]
- 41.Rønnestad A., Abrahamsen T. G., Gaustad P., Finne P. H. Blood culture isolates during 6 years in a tertiary neonatal intensive care unit. Scandinavian Journal of Infectious Diseases. 1998;30(3):245–251. doi: 10.1080/00365549850160873. [DOI] [PubMed] [Google Scholar]
- 42.Zaidi A. K. M., Huskins W. C., Thaver D., Bhutta Z. A., Abbas Z., Goldmann D. A. Hospital-acquired neonatal infections in developing countries. The Lancet. 2005;365(9465):1175–1188. doi: 10.1016/s0140-6736(05)71881-x. [DOI] [PubMed] [Google Scholar]
- 43.Sundaram V., Kumar P., Dutta S., et al. Blood culture-confirmed bacterial sepsis in neonates in a north Indian tertiary care center: changes over the last decade. Japanese Journal of Infectious Diseases. 2009;62(1):46–50. [PubMed] [Google Scholar]
- 44.Stoll B. J., Fanaroff A. Early-onset coagulase-negative staphylococcal sepsis in preterm neonate. National Institute of Child Health and Human Development (NICHD) Neonatal Research Network. The Lancet. 1995;345(8959):1236–1237. doi: 10.1016/s0140-6736(95)92017-x. [DOI] [PubMed] [Google Scholar]
- 45.Fahmey S. S. Early-onset sepsis in a neonatal intensive care unit in beni suef, Egypt: bacterial isolates and antibiotic resistance pattern. Korean Journal of Pediatrics. 2013;56(8):332–337. doi: 10.3345/kjp.2013.56.8.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macharashvili N., Kourbatova E., Butsashvili M., Tsertsvadze T., McNutt L.-A., Leonard M. K. Etiology of neonatal blood stream infections in Tbilisi, Republic of Georgia. International Journal of Infectious Diseases. 2009;13(4):499–505. doi: 10.1016/j.ijid.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naher H. S., Khamael A. B. Neonatal sepsis; the bacterial causes and the risk factors. International Journal of Research in Medical Sciences. 2013;1:19–22. [Google Scholar]
- 48.El Badawy A., El Sebaie D., Khairat S., Fouad S. A study of microbiological pattern of neonatal sepsis. Alexandria Journal of Pediatrics. 2005;19:357–367. [Google Scholar]
- 49.Koksal F., Yasar H., Samasti M. Antibiotic resistance patterns of coagulase-negative staphylococcus strains isolated from blood cultures of septicemic patients in Turkey. Microbiological Research. 2009;164(4):404–410. doi: 10.1016/j.micres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Piette A., Verschraegen G. Role of coagulase-negative staphylococci in human disease. Veterinary Microbiology. 2009;134(1-2):45–54. doi: 10.1016/j.vetmic.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Kapoor L., Randhawa V. S., Deb M. Microbiological profile of neonatal septicemia in a pediatric care hospital in Delhi. Journal of Communicable Diseases. 2005;37(3):227–232. [PubMed] [Google Scholar]
- 52.Aftab R., Iqbal I. Bacteriological agents of neonatal sepsis in Nicu at Nishtar Hospital Multan. Journal of the College of Physicians and Surgeons Pakistan. 2006;16(3):216–219. [PubMed] [Google Scholar]
- 53.Movahedian A. H., Moniri R., Mosayebi Z. Bacterial culture of neonatal sepsis. Iranian Journal of Public Health. 2006;35(4):84–89. [Google Scholar]
- 54.Hafsa A., Fakruddin M., Hakim M. A., Sharma J. D. Neonatal bacteremia in a neonatal intensive care unit: analysis of causative organisms and antimicrobial susceptibility. Bangladesh Journal of Medical Science. 2011;10(3):187–194. [Google Scholar]
- 55.Sarvikivi E., Lyytikäinen O., Salmenlinna S., et al. Clustering of Serratia marcescens infections in a neonatal intensive care unit. Infection Control and Hospital Epidemiology. 2004;25(9):723–729. doi: 10.1086/502467. [DOI] [PubMed] [Google Scholar]
- 56.MacDonald T. M., Langley J. M., Mailman T., et al. Serratia marcescens outbreak in a neonatal intensive care unit related to the exit port of an oscillator. Pediatric Critical Care Medicine. 2011;12(6):e282–e286. doi: 10.1097/pcc.0b013e31820ac42a. [DOI] [PubMed] [Google Scholar]
- 57.Voelz A., Müller A., Gillen J., et al. Outbreaks of Serratia marcescens in neonatal and pediatric intensive care units: clinical aspects, risk factors and management. International Journal of Hygiene and Environmental Health. 2010;213(2):79–87. doi: 10.1016/j.ijheh.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 58.Polilli E., Parruti G., Fazii P., et al. Rapidly controlled outbreak of serratia marcescens infection/colonisations in a neonatal intensive care unit, Pescara General Hospital, Pescara, Italy, April 2011. Eurosurveillance. 2011;16(24) doi: 10.2807/ese.16.24.19892-en. [DOI] [PubMed] [Google Scholar]
- 59.Kristóf K., Kocsis E., Nagy K. Clinical microbiology of early-onset and late-onset neonatal sepsis, particularly among preterm babies. Acta Microbiologica et Immunologica Hungarica. 2009;56(1):21–51. doi: 10.1556/amicr.56.2009.1.2. [DOI] [PubMed] [Google Scholar]
- 60.Cisse C. T., Mbengue-Diop R., Moubarek M., et al. Neonatal bacterial infections at the CUH of Dakar. Gynécologie Obstétrique & Fertilité. 2001;29(6):433–439. doi: 10.1016/S1297-9589(01)00157-6. [DOI] [PubMed] [Google Scholar]
- 61.Chiew Y. F., Charles M., Johnstone M. C., Thompson K. M., Parnell K. D., Penno E. C. Detection of vancomycin heteroresistant Staphylococcus haemolyticus and vancomycin intermediate resistant Staphylococcus epidermidis by means of vancomycin screening agar. Pathology. 2007;39(3):375–377. doi: 10.1080/00313020701330441. [DOI] [PubMed] [Google Scholar]
- 62.Yu M.-H., Chen Y.-G., Yu Y.-S., Chen C.-L., Li L.-J. Antimicrobial resistance and molecular characterization of Staphylococcus haemolyticus in a Chinese hospital. European Journal of Clinical Microbiology & Infectious Diseases. 2010;29(5):613–616. doi: 10.1007/s10096-010-0893-3. [DOI] [PubMed] [Google Scholar]
- 63.Bhat Y. R., Lewis L. E. S., Vandana K. E. Bacterial isolates of early-onset neonatal sepsis and their antibiotic susceptibility pattern between 1998 and 2004: an audit from a center in India. Italian Journal of Pediatrics. 2011;37, article 32 doi: 10.1186/1824-7288-37-32. [DOI] [PMC free article] [PubMed] [Google Scholar]