Abstract
Genes implicated in neurodevelopmental disorders (NDDs) important in cognition and behavior may have convergent function and several cellular pathways have been implicated, including protein translational control, chromatin modification, and synapse assembly and maintenance. Here, we test the convergent effects of methyl-CpG binding domain 5 (MBD5) and special AT-rich binding protein 2 (SATB2) reduced dosage in human neural stem cells (NSCs), two genes implicated in 2q23.1 and 2q33.1 deletion syndromes, respectively, to develop a generalized model for NDDs. We used short hairpin RNA stably incorporated into healthy neural stem cells to supress MBD5 and SATB2 expression, and massively parallel RNA sequencing, DNA methylation sequencing and microRNA arrays to test the hypothesis that a primary etiology of NDDs is the disruption of the balance of NSC proliferation and differentiation. We show that reduced dosage of either gene leads to significant overlap of gene-expression patterns, microRNA patterns and DNA methylation states with control NSCs in a differentiating state, suggesting that a unifying feature of 2q23.1 and 2q33.1 deletion syndrome may be a lack of regulation between proliferation and differentiation in NSCs, as we observed previously for TCF4 and EHMT1 suppression following a similar experimental paradigm. We propose a model of NDDs whereby the balance of NSC proliferation and differentiation is affected, but where the molecules that drive this effect are largely specific to disease-causing genetic variation. NDDs are diverse, complex and unique, but the optimal balance of factors that determine when and where neural stem cells differentiate may be a major feature underlying the diverse phenotypic spectrum of NDDs.
Introduction
Neurodevelopmental disorders (NDDs) that affect behavior and cognition are caused by a wide variety of mutations, and several hypotheses have been put forward to understand the underlying features of the disease. The identification of mutations in MECP21 and FMR12 in the 1990s suggested a role for chromatin modification and genomic regulation. The chromatin modification hypothesis3 has continued to receive support, and mutations in other genes4 related to genomic regulation have been identified in several cases.5 Discoveries in 2003 and 2004 led to a different theory about the underlying biology of NDDs, specifically autistic disorders, based on the identification of mutations in NLGN3 and NLGN4.6 This paved the way for the synaptic hypothesis of NDDs,7, 8 which states that NDDs are caused by dysfunctional assembly or maintenance of synapses, a hypothesis which continues to be supported by the discovery of more mutations in non-NLGN genes involved in synapse formation or stability.9, 10 Protein translation has also been associated to NDDs,11, 12 and in conjunction with the chromatin modification hypothesis, suggests that regulation of major cell processes in neurons may predispose to NDDs. The degree to which these disparate hypotheses may be connected is not known, although there is evidence to support WNT signaling as a potential convergence point.13, 14
The purpose of the current work is to identify a convergence point of NDDs on the basis of the two genes studied here, and to propose a molecular model that might apply to neurodevelopmental disorders more generally. We selected two genes associated with neurodevelopment disorders to identify convergence points, largely due to previous gene discovery studies5, 15 in which we participated. Methyl-CpG binding domain (MBD) 5 is a member of the MBD family characterized by a 70 amino-acid region thought to mediate association with methylated residues. MBD5 protein contains a PWWP (pro-trp-trp-pro) domain, thought to be important in cell division, growth and differentiation.16, 17 Mutations in MBD5 are thought to be important in the clinical phenotype of 2q23.1 deletion syndrome,18, 19 where subjects with a deletion in this region of chromosome 2 show intellectual disability and autistic-like features. Other studies also support the association of mutations in MBD5 and autism spectrum disorders (ASD),15, 20 though there is a large heterogeneity in phenotype. Special AT-rich binding protein 2 (SATB2) is a transcription factor that associates with AT-rich regions of the genome and the nuclear matrix,21, 22 and is thought to be involved in chromatin modification in neurons.23 SATB2 is found on 2q33.1 and reduced dosage of SATB2 is thought to be a major cause of 2q33.1 deletion syndrome,24 characterized by cleft palate,25 severe speech delay, intellectual disability and behavioral problems, including ASD.24, 26
Materials and methods
This work was reviewed and approved by the ethical review board of the Douglas Hospital Research Institute of McGill University.
Cell culture
Fetal brain cells (FBCs) are ReNcells derived form the ventral mesencephalon of human fetal brain (Millipore SCC008). Cells were grown on poly-L-ornithine/laminin (Sigma, St. Louis, MO, USA; P3655-50MG and l2020) coated six-well plates. Cells were maintained in 70% DMEM, 2% B27, 1% Pen/Strep (Life Technologies, Foster City, CA, USA) 30% Ham's F12 (Mediatech, Herndon, VA, USA) and 20 ng ml−1 bFGF (R&D Systems, Minneapolis, MN, USA, 233-FB-025), 20 ng ml−1 EGF (Sigma E9644) and 5 μg ml−1 heparin (Sigma). Differentiation is triggered by removing growth factors from media and letting cells grow for 30 days, with media changes every 3 days; proliferating cells are those maintained in bFGF and EGF.
Generation of stable knockdown cell lines
Short-hairpin RNAs (shRNAs) used in this study were designed, cloned into the pLKO.1 vector and packaged into lentivirus at the Broad Institute (Cambridge, MA, USA). To create stable cell lines, we transfected cells with lentivirus, then selected for cells where genomic integration occurred. For lentiviral transfection, FBCs were maintained at 30% confluency (~400 000 cells per well) in a six-well plate, then 20 μl viral media in 2 ml cell culture media without penicillin and streptomycin was added. Puromycin (Sigma; P8833; 0.8 μl ml−1), resistance to which is produced by the pLKO.1 vector, was added to cultures 48 h after infection and this followed an initial media change 24 h after transfection. Stable cell lines were selected by continuous maintenance of low-dose puromycin in culture media (0.2 μl ml−1). For controls, we used shRNAs targeting LacZ, GFP, RFP and Luc messenger RNA (mRNA). We refer to these controls as non-target (NT) controls because they were generated following identical procedures to MBD5 and SATB2 knockdown (KD) lines, but target mRNA was not present in the human genome.
RNAseq
RNAseq libraries were prepared from high quality (RIN>9; Agilent 2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA, USA) RNA, where RNA was extracted with the Qiagen (Hilden, Germany) RNeasy kit following manufacturer's instructions for RNA isolation from cells. For bioinformatic processing, we first used FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) for adapter trimming. We used an average read quality >30 for the total read and retained only those reads whose length ranged from 30 bp to 85 bp, and removed all the duplicate reads. Alignment to the human genome was done with TopHat27 and Bowtie2.28 We allowed two mismatches, an insert size of 300 000, a standard deviation of 100 and an expected (mean) inner distance between mate pairs of 150. We used Cufflinks2 (ref. 29) with default parameters to assemble aligned RNA-Seq reads into transcripts, to estimate their abundances, and to test differential expression. Gene ontology (GO) analyses were done under default conditions using DAVID.
Quantitative PCR
Reverse transcriptions were done on total RNA fraction to obtain complementary DNA in 40 μl volume containing 1 μg of total RNA; we used 0.5 μg random hexamer primers, 0.5 mM dNTPs, 0.01 M DTT and 400 U M-MLV RT (Carlsbad, CA, USA). For gene quantification, we ordered pre-designed Taqman primers, where each well included 6 μl of 2X gene-expression mastermix, 0.6 μl of 20X primer mix, 3.4 μl of RNase free water and 2 μl of complementary DNA. β-Actin and GAPDH were used as internal controls for normalization. We used the ABI 7900 light cycler for all the experiments and manufacturer's software for processing and analysis.
MicroRNA analysis
MicroRNAs were processed using the nCounter Human miRNA expression kit (NanoString Technologies, Seattle, WA, USA) at the NanoString facility at the Jewish General hospital, and all samples were run in duplicate. The nCounter data were processed using NanoStringNorm in R, with all data normalized to the geometric mean and microRNA spike-in controls, according to manufacturer's instructions. All the data were analyzed in R. For microRNA quantitative PCR (qPCR), the microRNA reverse transcription kit (Applied Biosystems, Burlington, ON, Canada) and specific primers to endogenous control and mir targets (#001093: RNU6B; Applied Biosystems) were used to generate complementary DNA from the same microRNA extracted for the NanoString analysis. For each reaction, the PCR mix included 10 μl of 2X NoAmperaseUNG mastermix (Applied Biosystems), 1 μl of primers/probe mix and 2 μl of complementary DNA, H20 20 μl.
Reduced representation bisulfite sequencing
We followed BisQC, a multiplexed bisulfite sequencing pipeline that we developed30, 31 for reduced representation bisulfite sequencing library preparation stages. We used TrimGalore followed by Bismark32 to align bisulfite-treated reads to a reference genome with Bowtie 2 (with zero mismatches allowed in a seed alignment during multi-seed alignment). We also used BisSNP, a package based on the Genome Analysis Toolkit map-reduce framework for genotyping and accurate DNA methylation calling in bisulfite-treated massively parallel sequencing with the Illumina directional library protocol, to identify those methylated regions that may be confounded by genetic variation—these positions were removed from all the analyses. Only CpG sites with coverage >5X were included, and we excluded the 0.1% of CpGs that showed the highest coverage for each sample. We defined CpG clusters as any contiuous set of CpG sites within 50 bp of another CpG site, and used methylation frequency at each CpG site within a cluster in MBD5 KD or SATB2 KD compared with NT controls as values in a t-test.
Results
Generation of NSC models of reduced dosage associated with neurodevelopmental disorders
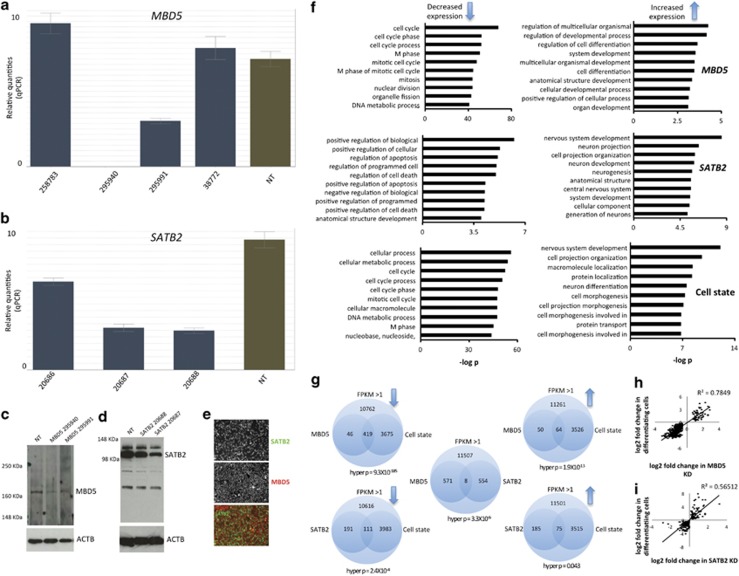
To model genomic dosage disorders, we used KD technology in a human FBC line derived from a healthy fetus, where this cell line has been thoroughly characterized.33 We used multiple shRNA vectors targeting different regions of either MBD5 or SATB2 mRNA packaged into lentivirus and infected FBCs with either of these constructs or any of four NT control constructs. After KD of either gene followed by puromycin selection, we extracted RNA from each cell and performed qPCR using primers targeting MBD5 or SATB2 mRNA. For MBD5 KD, we found that two of four constructs show significant decrease in expression of MBD5 (Figure 1a) of 64% and 99% (s.d.=25%). For SATB2, we found that all the three shRNA constructs were able to knock down SATB2 (Figure 1b) at levels ranging from 31, 61 and 70% knockdown (s.d.=21%). We selected two shRNAs per gene for western blot validation and confirmed decreased expression at the protein level (Figures 1c and d), with knockdown levels of MBD5 protein down 65 and 61% (s.d.=3%), and for SATB2 protein was down 31 and 54% (s.d.=16%). We also performed immunocytochemistry to ensure that proliferating FBCs express both genes in all cells, as expected. We observed nuclear localization of SATB2 and cytoplasmic localization of MBD5 in all neural stem cells (NSCs; Figure 1e). We proposed that MBD5 might function as a methylated RNA binding molecule in the cytoplasm, where methylated RNA may be important in stem cell proliferation.34
Figure 1.
Generation of human neural stem cell (NSC) models of MBD5 and SATB2 suppression, and RNAseq comparative analysis. (a) MBD5 gene-expression analysis in four cell lines that underwent MBD5 shRNA lentiviral infection (blue bars). Numbers represent RNAi consortium (TRC) identifiers. Green bar represents mean MBD5 expression across four independent non-target (NT) control cell lines. (b) SATB2 gene-expression analysis in three independent cell lines (blue bar) and four independent non-target controls (green bar). (c) Western blot experiment showing the two MBD5 KD cell lines with greatest degree of KD from a and one NT control, targeting LacZ mRNA. MBD5 is detected at ~168 KDa. (d) Western blot analysis of SATB2 knockdown in LacZ and two SATB2 KD cell lines. (e) Immunocytochemical analysis of MBD5 and SATB2 protein demonstrating presence of both proteins in all cells, with a cytoplasmic distribution of MBD5 and nuclear localization of SATB2 protein. (f) Gene ontology analysis of differentially expressed mRNA in MBD5 KD, SATB2 KD and the cell state experiment (non-target proliferating cells compared with non-target differentiating cells). (g) Statistical analysis of the probability of observing overlapping significantly differentially expressed mRNA across each experiment. (h) mRNAs that overlap in MBD5 KD proliferating cells and non-target differentiating cells are correlated. (i) mRNAs that overlap in SATB2 KD proliferating cells and non-target differentiating cells are correlated. KD, knockdown; MBD, methyl-CpG binding domain; mRNA, messenger RNA; SATB, special AT-rich binding protein.
Reduced dosage of MBD5 or SATB2 leads to mRNA expression patterns more characteristic of differentiating than proliferating neural stem cells
Our previous work investigating reduced dosage of TCF4 and EHMT1 suggested that modeling genomic dosage disorders in NSCs made them more characteristic of a differentiating cell state compared with their actual proliferating state, implying a convergence point for at least some neurodevelopmental disorders.33 To test this hypothesis in the MBD5 KD and SATB2 KD models, we performed RNAseq using two shRNA constructs per gene and four non-target control shRNA, then generated a list of differentially expressed genes for each reduced dosage model compared with control shRNA. For MBD5 KD decreased expression genes, we found an overrepresentation of genes implicated in cell division and proliferation (Figure 1f—MBD5), whereas for increased expression genes, there was an overrepresentation of genes involved in neural differentiation. For SATB2 KD decreased expression genes, we observed an overrepresentation of genes implicated in apoptosis or cell death (Figure 1f—SATB2), whereas increased expression genes were implicated in neurodevelopment. These data suggest that reduced dosage of either MBD5 or SATB2 leads to increased expression of genes related to neural differentiation, consistent with previous results from reduced dosage models of TCF4 and EHMT1. With respect to repression of cell proliferation markers, we find that only MBD5 KD had repressed markers associated with cell proliferation, likely consistent with the function of the PWWP domain, and suggesting that genes implicated in NDDs can affect NSC differentiation, affect NSC proliferation or do both.
Before empirically testing whether MBD5 KD or SATB2 KD were in a state more characteristic of differentiating cells, we grew NT control cells and differentiated them for 30 days in the absence of growth factors. We extracted RNA and performed RNAseq on these differentiating cells, and compared them with the same NT control cells in the proliferating state. We then performed GO analyses for all genes that showed differential expression in this experiment and found, as expected, GO terms related to the cell cycle for decreased expression genes and GO terms related to neurodevelopment for increased expression genes (Figure 1f—cell state). This sets a baseline for what is expected with respect to ‘proliferating' genes and ‘differentiating' genes in control conditions.
If RNA expression differences detected in the MBD5 KD and SATB2 KD experiments are characteristic of differentiating NT control cells, than the same genes that show differential expression in the reduced dosage models should show differential expression in the cell state experiment. To test this hypothesis, we intersected all genes that were significantly differentially expressed across each analysis and computed the probability of the intersection occurring by chance (Figure 1g), using the hypergeometric probability.35 First, we calculated the total number of tests by determining the maximum number of mRNA that could be identified in these RNAseq experiments. We found that 12 640–14 901 different mRNA could be detected (FPKM >1) across different experiments, and we use these numbers as our global pool of mRNA. For genes that showed significantly decreased expression in MBD5 KD, we found that 90.3% were also significantly decreased in differentiating NSCs, whereas 56.1% of genes that showed increased expression in MBD5 KD were common with those in the cell state experiment, and this overlap is strongly unlikely to occur by chance (Figure 1g). For mRNA from SATB2 KD that showed decreased expression, we found that 36.8% were also significantly differentially expressed in the cell state experiment (Figure 1g), whereas, for SATB2 KD mRNA that showed increased expression, 28.9% were common to cell state. For both MBD5 KD and SATB2 KD, we found a strong correlation between differentially expressed genes and expression differences specific to differentiating cells (Figures 1h and i, P<0.01).
We next asked what the probability was for the same genes that were significantly differentially expressed in both MBD5 KD and SATB2 KD cells to determine the degree of convergence between these two disease models. We found that eight genes were common to both analyses (Figure 1g), an event unlikely to occur by chance (hyper P=3.3X10−6), all of which were directionally identical between cell models. Differentially expressed genes common to both reduced dosage models that were increased were NCAN,36GPR56,37TTYH138 and PLP1,39 all of which are implicated in cell differentiation. Genes that were downregulated in both cell models included MSMO1, FDFT1, SERPINE1 and ANXA2, where MSMO1 and FDFT1 are involved in cholesterol biosynthesis. We selected three genes known to be involved in neural differentiation and ANXA2, which is reported to have a role in psychiatric disorders40 and neuritogenesis41 for RNAseq validation (Supplementary Figure S1).
Because of the effect in MBD5 KD of decreased expression of genes associated with the cell cycle, we performed both cell proliferation and cell cycle assays (Supplementary Information and Supplementary Figure S2). We found no deficits of cell proliferation or cell cycle progression in MBD5 KD cells compared with non-target controls (Supplementary Figure S2). We also performed targeted qPCR analysis of two genes implicated in neural differentiation (HES6 and MALAT1) and two genes implicated in cell proliferation (CDK1 and CKS2; Supplementary Figure S2). All RNAseq data were confirmed, strongly supporting our conclusion from the GO analysis.
Reduced dosage of MBD5 or SATB2 leads to microRNA expression patterns more characteristic of differentiating than proliferating cells
We wanted to further pursue our hypothesis by looking at other measurable cell features and following the similar experimental paradigm to RNAseq experiments. To this end, we performed genome-wide microRNA experiments in MBD5 KD, SATB2 KD, proliferating non-target controls and differentiating non-target controls.
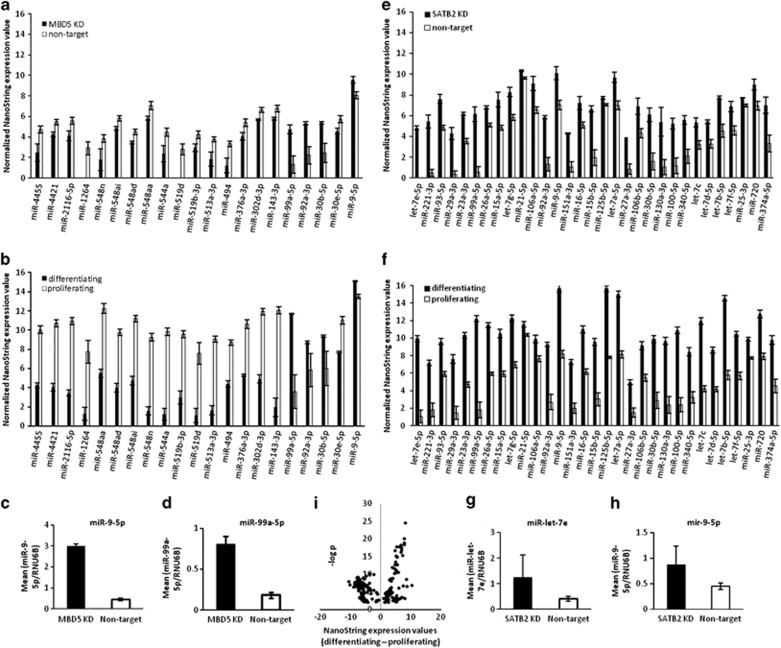
We performed global microRNA analysis using four non-target controls and the two MBD5 KD cell lines, all of which was performed in replicate. We excluded any microRNA not expressed in either 6/7 controls or in 3/4 MBD5 KD, which left 254 microRNAs for analysis. We found 21 differentially expressed microRNA (P<0.05), though none passed FDR correction (Figure 2a). For differentiating and proliferating non-target controls cells, we extracted microRNA in both states (n=4 samples per cell state, in duplicate) and performed global NanoString analysis. There were 395 microRNAs that were detectable, and many microRNAs that showed significant differences between cell states (259 microRNAs with P-values <0.05). To test whether the same microRNAs that were differentially expressed in MBD5 KD were also differentially expressed in the cell state experiment, we plotted data from the cell state experiment for only those microRNAs that were also differentially expressed in the MBD5 KD experiment (Figure 2b). Strikingly, we found that all microRNA changes were directionally identical: there were four microRNAs with increased expression in the MBD5 KD experiment and these were identical to the cell state experiment. These four microRNAs, (mir99,42 mir9,43 mir30b44 and mir92a-3p45) are associated with differentiation or suppression of proliferation, further supporting our hypothesis. The relationship between MBD5 KD and differentiating cells is reflected in Pearson correlation coefficient of the log2 fold-change differences between MBD5 KD and non-target differentiating cells (Pearson R=0.66, P=0.0011). Immediately apparent is that the magnitude of the expression differences are much more pronounced in NT differentiating cells than in MBD5 KD cells (compare heights of the bars in Figures 2a and b). To confirm the validity of the NanoString results, we performed targeted microRNA qPCR on two targets (Figure 2c: miR-99-5p, P=3.96 × 10−5; Figure 2d: mir-9-5p, P=6.23X10−9).
Figure 2.
MicroRNA expression patterns in neural stem cell models of gene dosage disorders are more characteristic of differentiating non-target cells than proliferating non-target cells. (a) NanoString expression values for all microRNA with single point P-values <0.05 in the MBD5 KD experiment compared with non-target proliferating cells. (b) NanoString expression values for the same microRNAs identified in the MBD5 KD experiment but showing values for the cell state (differentiating non-target cells compared with proliferating non-target cells) experiment. (c and d) Quantitative PCR (qPCR) validation of two microRNAs identified in the MBD5 KD experiment. (e) NanoString expression values for all microRNA with single point P-values <0.05 in the SATB2 KD experiment (f) NanoString expression values for the same microRNAs identified in the SATB2 KD experiment, but showing values for the cell state (differentiating non-target cells compared with proliferating non-target cells) experiment. (g and h) qPCR validation of two microRNAs identified in the SATB2 KD experiment. (i) Distribution of microRNAs that are up- or downregulated in the cell state experiment plotted as a function of P-value. KD, knockdown; MBD, methyl-CpG binding domain; SATB, special AT-rich binding protein.
For SATB2 KD, we identified 31 microRNAs with nominal P-values <0.05 with mir-let7e, mir-221-3p and mir-93-5p showing FDR significant q-values <0.10. Comparing the microRNAs identified as differentially expressed in the SATB2 KD experiment (Figure 2e) to those same microRNAs present in the cell state experiment (Figure 2f) revealed a significant correlation (Pearson correlation of log2 fold-change differences=0.59, P=0.01, where the direction of change was identical for each microRNA), with a more extreme extent of change in the cell state experiment, similar to findings to MBD5 KD. We performed qPCR on two microRNAs from this analysis (Figures 2g and h: miR-let-7e; P=0.11; miR-9-5p; P=0.11). For comparison, we provide the expression changes of all significantly differentially expressed microRNA in the cell state experiment (Figure 2i), where 106/259 microRNAs showed increased expression in differentiating cells.
Reduced dosage of MBD5 or SATB2 leads to DNA methylation patterns more characteristic of differentiating than proliferating cells
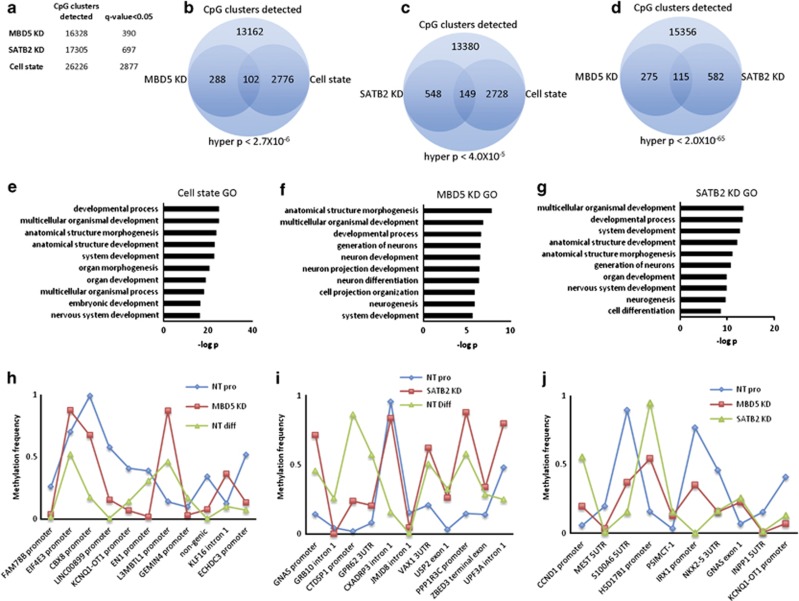
To determine whether DNA methylation patterns in MBD5 KD or SATB2 KD have taken on characteristics of differentiating cells, we performed whole-genome methylation experiments in reduced genomic space. First, we compared MBD5 KD (two shRNA and two replicates each) to non-target control proliferating cells (four shRNAs) and found 390 genome-wide significant CpG clusters (Figure 3a), where a cluster is defined as two or more CpGs that are <50 bp apart. For SATB2 KD (two shRNAs and two replicates each), we found 697 genome-wide significant CpG clusters; whereas, in the non-target cell state experiment, we found 2877 genome-wide significant CpG clusters. We calculated the hypergeometric probability for both reduced dosage models with cell state and we found that overlapping CpG clusters were significantly unlikely to occur by chance (Figures 3b and c), suggesting that these particular clusters may be important in cell differentiation and/or proliferation. To test whether MBD5 and SATB2 reduced dosage model methylation regions converged, we intersected data from each analysis and calculated the hypergeometric probability of the overlap, which was highly significant (Figure 3d). To support the idea that differential methylation is relevant to NSC proliferation or differentiation, we mapped all differentially methylated clusters to genes (within 5Kb 5' and 2Kb 3' Figures 3g and i), then performed GO analysis with these gene lists. Across each GO analysis, we found terms related to neurodevelopment to be overrepresented (Figures 3e and g).
Figure 3.
DNA methylation patterns in neural stem cell models of gene dosage disorders are more characteristic of differentiating cells than proliferating cells. (a) Total number of CpG clusters detected and the total number of genome-wide significant CpG clusters that show differential methylation. (b) Graphical representation of the likelihood of observing overlapping CpG clusters in the MBD5 KD, (c) SATB2 KD and (d) cell state experiment. (e–g) Significant GO terms associated with CpG clusters near genes for the cell state (e), MBD5 KD (f) and SATB2 KD (g) experiments. (h–j) Traces showing methylation differences for the most significantly differentially methylated CpG clusters in (h) MBD5 KD, (i) SATB2 KD and (j) most significant CpG clusters that overlap in both SATB2 KD and MBD5 KD. GO, gene ontology; KD, knockdown; MBD, methyl-CpG binding domain; SATB, special AT-rich binding protein.
We calculated the Pearson correlation for all CpG clusters that were significant in both a reduced-dosage model and in the cell state experiment. For MBD5 KD, there were 102 clusters that met this criteria and we found a Pearson correlation of 0.49 when comparing MBD5 KD to the mean methylation frequency in proliferating control cells, and a Pearson value of 0.76 when the same MBD5 KD methylation frequencies were correlated with mean per cluster methylation frequencies in NT differentiating cells, implying that the methylation in MBD5 KD is more similar to differentiating NSCs than to proliferating NSCs. For SATB2 KD, we found Pearson values of 0.51 and 0.53, where the higher Pearson value was with differentiating cells, though these two values are essentially identical.
We observed a much stronger Pearson value between each KD line than with either KD line with NT cells (MBD5 KD and SATB2 KD: Pearson=0.87; MBD5 KD and NT pro: 0.37; SATB2 KD and NT pro: 0.29). This Pearson correlation between SATB2 KD and MBD5 KD suggests that the directionality of methylation frequency between KD lines is similar for those clusters that are significantly different from proliferating non-target control cells. We show methylation frequency data for the top 10 most significant CpG clusters for each reduced expression analysis (Figures 3h and j).
Discussion
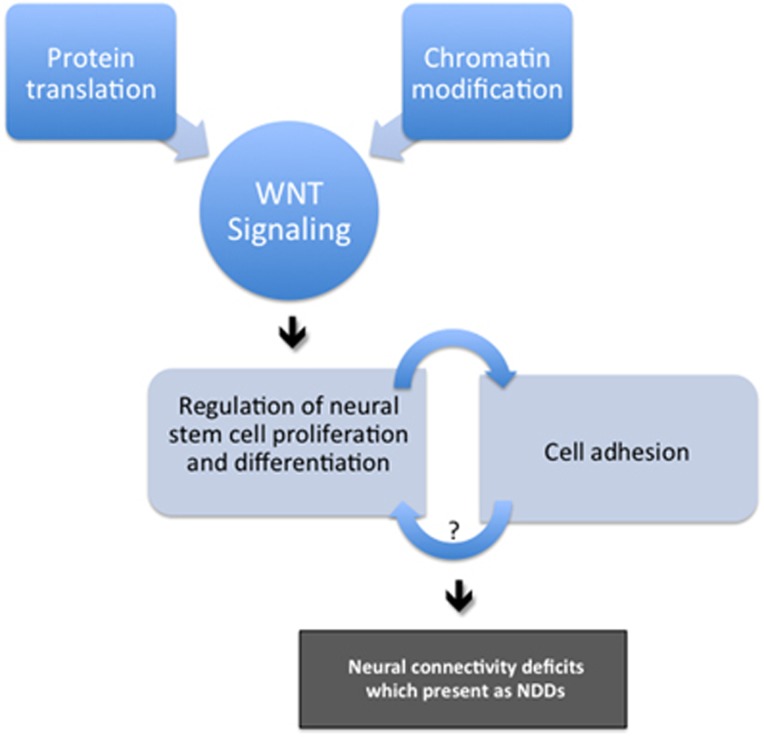
We tested the hypothesis that reduced dosage of two different genes made neural stem cells more characteristic of differentiating cells than their actual proliferating state, an idea that is well supported by many other genes associated with NDDs (Table 1). We suggest that both protein translation and chromatin modification hypotheses of NDDs important in behavior and cognition converge on this pathway, an idea in line with the WNT hypothesis of ASDs.14 Specifically, we suggest that it is not chromatin modification or protein translation per se, but rather the functional effects of genes in these categories that impact neural differentiation. Our data suggest that there is significant molecular overlap between NDDs caused by different genetic variation, but that this is not overwhelming (5–15% across cell features)—it appears that outcome of these different genetic variation (regulation of NSC differentiation and/or proliferation) is the crucial factor, and that many different molecules drive this effect. A mutation in any element of this pathway could lead to an NDD, with the degree of overlap across different genetic variation dependent on how close two genes are in the regulation of cell proliferation or differentiation. We suggest that aberrant neural differentiation may lead to inappropriate neural connections, which may explain why so many different synaptic-associated genes are also observed in ASD cases46 (Figure 4).
Table 1. Examples of well-known genes and genetic loci implicated in ASDs and/or NDDs with reported effects on proliferation or differentiation in NSCs.
| Gene | Functional effect |
|---|---|
| FMR1 | Regulation of cell differentiation47, 48, 49, 50, 51 |
| MECP2 | Modulates the balance between proliferation and neural differentiation through the Notch signaling pathway52 |
| TSC1/2 | Mutations cause premature differentiation and impaired maturation of neural precursor cells during both embryonic and postnatal development53 |
| ADNP | Role in neuronal differentiation and maintenance54, 55 |
| DLL1 | Promotes neuronal differentiation in the telencephalon56 |
| CTNNB1 | Functions in the decision of precursors to proliferate or differentiate during mammalian neuronal development57 |
| SMARCC2 | Promotes indirect neurogenesis by increasing the pool of progenitors58 |
| TBR1 | Promotes neuronal differentiation59 |
| CDKL5 | Mutation blocks cell cycle and promotes differentiation in neurons60 |
| PTEN | Deletion causes neuroblast differentiation through mTORC161 |
| CHD8 | Negative regulator of the Wnt-β-catenin signaling pathway62 |
| ARID1B | Part of the SWI/SNF complex, a cell cycle control complex63 |
| POGZ | Regulation of mitosis and proliferation in neurons64, 65 |
| SUV420H1 | Promotes neuroectodermal differentiation66 |
| EIF4E | Suppresses a pro-neurogenic program in neural progenitor cells67 |
| SHANK3 | Mediates sustained MAPK and PI3K signaling68 |
| NRXN1 | Reduced expression alters neuron differentiation69 |
| NLGN4X | Reduced expression delays neurodevelopment70 |
| 16p11.2 CNV | Reciprocal deletion and duplication CNV implicated in macrocephaly and microcephaly, respectively.71 May be caused by MAPK3 dosage effects |
| 1q21.1 CNV | Reciprocal deletion and duplication CNV implicated in microcephaly and macrocephaly, respectively.72 May be caused by CHD1L and/or BCL9 dosage effects |
Abbreviations: ASD, autism spectrum disorder; CNV, copy number variant; NDD, neurodevelopmental disorder; NSC, neural stem cell.
Many genes on this list have several functions (for example, NRXN1, NLGN4X and SHANK3 in cell adhesion), and here we have purposely shown only those functions associated with cell proliferation and differentiation, providing evidence that it is this pathway that unifies genes and loci associated with NDDs important to behavior and cognition. Our model predicts that genes that function to promote differentiation will show increased markers (microRNA, messenger RNA, DNA methylation patterns) of proliferation under disease conditions, whereas those genes that function to repress differentiation or allow NSCs to proliferate, will show increased markers of differentiation under disease conditions.
Figure 4.
Molecular model for neurodevelopmental disorders. Genes with mutations associated with NDDs might affect specific cell processes such as protein translation or chromatin modification in such a way as to impact pathways important in NSC proliferation or differentiation, such as the WNT signaling pathway. The measureable outcome of different genetic variation associated with NDDs may be NSCs with altered regulation of the balance between NSC proliferation and differentiation. These vulnerabilities, specific to each mutation associated with NDDs, might affect the timing of neural stem cell differentiation causing neurons to connect inappropriately in a neural circuit or respond uncharacteristically to attractant or repellent cues. NDD, neurodevelopmental disorder; NSC, neural stem cell.
We provide several lines of supporting evidence that reduced dosage of two different genes leads neural stem cells to adopt features more characteristic of differentiating than proliferating neural stem cells. We show that mRNAs, microRNAs and DNA methylation clusters that are significantly different in SATB2 KD or MBD5 KD compared with non-target controls, significantly overlap with the same features that are significantly different when we compare control proliferating NSCs to differentiating NSCs. This leads us to suggest that reduced dosage of either MBD5 or SATB2 causes neural stem cells to be more characteristic of differentiating cells than proliferating cells, and that this may be a unifying feature of neurodevelopmental disorders more generally. Together with our previous work investigating EHMT1 and TCF4 suppression,33 two other repressive factors thought to regulate the expression of many genes, we suggest that all four molecules act as a brake to repress a cell differentiation program (that is, maintain a proliferative state) in NSCs functioning to ensure the proper timing of neural differentiation. Independent suppression of all four factors in NSCs leads cells to take on characteristics of differentiating cells, though all four NSC models appear to continue to proliferate normally. When we looked at the function of several genes and loci associated with NDDs, we find that they all affect the balance of proliferation and differentiation in neural stem cells (Table 1). This suggests that the current data may not be specific to MBD5 and SATB2 reduced dosage, but may apply to many NDDs; thus we propose the current model (Figure 4).
Predictions of the model
Repression or enhancement of NSC proliferation or differentiation underlie ASDs/NDDs
All genes (TCF4, EHMT1,33 SATB2 and MBD5) that we have studied to develop the current model appear to enhance NSC differentiation, suggesting that NSCs may be primed to differentiate too early; however, a ‘positive control' example of an NDD, Fragile X syndrome caused by a trinucleotide repeat expansion and leading to supressed expression of FMR1, shows deficits in differentiation in iPSC-neural stem cell models,73 possibly suggesting Fragile X syndrome is caused by increased proliferation markers and decreased differentiation markers. In other words, FMR1's role might be to propel NSC differentiation. Similarly, iPSC models of Rett's Syndrome, caused by mutations in MECP2, suggest an impaired neuronal maturation phenotype.74 Together, this implies no ‘directionality' but rather an altered balance of proliferation or differentiation in NSCs as the underlying feature in NDDs.
CNVs implicated in NDDs will show reciprocal effects on NSC differentiation and proliferation (16p11.2 and 1q21.1 as examples)
The structural variants at 16p11.2 are among the most common genetic causes of NDDs and ASDs71 and occur either as a genomic deletion or duplication affecting ~26 genes. Clinical spectrum of the 16p11.2 deletions includes ASDs, language impairment, intellectual disability increased brain volume and body mass index.75 The 16p11.2 duplication is associated with a less severe and more variable phenotype including ASDs, intellectual disability and a decrease in brain volume.71 The 1q21.1 deletion or duplication occur in ~0.7% of all ID/ASD cases.72 The copy number variant (CNV, copy gain or loss) is 1.35 Mb encompassing seven genes, where deletion cases have mild-to-moderate developmental delay, and ~65% show microcephaly.72 Duplication cases also show mild-to-moderate developmental delay, and macrocephaly has been detected in >60% of duplication cases.76
CNVs at 1q21.1 and 16p11.2 may be implicated in neural stem cell proliferation and differentiation. For 16p11.2, a recent paper found that, ‘microcephaly is caused by decreased proliferation of neuronal progenitors with concomitant increase in apoptosis in the developing brain, whereas macrocephaly arises by increased proliferation and no changes in apoptosis'.77 This is direct evidence for our model, though one which has not been assessed in human cells. Evidence from a mouse model of 16p11.2 deletion in mouse also supports these findings—a recent study showed enhanced differentiation and supressed proliferation of neural stem cells.78 Further, ERK1 (aka MAPK3) is one of the 26 genes in the 16p11.2 region, and there are many studies documenting its role in neural differentiation and proliferation.79 For 1q21.1 syndrome, CHD1L is one of seven genes in the CNV regions, and is capable of inducing spontaneous tumors in tissue where it is injected in mice80—strongly suggesting a functional role in cell proliferation. Similarly, BCL9, a gene important in apoptosis and also one of seven genes in the 1q21.1 region, has a well-known role in cell proliferation in tumors.81 The model of NDD proposed here predicts that human NSC models of either 16p11.2 or 1q21.1 CNVs will have reciprocal effects (for example, increased differentiation markers for the deletion and decreased differentiation markers for the duplication).
Cell membrane proteins associated with ASDs/NDDs have a function in NSC proliferation and differentiation
Several genes associated with neurodevelopmental disorders include genes coding for channels (for example, GRIN2B, SCN2A) and synaptic adhesion proteins such as NRXN1 or NLGN4X. All of the genes that we have tested to date (TCF4, EHMT1,33 SATB2 and MBD5) are thought to act as transcriptional repressors either through direct interaction with DNA or indirectly through protein complex intermediaries, and lead to increased NSC differentiation when gene dosage is suppressed. One might suggest that this model might be restricted to repressor molecules that directly interact with the genome; however, we suggest that genes that code for channels or cell adhesion molecules may also be important in maintaining the balance of cell proliferation and differentiation in neural stem cells. We propose that altered electrical balance and cell–cell or cell–matrix contacts may also be important to determining when and where NSCs differentiate, and that this is the primary cause of NDDs. Our model predicts, controversially, that synaptic adhesion may be an important feature of NSC proliferation/differentiation and that altered synaptic connectivity is a secondary effect of altered NSC differentiation, rather than a primary cause of NDDs important to behavior.
The mechanistic property of proteins most associated with ASD is that of synapse assembly, maintenance or connectivity, including such genes as NLGN4, NRXN1, SHANK1/3, SYNGAP1 and many others. We propose that deficits in regulating the proper timing of neural differentiation may lead to inappropriate connections between neurons (Figure 4). NSCs must balance several external signaling cues (attractant or repellent cues, for example) and intrinsic ones (such as birth date) before making a commitment to differentiate. Slight changes to this program may lead to subtle connectivity problems in the brain, which may be expressed later in life as a social communication disorder.
Open questions
The development of this model is based on the genes studied here, our previous work,33 and functional studies of genes implicated in ASD/NDDs with a role in behavior (Table 1). Several questions remain to fully understand the relevance of this model. (1) Are deficits in NSC proliferation and differentiation the primary deficit causing NDDs or a secondary defect? This is testable using assays developed here for studies using iPSC models of disease—a practice that is occurring for almost all genetically defined NDDs. (2) What cell type should be used to study these deficits and are the effects cell autonomous? Modeling NDDs in stem cells is in its infancy and several concerns remain about derivation, differentiation and characterization. It is not immediately clear how these models recapitulate human brain development nor is it clear what cell type should be used (or what, precisely, defines a cell type). Experiments in culture also suffer from an inability to assess context, meaning that NSCs in vivo may behave very differently than cells in a dish. Further, effects observed in one cell type may not be recapitulated in a different cell type, even from the same donor. (3) How does the model apply to NDDs associated with metabolic disorders?
Acknowledgments
This work was funded by a grant from the Scottish Rite Charitable Foundation of Canada to CE, who receives salary support from the Canada Research Chairs program. COG was supported by a CNPQ (Conselho Nacional de Desenvolvimento Científico e Tecnológico) scholarship, ESC and VKO were supported by a fellowship from FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo), KV and JPL are supported by the Canadian Institute of Health Research, and LC is supported by the Fonds de Recherche de Quebec Sante.
The authors declare no conflicts of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Hinds HL, Ashley CT, Sutcliffe JS, Nelson DL, Warren ST, Housman DE, et al. Tissue specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat Genet. 1993;3:36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- Schanen NC.Epigenetics of autism spectrum disorders Hum Mol Genet 200615Spec No 2R138–R150. [DOI] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline H. Synaptogenesis: a balancing act between excitation and inhibition. Curr Biol. 2005;15:R203–R205. doi: 10.1016/j.cub.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Sahin M. Autism and the synapse: emerging mechanisms and mechanism-based therapies. Curr Opin Neurol. 2015;28:91–102. doi: 10.1097/WCO.0000000000000186. [DOI] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bena F, Bruno DL, Eriksson M, van Ravenswaaij-Arts C, Stark Z, Dijkhuizen T, et al. Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:388–403. doi: 10.1002/ajmg.b.32148. [DOI] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation. Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Kalkman HO. A review of the evidence for the canonical Wnt pathway in autism spectrum disorders. Mol Autism. 2012;3:10. doi: 10.1186/2040-2392-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, O'Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Mullegama SV, Rosenfeld JA, van Bon BW, Shen Y, Repnikova EA, et al. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am J Hum Genet. 2011;89:551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zeng H, Lam R, Tempel W, Amaya MF, Xu C, et al. Structural and histone binding ability characterizations of human PWWP domains. PLoS One. 2011;6:e18919. doi: 10.1371/journal.pone.0018919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation. FEBS Lett. 2000;473:1–5. doi: 10.1016/s0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- van Bon BW, Koolen DA, Brueton L, McMullan D, Lichtenbelt KD, Ades LC, et al. The 2q23.1 microdeletion syndrome: clinical and behavioural phenotype. Eur J Hum Genet. 2010;18:163–170. doi: 10.1038/ejhg.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillard S, Dubourg C, Gerard-Blanluet M, Delahaye A, Pasquier L, Dupont C, et al. 2q23.1 microdeletion identified by array comparative genomic hybridisation: an emerging phenotype with Angelman-like features. J Med Genet. 2009;46:847–855. doi: 10.1136/jmg.2008.058156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR, Mullegama SV, Rosenfeld JA, Dagli AI, Hatchwell E, Allen WP, et al. Haploinsufficiency of MBD5 associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures. Eur J Hum Genet. 2010;18:436–441. doi: 10.1038/ejhg.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21:658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Gyorgy AB, Szemes M, de Juan Romero C, Tarabykin V, Agoston DV. SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur J Neurosci. 2008;27:865–873. doi: 10.1111/j.1460-9568.2008.06061.x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Ballif BC, Lucas A, Spence EJ, Powell C, Aylsworth AS, et al. Small deletions of SATB2 cause some of the clinical features of the 2q33.1 microdeletion syndrome. PLoS One. 2009;4:e6568. doi: 10.1371/journal.pone.0006568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, et al. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet. 2003;12:2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- Usui D, Shimada S, Shimojima K, Sugawara M, Kawasaki H, Shigematu H, et al. Interstitial duplication of 2q32.1-q33.3 in a patient with epilepsy, developmental delay, and autistic behavior. Am J Med Genet A. 2013;161A:1078–1084. doi: 10.1002/ajmg.a.35679. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GG, Diallo AB, Poujol R, Nagy C, Staffa A, Vaillancourt K, et al. BisQC: an operational pipeline for multiplexed bisulfite sequencing. BMC Genomics. 2014;15:290. doi: 10.1186/1471-2164-15-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C, McGowan PO, Deleva V, Meaney MJ, Szyf M, Turecki G. The effects of pH on DNA methylation state: in vitro and post-mortem brain studies. J Neurosci Methods. 2008;174:123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Gigek CO, Rosenfeld JA, Diallo AB, Maussion G, Chen GG, et al. Molecular convergence of neurodevelopmental disorders. Am J Hum Genet. 2014;95:490–508. doi: 10.1016/j.ajhg.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Fury W, Batliwalla F, Gregersen PK, Li W. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf Proc IEEE Eng Med Biol Soc. 2006;1:5531–5534. doi: 10.1109/IEMBS.2006.260828. [DOI] [PubMed] [Google Scholar]
- Friedlander DR, Milev P, Karthikeyan L, Margolis RK, Margolis RU, Grumet M. The neuronal chondroitin sulfate proteoglycan neurocan binds to the neural cell adhesion molecules Ng-CAM/L1/NILE and N-CAM, and inhibits neuronal adhesion and neurite outgrowth. J Cell Biol. 1994;125:669–680. doi: 10.1083/jcb.125.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae BI, Tietjen I, Atabay KD, Evrony GD, Johnson MB, Asare E, et al. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science. 2014;343:764–768. doi: 10.1126/science.1244392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefaniuk M, Swiech L, Dzwonek J, Lukasiuk K. Expression of Ttyh1, a member of the Tweety family in neurons in vitro and in vivo and its potential role in brain pathology. J Neurochem. 2010;115:1183–1194. doi: 10.1111/j.1471-4159.2010.07023.x. [DOI] [PubMed] [Google Scholar]
- Shiihara T, Watanabe M, Moriyama K, Uematsu M, Sameshima K. A novel PLP1 frameshift mutation causing a milder form of Pelizaeus-Merzbacher disease. Brain Dev. 2014;37:455–458. doi: 10.1016/j.braindev.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Schulz DM, Kalkhof S, Schmidt A, Ihling C, Stingl C, Mechtler K, et al. Annexin A2/P11 interaction: new insights into annexin A2 tetramer structure by chemical crosslinking, high-resolution mass spectrometry, and computational modeling. Proteins. 2007;69:254–269. doi: 10.1002/prot.21445. [DOI] [PubMed] [Google Scholar]
- Jacovina AT, Zhong F, Khazanova E, Lev E, Deora AB, Hajjar KA. Neuritogenesis and the nerve growth factor-induced differentiation of PC-12 cells requires annexin II-mediated plasmin generation. J Biol Chem. 2001;276:49350–49358. doi: 10.1074/jbc.M106289200. [DOI] [PubMed] [Google Scholar]
- Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, Evans C, et al. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71:1313–1324. doi: 10.1158/0008-5472.CAN-10-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, Deng PY, et al. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84:311–323. doi: 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY, Cui YM, et al. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol. 2014;232:415–427. doi: 10.1002/path.4309. [DOI] [PubMed] [Google Scholar]
- Sharifi M, Salehi R, Gheisari Y, Kazemi M. Inhibition of microRNA miR-92a induces apoptosis and inhibits cell proliferation in human acute promyelocytic leukemia through modulation of p63 expression. Mol Biol Rep. 2014;41:2799–2808. doi: 10.1007/s11033-014-3134-5. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SJ, Kim JW, Kim KC, Han SM, Go HS, Seo JE, et al. Translational regulation of NeuroD1 expression by FMRP: involvement in glutamatergic neuronal differentiation of cultured rat primary neural progenitor cells. Cell Mol Neurobiol. 2014;34:297–305. doi: 10.1007/s10571-013-0014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Zhang K, Wang Y, Wang J, Cui Y, Li S, et al. MicroRNA-130b targets Fmr1 and regulates embryonic neural progenitor cell proliferation and differentiation. Biochem Biophys Res Commun. 2013;439:493–500. doi: 10.1016/j.bbrc.2013.08.096. [DOI] [PubMed] [Google Scholar]
- Scotto-Lomassese S, Nissant A, Mota T, Neant-Fery M, Oostra BA, Greer CA, et al. Fragile X mental retardation protein regulates new neuron differentiation in the adult olfactory bulb. J Neurosci. 2011;31:2205–2215. doi: 10.1523/JNEUROSCI.5514-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan MA, Cabernard C, Heck J, Luois S, Doe CQ, Zarnescu DC. Fragile X protein controls neural stem cell proliferation in the Drosophila brain. Hum Mol Genet. 2010;19:3068–3079. doi: 10.1093/hmg/ddq213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castren M, Tervonen T, Karkkainen V, Heinonen S, Castren E, Larsson K, et al. Altered differentiation of neural stem cells in fragile X syndrome. Proc Natl Acad Sci USA. 2005;102:17834–17839. doi: 10.1073/pnas.0508995102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhong X, Chau KF, Santistevan NJ, Guo W, Kong G, et al. Cell cycle-linked MeCP2 phosphorylation modulates adult neurogenesis involving the Notch signalling pathway. Nat Commun. 2014;5:5601. doi: 10.1038/ncomms6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri L, Cambiaghi M, Cominelli M, Alfaro-Cervello C, Cursi M, Pala M, et al. Sustained activation of mTOR pathway in embryonic neural stem cells leads to development of tuberous sclerosis complex-associated lesions. Cell Stem Cell. 2011;9:447–462. doi: 10.1016/j.stem.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Mandel S, Spivak-Pohis I, Gozes I. ADNP differential nucleus/cytoplasm localization in neurons suggests multiple roles in neuronal differentiation and maintenance. J Mol Neurosci. 2008;35:127–141. doi: 10.1007/s12031-007-9013-y. [DOI] [PubMed] [Google Scholar]
- Pascual M, Guerri C. The peptide NAP promotes neuronal growth and differentiation through extracellular signal-regulated protein kinase and Akt pathways, and protects neurons co-cultured with astrocytes damaged by ethanol. J Neurochem. 2007;103:557–568. doi: 10.1111/j.1471-4159.2007.04761.x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi D, Yoshimatsu T, Hozumi K, Gotoh Y. Selection of differentiating cells by different levels of delta-like 1 among neural precursor cells in the developing mouse telencephalon. Development. 2008;135:3849–3858. doi: 10.1242/dev.024570. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, et al. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Mendez-Gomez HR, Vergano-Vera E, Abad JL, Bulfone A, Moratalla R, de Pablo F, et al. The T-box brain 1 (Tbr1) transcription factor inhibits astrocyte formation in the olfactory bulb and regulates neural stem cell fate. Mol Cell Neurosci. 2011;46:108–121. doi: 10.1016/j.mcn.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Valli E, Trazzi S, Fuchs C, Erriquez D, Bartesaghi R, Perini G, et al. CDKL5, a novel MYCN-repressed gene, blocks cell cycle and promotes differentiation of neuronal cells. Biochim Biophys Acta. 2012;1819:1173–1185. doi: 10.1016/j.bbagrm.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Chow LM, Bayazitov IT, Tong Y, Gilbertson RJ, Zakharenko SS, et al. Pten deletion causes mTorc1-dependent ectopic neuroblast differentiation without causing uniform migration defects. Development. 2012;139:3422–3431. doi: 10.1242/dev.083154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Skoultchi AI, Nakayama KI. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-beta-catenin signaling pathway. Mol Cell Biol. 2012;32:501–512. doi: 10.1128/MCB.06409-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Alcantar A, Gonzalez-Sandoval A, Escalante-Alcalde D, Lomeli H. Dynamics of expression of ARID1A and ARID1B subunits in mouse embryos and in cells during the cell cycle. Cell Tissue Res. 2011;345:137–148. doi: 10.1007/s00441-011-1182-x. [DOI] [PubMed] [Google Scholar]
- Fukai R, Hiraki Y, Yofune H, Tsurusaki Y, Nakashima M, Saitsu H, et al. A case of autism spectrum disorder arising from a de novo missense mutation in POGZ J Hum Genet 2015. doi: 10.1038/jhg.2015.13e-pub ahead of print 19 February 2015. [DOI] [PubMed]
- Nozawa RS, Nagao K, Masuda HT, Iwasaki O, Hirota T, Nozaki N, et al. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol. 2010;12:719–727. doi: 10.1038/ncb2075. [DOI] [PubMed] [Google Scholar]
- Nicetto D, Hahn M, Jung J, Schneider TD, Straub T, David R, et al. Suv4-20 h histone methyltransferases promote neuroectodermal differentiation by silencing the pluripotency-associated Oct-25 gene. PLoS Genet. 2013;9:e1003188. doi: 10.1371/journal.pgen.1003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Smibert CA, Kaplan DR, Miller FD. An eIF4E1/4E-T complex determines the genesis of neurons from precursors by translationally repressing a proneurogenic transcription program. Neuron. 2014;84:723–739. doi: 10.1016/j.neuron.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Schuetz G, Rosario M, Grimm J, Boeckers TM, Gundelfinger ED, Birchmeier W. The neuronal scaffold protein Shank3 mediates signaling and biological function of the receptor tyrosine kinase Ret in epithelial cells. J Cell Biol. 2004;167:945–952. doi: 10.1083/jcb.200404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhang P, Shi L, Yamamoto V, Lu W, Wang K. Functional impacts of NRXN1 knockdown on neurodevelopment in stem cell models. PLoS One. 2013;8:e59685. doi: 10.1371/journal.pone.0059685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Chang X, Zhang P, Coba MP, Lu W, Wang K. The functional genetic link of NLGN4X knockdown and neurodevelopment in neural stem cells. Hum Mol Genet. 2013;22:3749–3760. doi: 10.1093/hmg/ddt226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuric U, Cheung AY, Zhang W, Mok RS, Lai W, Piekna A, et al. MECP2e1 isoform mutation affects the form and function of neurons derived from Rett syndrome patient iPS cells. Neurobiol Dis. 2015;76C:37–45. doi: 10.1016/j.nbd.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson E, Nasir RH, Fong A, Lian A, Hundley R, Shen Y, et al. Cognitive and behavioral characterization of 16p11.2 deletion syndrome. J Dev Behav Pediatr. 2010;31:649–657. doi: 10.1097/DBP.0b013e3181ea50ed. [DOI] [PubMed] [Google Scholar]
- Dolcetti A, Silversides CK, Marshall CR, Lionel AC, Stavropoulos DJ, Scherer SW, et al. 1q21.1 Microduplication expression in adults. Genet Med. 2013;15:282–289. doi: 10.1038/gim.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucilowska J, Vithayathil J, Tavares EJ, Kelly C, Karlo JC, Landreth GE. The 16p11.2 Deletion Mouse Model of Autism Exhibits Altered Cortical Progenitor Proliferation and Brain Cytoarchitecture Linked to the ERK MAPK Pathway. J Neurosci. 2015;35:3190–3200. doi: 10.1523/JNEUROSCI.4864-13.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Gao Y, Xiao Z, Chen B, Han J, Zhang J, et al. Erk1/2 promotes proliferation and inhibits neuronal differentiation of neural stem cells. Neurosci Lett. 2009;461:252–257. doi: 10.1016/j.neulet.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Chen M, Huang JD, Hu L, Zheng BJ, Chen L, Tsang SL, et al. Transgenic CHD1L expression in mouse induces spontaneous tumors. PLoS One. 2009;4:e6727. doi: 10.1371/journal.pone.0006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani M, Carrasco DE, Zhang Y, Takada K, Gatt ME, Dutta-Simmons J, et al. BCL9 promotes tumor progression by conferring enhanced proliferative, metastatic, and angiogenic properties to cancer cells. Cancer Res. 2009;69:7577–7586. doi: 10.1158/0008-5472.CAN-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.