Abstract
The transcription factor forkhead box F2 (FOXF2) is an evolutionarily conserved DNA-binding protein involved in embryogenesis and metabolism. Although recent studies prove that FOXF2 is a tumor suppressor in various human cancers, the role of FOXF2 in esophageal squamous cell carcinoma (ESCC) remains unknown. Therefore, samples were collected from 188 ESCC patients, including 33 pairs of tumor and non-tumor tissues, and FOXF2 mRNA expression was investigated by quantitative polymerase chain reaction. The results demonstrated that FOXF2 mRNA is downregulated in tumor tissues compared to paired non-tumor tissues (P=0.048). The receiver operating characteristic curve analysis indicated 1.2 as a cut-off point and, thus, 125 and 63 tumors were classified as low- and high-level FOXF2 mRNA expression, respectively. We observed that low-level FOXF2 mRNA expression in the tumors was associated with a higher frequency of lymph node metastasis (P=0.044), an effect further suggested by the multivariate logistic regression analysis (P=0.060). According to the univariate Cox analysis, patients harboring tumors with low-level FOXF2 mRNA expression had a significantly increased mortality risk compared to those with high-level expression (hazard ratio=1.700, 95% confidence interval, 1.077–2.681), with 5-year survival rates of 41.1 and 61.9%, respectively. This negative prognostic effect of low-level FOXF2 mRNA expression was further validated in the multivariate Cox analysis (P=0.021). The subgroup analysis demonstrated that the effect of FOX2 mRNA expression was limited to male patients and those with advanced-stage disease. Taken together, these findings suggest that FOXF2 may be an anti-oncogene for ESCC and decreased FOXF2 mRNA expression is associated with a poor prognosis in patients with ESCC.
Keywords: biomarker, gene expression, transcription factor forkhead box F2, esophageal cancer, outcomes
Introduction
Esophageal squamous cell carcinoma (ESCC) is a common type of cancer worldwide. Despite the significant improvements in diagnostic methods, surgical resection and multidisciplinary therapy, the outcome remains dismal, with a 5-year survival rate of <40% (1,2). Although the tumor-node-metastasis staging system is well established (3), patients with the same stage commonly present with different survival rates. Therefore, it is crucial to evaluate the molecular and genetic characteristics of ESCC.
The forkhead box family of transcription factors comprises evolutionarily conserved DNA-binding proteins that are present in several organisms (4,5). As a member of this family, transcription factor forkhead box F2 (FOXF2) has been well described as an essential signaling molecule for embryogenesis and metabolism (6–12). Experiments in vivo have revealed cleft palate and gastrointestinal defects in FOXF2 knockout mice (6,7). Studies in humans also demonstrated associations between FOXF2 mutations and congenital diseases (8,9). In addition, a recent study proved that FOXF2 is involved in the process of glycose metabolism (10).
Recently, decreased FOXF2 expression was shown to promote tumor development (13) and display several functions critical for cancer initiation and progression. Moreover, reduced FOXF2 mRNA expression was found to be associated with early-onset metastasis and poor prognosis in breast cancer patients (14). It was also reported that FOXF2 may be downregulated by microRNA-182 (15), which has been proved to accelerate metastasis and promote cell invasion (16,17). However, the clinicopathological and prognostic significance of FOXF2 in human ESCC remains unknown.
To elucidate the clinicopathological and prognostic value of FOXF2 in ESCC, we determined FOXF2 mRNA expression by quantitative polymerase chain reaction (qPCR) and evaluated its feasibility as a biomarker for ESCC patients.
Materials and methods
Patient selection
Following approval by the local Institutional Ethics Committee, a series of 188 consecutive patients with ESCC who underwent esophagectomy with extended two-field lymphadenectomy at the Department of Thoracic Oncology of Sun Yat-sen University Cancer Center between January, 2002 and December, 2008, were enrolled in this study. Written informed consent was provided by the participants for their clinical records to be used in this study. All patient data were anonymized and de-identified in a confidential manner.
The inclusion criteria were as follows: i) Pathologically diagnosed ESCC; ii) complete surgical resection; iii) no distant metastasis; iv) no preexisting/concurrent malignant disease or a second primary tumor; v) no perioperative mortality; and vi) availability of fresh samples. Patients receiving neoadjuvant or adjuvant treatment were excluded. The pretreatment evaluation included a complete history and physical examination, complete blood cell count, serum biochemistry, chest radiography, esophageal barium meal, computed tomography scan of the cervical region, chest and abdomen, endoscopy and ultrasonography scan of the abdomen. The pathological staging was reverified based on the 7th American Joint Committee on Cancer (AJCC) staging system (3).
Following primary treatment, the majority of the patients were followed up in the outpatient clinic every 3 months during the first 2 years, every 6 months during years 3–5 and every 12 months thereafter. The survival status was reverified using the best available method in June, 2014. The median time from surgery to the last censoring date for the entire cohort was 68.5 months.
qPCR assays
Fresh tumor and non-tumor samples were collected from regions that were macroscopically assessed as neoplastic and normal, respectively. The samples were immediately stored on dry ice after resection and then frozen at −80°C. Total RNA was extracted from the specimens using the TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Each cDNA was synthesized from 1 µg of total RNA using RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) and stored at −80°C. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene, was selected as an internal standard to control for amplification variability. The following primers were used: FOXF2, forward 5′-CACTACTGGACCATCGACCC-3′; and reverse 5′-CTCACCACGCGGTGGTACAT-3′ (NCBI: NM_001452.1); and GAPDH, forward 5′-ACT TCAACAGCGACACCCACTC-3′ and reverse 5′-TACCAGGAAATGAGCTTGACAAAG-3′ (NCBI: NM_001256799.1). As the reverse transcription (RT)-qPCR assays were not performed at the same time, we utilized FOXF2 mRNA expression of the EC109 ESCC cell line as an internal control (calibrator) to adjust variation. The PCR mixture of each PCR analysis included 0.12 µl of cDNA, 5 µl of 2x Power SYBR® Green PCR Master Mix (Applied Biosystems, Life Technologies, Grand Island, NY, USA), 0.25 µl of 20 mmol/l forward primer, 0.25 µl of reverse primer and 4.38 µl of distilled water. RT-qPCR was performed using LightCycler 480 (Roche Applied Science, Penzberg, Germany) with the following thermal cycling profile: 95°C for 10 min, followed by 40 cycles of amplification (95°C for 10 sec and 60°C for 20 sec) and then 72°C for 30 sec. Each assay was performed at least three times. Any samples with a coefficient of variance >10% were retested. The relative expression level of FOXF2 mRNA for each sample was calculated as 2−ΔΔCtsample as follows: ΔΔCtsample = ΔCtcalibrator - ΔCtsample, where ΔCtcalibrator of FOXF2 mRNA = ΔCtcalibrator of FOXF2 - Ctcalibrator of GAPDH; ΔCtsample = Ctsample of FOXF2 - Ctsample of GAPDH. The value of the Ct difference is equal to a 2n-fold difference.
Statistical analysis
The statistical analysis was performed using SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA). A paired two-tailed t-test was employed to analyze the difference in FOXF2 mRNA expression between the paired tumor and non-tumor tissues. A receiver operating characteristic (ROC) curve was generated to select the optimal cut-off value and, therefore, divided the FOXF2 mRNA expression into two groups, namely low- and high-expression groups. The χ2 test was used to determine the associations between FOXF2 mRNA expression and categorized clinicopathological parameters. To determine the factors associated with an increased risk of lymph node metastasis (LNM), crude and adjusted analyses were performed using univariate and multivariate logistic regression.
The cancer-specific survival (CSS) was calculated from the date of surgery to either the date of death from ESCC or the last follow-up. The survival analysis was performed using the Kaplan-Meier method and the differences between curves were assessed by the log-rank test. Univariate and multivariate Cox regression analyses were used to identify the factors associated with prognosis. For selecting variables for the multivariate logistic/Cox regression model, a cut-off value of P=0.10 was used. P<0.05 was considered to indicate a statistically significant difference.
Results
Human FOXF2 mRNA expression in tumor and non-tumor tissues
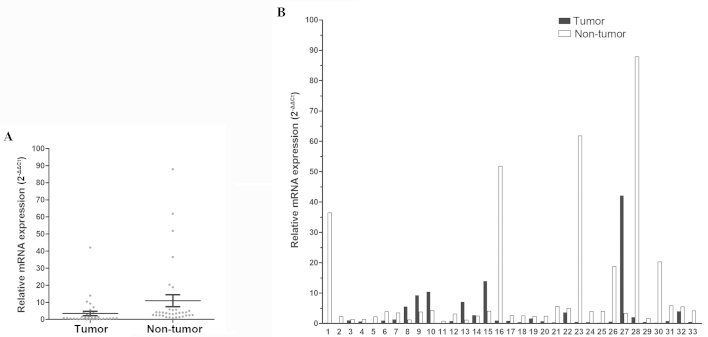
In total, 33 patients with paired tumor and non-tumor tissues were enrolled in this study. FOXF2 was found to be significantly downregulated in tumor compared to non-tumor tissues from the same patient (P=0.048, Fig. 1A). The median value of the normal/tumor (N/T) ratio of FOXF2 mRNA expression was 4.1; the N/T ratio was >two-fold in 66.7% of the patients (22/33) and the highest ratio was ≤165.91-fold (Fig. 1B).
Figure 1.
Analysis of FOXF2 mRNA expression in esophageal tissues. (A) FOXF2 mRNA expression was significantly downregulated in tumor tissue (P=0.048). (B) FOXF2 mRNA levels in 33 paired non-tumor and tumor tissues were compared individually. The gene expression results were normalized to the internal control glyceraldehyde-3-phosphate dehydrogenase. FOXF2, forkhead box F2.
Association between FOXF2 mRNA expression level and clinicopathological parameters
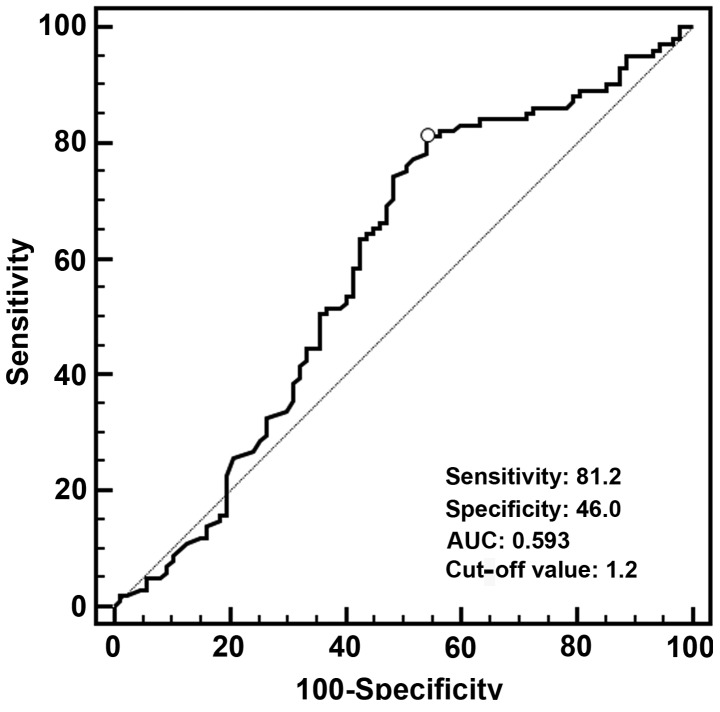
A total of 188 patients with available tumor tissue samples were enrolled in this study, with a median age of 59 years (range, 34–88 years). The patient characteristics are summarized in Table I. The median value of FOXF2 mRNA expression was 0.70 in the tumor tissue samples (range, 0.02–42.10). According to the ROC curve (Fig. 2), the optimal cut-off value of FOXF2 mRNA with the best discriminatory power was 1.2. Using this cut-off value, the entire cohort was classified into two groups, namely high-level (>1.2; n=125) and low-level (≤1.2; n=63) FOXF2 mRNA expression. There was no significant association between FOXF2 mRNA expression and age, gender, tumor location, tumor length, tumor cell differentiation, pathological T stage, AJCC stage and the number of resected lymph nodes. However, we observed that the rate of LNM was significantly associated with a low level of FOXF2 mRNA expression (P=0.044) (Table I).
Table I.
Correlations between FOXF2 expression and clinicopathologicalcharacteristics.
| Characteristics | Total (n=188) | FOXF2 mRNA expression | P-valuea | |
|---|---|---|---|---|
| High (n=63) | Low (n=125) | |||
| Age, years | 0.198 | |||
| ≤59b | 98 | 37 (58.7) | 61 (48.8) | |
| >59 | 90 | 26 (41.3) | 64 (51.2) | |
| Gender | 0.468 | |||
| Female | 51 | 15 (23.8) | 36 (28.8) | |
| Male | 137 | 48 (76.2) | 89 (71.2) | |
| Tumor location | 0.374 | |||
| Upper | 36 | 15 (23.8) | 21 (16.8) | |
| Middle | 104 | 35 (55.6) | 69 (55.2) | |
| Lower | 48 | 13 (20.6) | 35 (28.0) | |
| Tumor length, cm | 0.380 | |||
| ≤4.2b | 90 | 33 (52.4) | 57 (45.6) | |
| >4.2 | 98 | 30 (47.6) | 68 (54.4) | |
| Tumor cell differentiation | 0.888 | |||
| High | 45 | 14 (22.2) | 31 (24.8) | |
| Moderate | 97 | 34 (54.0) | 63 (50.4) | |
| Poor | 46 | 15 (23.8) | 31 (24.8) | |
| pT stage | 0.353 | |||
| T1b-T2 | 46 | 18 (28.6) | 28 (22.4) | |
| T3-T4a | 142 | 45 (71.4) | 97 (77.6) | |
| pN stage | 0.044 | |||
| N0 | 100 | 40 (63.5) | 60 (48.0) | |
| N1/N2/N3 | 88 | 23 (36.5) | 65 (52.0) | |
| AJCC stage | 0.088 | |||
| I–II | 106 | 41 (65.1) | 65 (52.0) | |
| III | 82 | 22 (34.9) | 60 (48.0) | |
| Resected lymph | 0.656 | |||
| nodes, no. ± SD | 24.0±11.8 | 23.8±11.9 | 24.6±11.9 | |
χ2 test
Median. FOXF2, forkhead box F2; AJCC, American Joint Committee on Cancer; SD, standard deviation.
Figure 2.
Receiver operating characteristic curve using FOXF2 mRNA expression levels. The optimal cut-off value was 1.20, with a sensitivity of 81.2% and a specificity of 46.0%. AUC, area under the curve; FOXF2, forkhead box F2.
Evaluation of FOXF2 mRNA expression as a risk factor for LNM
In the univariate logistic regression analysis, patients with a low level of FOXF2 mRNA expression exhibited a significantly higher risk of LNM compared to those with a high level of expression, with a hazard ratio (HR) of 1.884 [95% confidence interval (CI): 1.012–3.507]. This effect was further observed in the multivariate logistic analysis, with a marginal significance (P=0.060) and an adjusted HR of 1.828 (95% CI: 0.975–3.430). The independent risk factor for LNM was advanced pathological T stage (P=0.028) (Table II).
Table II.
Univariate and multivariate logistic regression analyses of factors associated with lymph node metastasis.
| Factors | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age, years (≤59/>59) | 1.078 (0.607–1.912) | 0.799 | – | NAa |
| Gender (F/M) | 1.915 (0.985–3.724) | 0.055 | 1.834 (0.921–3.655) | 0.085 |
| Tumor location (U/Md/L) | 1.178 (0.765–1.814) | 0.458 | – | NAa |
| Tumor differentiation (high/moderate/poor) | 1.337 (0.882–2.027) | 0.171 | – | NAa |
| Tumor length, cm (≤4.2/>4.2) | 1.425 (0.801–2.546) | 0.228 | – | NAa |
| pT stage (T1b-T2/T3-T4a) | 2.186 (1.087–4.398) | 0.028 | 2.186 (1.087–4.398) | 0.028 |
| resected lymph nodes, no. (≤21/>21) | 0.984 (0.554–1.747) | 0.955 | – | NAa |
| FOXF2 mRNA expression (high/low) | 1.884 (1.012–3.507) | 0.046 | 1.828 (0.975–3.430) | 0.060 |
Not assessed due to an insignificant result in the univariate analysis (P>0.1). F, female; M, male; U, upper; Md, middle; L, lower; HR, hazard ratio; CI, confidence interval; FOXF2, forkhead box F2.
Association between FOXF2 mRNA expression level and CSS
In this study, the median survival time was 54.0 months, with an estimated 5-year CSS of 48.0%. In the univariate Cox analysis, patients with low-level FOXF2 mRNA expression exhibited a significantly enhanced mortality risk compared to those with high-level expression (HR=1.700, 95% CI: 1.077–2.681), with a 5-year CSS of 41.1 and 61.9%, respectively (Fig. 3A). This effect was further verified in the multivariate Cox analysis, with an adjusted HR of 1.714 (95% CI: 1.085–2.708). Other negative prognostic factors with independent significance included advanced AJCC stage (P<0.001) and a resected lymph node number of ≤21 (P=0.016). The details of the univariate and multivariate analyses are shown in Table III.
Figure 3.
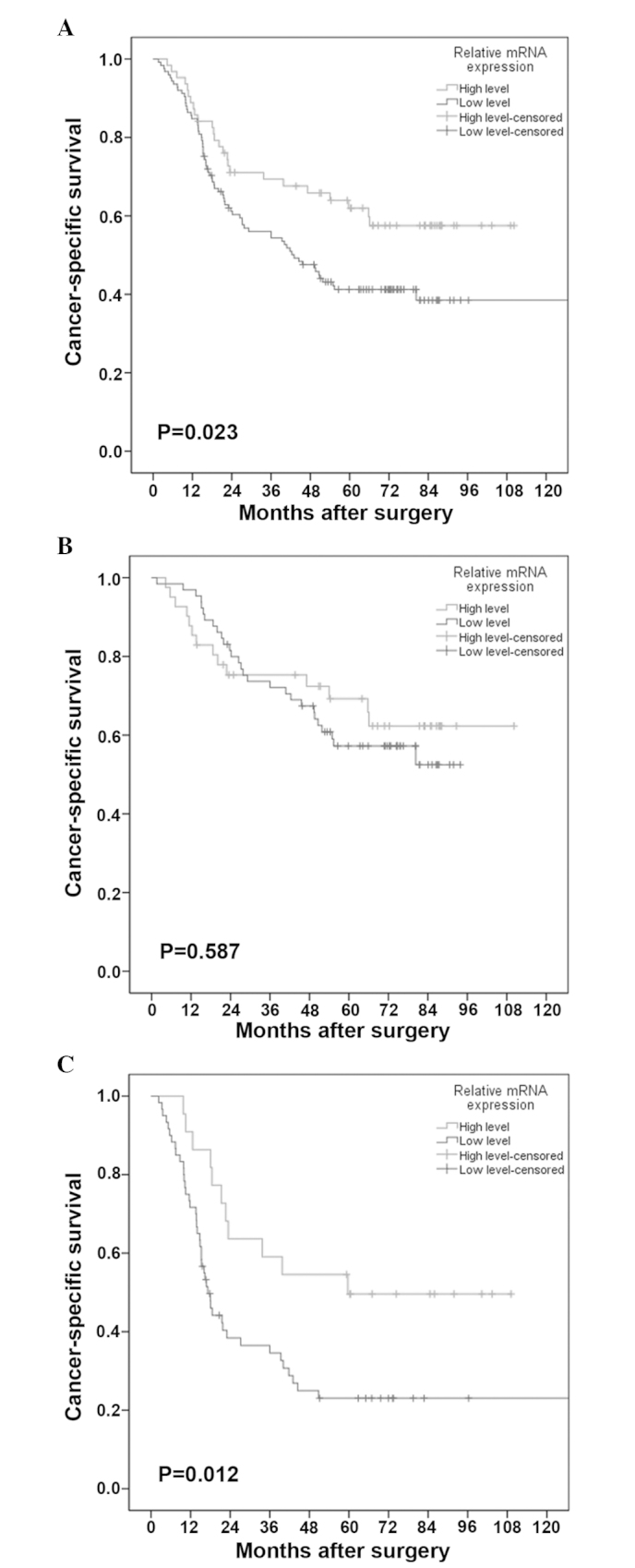
Cancer-specific survival curves stratified by FOXF2 mRNA expression levels in (A) the entire cohort, (B) patients with stage I/II disease and (C) patients with stage III disease. FOXF2, forkhead box F2.
Table III.
Univariate and multivariate Cox analyses of factors associated with cancer-specific survival.
| Factors | Univariate Cox analysis | Multivariate Cox analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age, years (≤59/>59) | 1.300 (0.872–1.936) | 0.198 | – | NAa |
| Gender (F/M) | 1.119 (0.717–1.745) | 0.621 | – | NAa |
| Tumor location (U/Md/L) | 1.131 (0.837–1.527) | 0.422 | – | NAa |
| Tumor differentiation (high/moderate/poor) | 1.352 (1.009–1.812) | 0.043 | 1.276 (0.939–1.735) | 0.119 |
| Tumor length, cm (≤4.2/>4.2) | 0.913 (0.613–1.360) | 0.654 | – | NAa |
| AJCC stage (I/II/III) | 2.384 (1.660–3.424) | <0.001 | 2.482 (1.728–3.566) | <0.001 |
| resected lymph nodes, no. (≤21/>21) | 0.646 (0.431–0.971) | 0.035 | 0.606 (0.403–0.910) | 0.016 |
| FOXF2 mRNA expression (high/low) | 1.700 (1.077–2.681) | 0.023 | 1.714 (1.085–2.708) | 0.021 |
Not assessed due to an insignificant result in the univariate analysis (P>0.1). F, female; M, male; U, upper; Md, middle; L, lower; HR, hazard ratio; CI, confidence interval; FOXF2, forkhead box F2; AJCC, American Joint Committee on Cancer.
Prognostic significance of FOXF2 mRNA expression level in the subgroup analysis
The association between FOXF2 mRNA expression and CSS across strata of other potential predictors of patient outcome were assessed. As shown in Table IV, following adjustment for known prognostic factors, the increased risk of cancer-related mortality conferred by low-level FOXF2 mRNA expression was unchanged in male patients (adjusted HR=1.790, 95% CI: 1.044–3.072) and patients with advanced-stage disease (adjusted HR=2.924, 95% CI: 1.466–5.833). However, this association was insignificant in other subgroup analyses. The CSS curves stratified by FOXF2 mRNA expression level in patients with stage I/II and patients with stage III disease are shown in Fig. 3B and C, respectively.
Table IV.
Subgroup analysis for FOXF2 mRNA expression.
| Factors | 5-year OS (%) | FOXF2 mRNA expression (no. of events/no. at risk) | ||||
|---|---|---|---|---|---|---|
| Multivariate Cox analysis | ||||||
| High | Low | HR | 95% CI | P-value | ||
| Age, years | ||||||
| ≤59a | 53.1 | 13/37 | 34/61 | 1.862 | 0.980–3.539 | 0.058 |
| >59 | 42.4 | 12/26 | 38/64 | – | – | NAb |
| Gender | ||||||
| Female | 46.6 | 7/15 | 20/36 | – | – | NAb |
| Male | 49.9 | 18/48 | 52/89 | 1.790 | 1.044–3.072 | 0.034 |
| Tumor location | ||||||
| Upper | 56.4 | 5/15 | 11/21 | – | – | NAb |
| Middle | 46.0 | 15/35 | 41/69 | 1.667 | 0.921–3.017 | 0.091 |
| Lower | 47.1 | 5/13 | 20/35 | – | – | NAb |
| Tumor length, cm | ||||||
| ≤4.2a | 47.4 | 14/33 | 35/57 | – | – | NAb |
| >4.2 | 49.8 | 11/30 | 37/68 | 1.777 | 0.945–3.340 | 0.074 |
| AJCC stage | ||||||
| I–II | 61.3 | 14/41 | 28/65 | – | – | NAb |
| III | 20.7 | 11/22 | 44/60 | 2.924 | 1.466–5.833 | 0.002 |
| Resected lymph nodes, no. | ||||||
| ≤21a | 41.7 | 15/32 | 43/68 | – | – | NAb |
| >21 | 55.6 | 10/31 | 29/57 | 2.038 | 0.987–4.205 | 0.054 |
Median.
Not assessed due to an insignificant result in the univariate analysis (P>0.1). FOXF2, forkhead box F2; OS, overall survival; HR, hazard ratio; CI, confidence interval; AJCC, American Joint Committee on Cancer.
Discussion
It was previously demonstrated that FOXF2 acts as a tumor suppressor in breast and prostate cancer (14,15). In this study, we observed that FOXF2 mRNA expression was significantly higher in normal esophageal tissue compared to that in tumor specimens. Thus, FOXF2 may act as a tumor suppressor in ESCC. We hypothesized that FOXF2 maintains a normal expression level in normal esophageal epithelial cells and plays an important role in balancing cell behavior, such as proliferation, differentiation, mitotic cycle and apoptosis, which was also supported by previous studies (7,11–13). However, FOXF2 expression may be compromised in ESCC during the course of tumorigenesis, similar to prostate cancer (18,19). Although the precise mechanism is not known, one possible explanation involves FOXF2 downregulation by microRNA-182 (15), the overexpression of which has been observed in various human cancers, such as colorectal (20), cervical (21) and non-small-cell lung cancer (22). In addition, we observed that a low level of FOXF2 mRNA expression was associated with a high rate of LNM; the same result was obtained in patients with breast cancer (14).
When our analyses focused exclusively on patient survival, the level of FOXF2 mRNA expression was found to be an independent and significant predictive variable. The 5-year CSS was significantly higher in patients with high-level FOXF2 mRNA expression compared to that in patients with low-level expression. The effect of FOXF2 on prognosis may be attributed to its intrinsic nature as an anti-oncogene.
Several studies have been conducted to elucidate how FOXF2 exerts its tumor suppressor effect. It was reported that FOXF2 may downregulate matrix metalloproteinases (MMPs) and upregulate tissue inhibitor of metalloproteinase-3, a known inhibitor of MMPs (15,19). Due to their ability to degrade the extracellular matrix, activated MMPs accelerate metastasis and decrease survival in patients with ESCC (23), colorectal (24), breast (25) and gastric cancer (26). In addition, FOXF2 was reported to inhibit the Wnt pathway (7,13), the activation of which has been associated with a high risk of metastasis and a poor outcome in patients with pancreatic (27), breast (28) and prostate cancer (29). However, the potential mechanisms of FOXF2 as a tumor suppressor in ESCC remain unknown and require further investigation.
In our subgroup analysis, we observed that the increased risk conferred by low-level FOXF2 mRNA expression was limited to patients with stage III disease. A possible explanation may be associated with the fact that the survival rate is generally relatively low in patients with advanced ESCC; therefore, there may be a tendency to observe significant survival differences according to a certain factor. Second, the superior outcomes in the high FOXF2 mRNA expression group may be partially associated with the role of FOXF2 in suppressing metastasis (7,13,15,19); this effect is expected to be more prominent in patients with a high predisposition for metastasis, which was also indicated by Kong et al (14); in that study, the poor effect of decreased FOXF2 mRNA expression on survival in breast cancer was limited to patients with a triple-negative profile, a well-known subtype characterized by early-onset metastasis and a dismal outcome (30). Additionally, we found FOXF2 mRNA expression to be associated with survival in male patients. Since the interaction between gender and FOXF2 remains unknown, we hypothesized that this phenomenon may be partially attributed to the high risk of LNM in male patients (adjusted HR=1.834, 95% CI: 0.921–3.655) in this study.
To date, advances in molecular biology have led to the rapid development of individualized management in various human cancers. Based on a cohort of patients treated by surgery alone, we observed that patients with low-level FOXF2 mRNA expression had a significantly lower CSS compared to those with high-level expression. Furthermore, this effect was independent of the aggressiveness of lymphadenectomy and patient characteristics. In this sense, surgery alone may be insufficient for patients with a low level of FOXF2 mRNA expression and multidisciplinary therapy should be recommended. Future studies should focus on the interaction between chemotherapy/chemoradiotherapy and FOXF2 expression to verify our hypotheses.
In conclusion, FOXF2 may be an anti-oncogene in ESCC. Decreased FOXF2 mRNA expression was found to be associated with poor prognosis in patients with completely resected ESCC. However, the clinical value of the changes in FOXF2 mRNA levels in ESCC require further validation by large multicenter studies.
Acknowledgements
This study was supported by grants from the Fundamental Research Funds for the Central Universities (no. 13ykpy49) and the National Natural Science Foundation of China (no. 81402003).
References
- 1.Rice TW, Rusch VW, Apperson-Hansen C, et al. Worldwide esophageal cancer collaboration. Dis Esophagus. 2009;22:1–8. doi: 10.1111/j.1442-2050.2008.00901.x. [DOI] [PubMed] [Google Scholar]
- 2.Allum WH, Stenning SP, Bancewicz J, Clark PI, Langley RE. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J Clin Oncol. 2009;27:5062–5067. doi: 10.1200/JCO.2009.22.2083. [DOI] [PubMed] [Google Scholar]
- 3.Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. 7th. Springer; Chicago: 2010. pp. 67–72. [Google Scholar]
- 4.Carlsson P, Mahlapuu M. Forkhead transcription factors: Key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 5.Katoh M, Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- 6.Wang T, Tamakoshi T, Uezato T, Shu F, Kanzaki-Kato N, Fu Y, Koseki H, Yoshida N, Sugiyama T, Miura N. Forkhead transcription factor FoxF2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev Biol. 2003;259:83–94. doi: 10.1016/S0012-1606(03)00176-3. [DOI] [PubMed] [Google Scholar]
- 7.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- 8.Jochumsen U, Werner R, Miura N, Richter-Unruh A, Hiort O, Holterhus PM. Mutation analysis of FOXF2 in patients with disorders of sex development (DSD) in combination with cleft palate. Sex Dev. 2008;2:302–308. doi: 10.1159/000195679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu L, Wynn J, Ma L, et al. De novo copy number variants are associated with congenital diaphragmatic hernia. J Med Genet. 2012;49:650–659. doi: 10.1136/jmedgenet-2012-101135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westergren R, Nilsson D, Heglind M, Arani Z, Grände M, Cederberg A, Ahrén B, Enerbäck S. Overexpression of Foxf2 in adipose tissue is associated with lower levels of IRS1 and decreased glucose uptake in vivo. Am J Physiol Endocrinol Metab. 2010;298:E548–E554. doi: 10.1152/ajpendo.00395.2009. [DOI] [PubMed] [Google Scholar]
- 11.Aitola M, Carlsson P, Mahlapuu M, Enerbäck S, Pelto-Huikko M. Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epithelio-mesenchymal interactions. Dev Dyn. 2000;218:136–149. doi: 10.1002/(SICI)1097-0177(200005)218:1<136::AID-DVDY12>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 12.Nik AM, Reyahi A, Pontén F, Carlsson P. Foxf2 in intestinal fibroblasts reduces numbers of Lgr5+ stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology. 2013;144:1001–1011. doi: 10.1053/j.gastro.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 13.van den Brink GR, Rubin DC. Foxf2: A mesenchymal regulator of intestinal adenoma development. Gastroenterology. 2013;144:873–876. doi: 10.1053/j.gastro.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong PZ, Yang F, Li L, Li XQ, Feng YM. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PLoS ONE. 2013;8:e61591. doi: 10.1371/journal.pone.0061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirata H, Ueno K, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2, RECK and MTSS1 genes in human prostate cancer. PLoS ONE. 2013;8:e55502. doi: 10.1371/journal.pone.0055502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sachdeva M, Mito JK, Lee CL, et al. MicroRNA-182 drives metastasis of primary sarcomas by targeting multiple genes. J Clin Invest. 2014;124:4305–4319. doi: 10.1172/JCI77116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei R, Tang J, Zhuang X, et al. Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis. Oncogene. 2014;33:1287–1296. doi: 10.1038/onc.2013.65. [DOI] [PubMed] [Google Scholar]
- 18.van der Heul-Nieuwenhuijsen L, Dits NF, Jenster G. Gene expression of forkhead transcription factors in the normal and diseased human prostate. BJU Int. 2009;103:1574–1580. doi: 10.1111/j.1464-410X.2009.08351.x. [DOI] [PubMed] [Google Scholar]
- 19.van der Heul-Nieuwenhuijsen L, Dits N, Van Ijcken W, de Lange D, Jenster G. The FOXF2 pathway in the human prostate stroma. Prostate. 2009;69:1538–1547. doi: 10.1002/pros.20996. [DOI] [PubMed] [Google Scholar]
- 20.Yang MH, Yu J, Jiang DM, Li WL, Wang S, Ding YQ. microRNA-182 targets special AT-rich sequence-binding protein 2 to promote colorectal cancer proliferation and metastasis. J Transl Med. 2014;12:109. doi: 10.1186/1479-5876-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang T, Wong HK, Gu W, Yu MY, To KF, Wang CC, Wong YF, Cheung TH, Chung TK, Choy KW. MicroRNA-182 plays an onco-miRNA role in cervical cancer. Gynecol Oncol. 2013;129:199–208. doi: 10.1016/j.ygyno.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 22.Ning FL, Wang F, Li ML, Yu ZS, Hao YZ, Chen SS. MicroRNA-182 modulates chemosensitivity of human non-small cell lung cancer to cisplatin by targeting PDCD4. Diagn Pathol. 2014;9:143. doi: 10.1186/1746-1596-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu ZD, Li JY, Li M, Gu J, Shi XT, Ke Y, Chen KN. Matrix metalloproteinases expression correlates with survival in patients with esophageal squamous cell carcinoma. Am J Gastroenterol. 2005;100:1835–1843. doi: 10.1111/j.1572-0241.2005.50018.x. [DOI] [PubMed] [Google Scholar]
- 24.Murray GI, Duncan ME, O'Neil P, Melvin WT, Fothergill JE. Matrix metalloproteinase-1 is associated with poor prognosis in colorectal cancer. Nat Med. 1996;2:461–462. doi: 10.1038/nm0496-461. [DOI] [PubMed] [Google Scholar]
- 25.Pellikainen JM, Ropponen KM, Kataja VV, Kellokoski JK, Eskelinen MJ, Kosma VM. Expression of matrix metalloproteinase (MMP)-2 and MMP-9 in breast cancer with a special reference to activator protein-2, HER2 and prognosis. Clin Cancer Res. 2004;10:7621–7628. doi: 10.1158/1078-0432.CCR-04-1061. [DOI] [PubMed] [Google Scholar]
- 26.Koskensalo S, Mrena J, Wiksten JP, Nordling S, Kokkola A, Hagström J, Haglund C. MMP-7 overexpression is an independent prognostic marker in gastric cancer. Tumour Biol. 2010;31:149–155. doi: 10.1007/s13277-010-0020-1. [DOI] [PubMed] [Google Scholar]
- 27.Jiang H, Li Q, He C, et al. Activation of the Wnt pathway through Wnt2 promotes metastasis in pancreatic cancer. Am J Cancer Res. 2014;4:537–544. [PMC free article] [PubMed] [Google Scholar]
- 28.Dey N, Barwick BG, Moreno CS, et al. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer. 2013;13:537. doi: 10.1186/1471-2407-13-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto H, Oue N, Sato A, Hasegawa Y, Yamamoto H, Matsubara A, Yasui W, Kikuchi A. Wnt5a signaling is involved in the aggressiveness of prostate cancer and expression of metalloproteinase. Oncogene. 2010;29:2036–2046. doi: 10.1038/onc.2009.496. [DOI] [PubMed] [Google Scholar]
- 30.Uhm JE, Park YH, Yi SY, et al. Treatment outcomes and clinicopathologic characteristics of triple-negative breast cancer patients who received platinum-containing chemotherapy. Int J Cancer. 2009;124:1457–1462. doi: 10.1002/ijc.24090. [DOI] [PubMed] [Google Scholar]