Abstract
The significance of disseminated tumor cells (DTCs) in bone marrow (BM) on tumor recurrence has not been investigated in patients with hepatocellular carcinoma (HCC). The aim of te present study was to clarify the molecular characteristics of DTCs that affect postoperative recurrence based on microRNA (miRNA) expression profiles in clinical HCC patients undergoing curative resection. DTCs were prospectively collected from the BM of preoperative HCC patients using immunomagnetic beads and subjected to miRNA microarray analysis. Microarray analysis of nine HCC patients (n=5 patients with postoperative HCC recurrence, n=4 patients without HCC recurrence) demonstrated that miR-615-3p is significantly upregulated in the DTCs of patients with recurrence compared to the DTCs from patients without recurrence. In vitro experiments demonstrated that the miR-615-3p expression level is significantly correlated with malignant characteristics in HCC cells. These data suggest that miR-615-3p in DTCs may play an important role in postoperative HCC recurrence, which suggests that miR-615-3p is a potential target molecule for regulating postoperative HCC recurrence.
Keywords: hepatocellular carcinoma, disseminated tumor cells, bone marrow, microRNA, miR-615-3p
Introduction
Hepatocellular carcinoma (HCC) is a common cancer and the third most frequent cause of cancer-related mortalities worldwide (1,2). The prognosis of HCC patients remains poor even following curative treatment, owing to the high incidence of post-treatment recurrence. Additionally, no effective chemotherapy regimens have been established for treating HCC. Furthermore, mechanisms by which HCC recurs are not fully understood (3). To improve the prognosis of HCC patients, the mechanism underlying HCC recurrence requires clarification and new, effective therapies based on that mechanism require development.
Disseminated tumor cells (DTCs) represent the beginning of systemic disease arising from localized cancer (4,5). Over the past 20 years, evidence has been accumulating that the presence of DTCs in bone marrow (BM) is a prognostic indicator for patients with various types of cancer. However, recent studies indicate that all the patients with DTCs do not necessarily develop recurrence and/or metastasis (6,7). In a recent study we demonstrated that DTCs are observed in BM even in patients with early-stage gastric cancer (8). These findings indicate that DTCs from different individuals may have varying characteristics that influence the capacity to develop cancer recurrence and/or metastasis.
microRNAs (miRNAs) constitute a class of small non-coding RNAs that function as a novel class of global, post-transcriptional gene regulators by binding to partially complementary sequences in the 3′ untranslated regions of downstream target mRNAs (9–11). Studies have indicated that miRNAs determine cancer metastasis and recurrence through regulating multiple target genes in numerous types of cancer, including HCC (12–15). Furthermore, our recent study showed that miRNA profiles in DTCs in BM differ significantly between colorectal cancer patients with and without metastasis and that the expression level of specific miRNAs correlates significantly with malignant potential and long-term prognosis (16).
These findings led us to consider that an investigation of the miRNA expression profiles in DTCs from the BM of HCC patients may be useful to identify the mechanisms of HCC recurrence and metastasis based on miRNA-regulated DTC characteristics, which will in turn lead to the development of novel, mechanism-dependent effective therapies. To clarify the mechanism underlying postoperative HCC recurrence, BM samples were prospectively collected from preoperative HCC patients and their miRNA expression profiles were analyzed. Using patient clinical data, an attempt was made to identify the miRNA expression pattern responsible for postoperative recurrence by comparing miRNA expression profiles from the BM of patients with and without postoperative recurrence.
Materials and methods
Cell lines
The human HCC cell lines PLC/PRF/5, HuH7, HLE and HLF were obtained from the Japan Cancer Research Resources Bank (Tokyo, Japan). These cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C in a humidified incubator with 5% CO2.
HCC patients
The study enrolled nine HCC patients who had undergone curative hepatic resection for HCC at the Department of Surgery, Osaka University Hospital (Osaka, Japan) between May 2010 and August 2010. The patient characteristics are shown in Table I. Curative resection was defined as the complete removal of all the macroscopically evident tumors. All the patients were postoperatively followed at regular intervals of 3–4 months to check for postoperative recurrences. The Institutional Review Board of Osaka University Hospital approved the study protocol (IRB #09142) and a signed consent form was obtained from each patient.
Table I.
Characteristics of the HCC patients enrolled in the study.
| No. | Gender | Age, years | Risk factor | Child-Pugh classification | Multiplicity | Maximum tumor size, cm | Tumor thrombus | Postoperative recurrencea |
|---|---|---|---|---|---|---|---|---|
| 1 | M | 37 | HBV | A | Solitary | 1.1 | – | – |
| 2 | F | 69 | HBV | A | Solitary | 2.0 | – | + |
| 3 | M | 70 | HCV | A | Multiple | 1.7 | + | – |
| 4 | M | 72 | HCV | B | Solitary | 3.0 | – | – |
| 5 | M | 65 | – | A | Multiple | 5.0 | – | + |
| 6 | M | 60 | HCV | A | Solitary | 0.9 | – | – |
| 7 | M | 40 | HBV | A | Multiple | 8.0 | + | + |
| 8 | M | 76 | – | A | Solitary | 1.8 | – | + |
| 9 | M | 47 | HBV, HCV | A | Multiple | 3.5 | – | – |
Postoperative HCC recurrence was investigated during 2 years of observation after surgery. HCC, hepatocellular carcinoma; M, male; F, female; HBV, hepatitis B virus; HCV, hepatitis C virus.
Drugs
5-Fluorouracil (5-FU) and cisplatin (CDDP) were kindly supplied by Kyowa Hakko Kogyo Co., Ltd., (Tokyo, Japan) and Pfizer Japan, Inc. (Tokyo, Japan), respectively.
BM samples
BM was aspirated under general anesthesia immediately before surgery, as previously described (17). The BM aspirate was obtained from the ilium of each patient using a BM aspiration needle. The mononuclear cell fraction was isolated and RNA was extracted from the BM sample.
Magnetic-activated cell sorting (MACS)
Cell sorting was performed as previously described (16,18). Briefly, the mononuclear cell fraction was isolated using density-gradient centrifugation. Each fraction was separated by MACS. First, cluster of differentiation 14 (CD14)-allophycocyanin and CD45-fluorescein isothiocyanate antibodies (Miltenyi Biotec, Auburn, CA, USA) were applied to detect human monocytes and macrophages, respectively, and the sorted cells were removed as host-derived cells. Subsequently, using the epithelial cell adhesion molecule (EpCAM)-phycoerythrin antibody (Miltenyi Biotec.), EpCAM-positive cells were sorted from the remaining fraction and used as viable epithelial tumor cells.
Transfection
The oligonucleotide hsa-miR-615-3p (pre-miR-615-3p) and the antisense oligonucleotide inhibitor of hsa-miR-615-3p (anti-miR-615-3p) were purchased from Ambion Inc., (Austin, TX, USA). Pre/anti-miR-615-3p were transfected using Lipofectamine® RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Each scrambled oligonucleotide was transfected in the same way as a matched negative control.
RNA extraction
Total RNA was isolated from cell lines and BM samples with Trizol reagent (Invitrogen) as previously described (19). RNA quality was assessed with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) at 260 and 280 nm wavelengths.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for miRNA expression
RT was performed with the TaqMan MicroRNA RT kit (Applied Biosystems, Foster City, CA, USA) and real-time RT-qPCR was performed with TaqMan MicroRNA assays (Applied Biosystems) using the ABI7900HT system (Applied Biosystems). The expression of the target miRNA was normalized relative to that of the endogenous control, RNU48. Data were analyzed according to the comparative Ct method (20).
RT-qPCR for mRNA expression
RT was performed as previously described (21). Real-time RT-qPCR was performed using designed oligonucleotide primers and the LightCycler 480 Real-Time PCR system (Roche Diagnostics, Mannheim, Germany). To detect the amplification products, the LightCycler-DNA master SYBR-green I (Roche Diagnostics) was used as described previously and the amount of target gene expression was calculated (22). The expression of the target gene was normalized relative to the expression of GAPDH, which was used as an endogenous control. The designed PCR primers were as follows: E-cadherin forward, 5′-TGCCCAGAAAATGAAAAAGG-3′; and reverse primer, 5′-GTGTATGTGGCAATGCGTTC-3′; N-cadherin forward, 5′-TGAAACGCCGGATAAAGAACG-3′; and reverse primer, 5′-TGCTGCAGCTGGCTCAAGTCAT-3′; vimentin forward, 5′-TCTGGATTCACTCCCTCTGG-3′; and reverse primer, 5′-TGCACTGAGTGTGTGCAATTT-3′; fibronectin forward, 5′-CAGTGGGAGACCTCGAGAAG-3′; and reverse primer, 5′-TCCCTCGGAACATCAGAAAC-3′; and GAPDH forward, 5′-GTCGGAGTCAACGGATTTGGT-3′; and reverse primer, 5′-GCCATGGGTGGAATCATATTGG-3′.
miRNA microarray analysis
Extracted total RNA was labeled with Hy5 using the miRCURY LNA Array miR labeling kit (Exiqon, Vedbæk, Denmark). The labeled RNAs were hybridized onto 3D-Gene Human microRNA Oligo chips containing 941 antisense probes printed in duplicate spots (Toray, Kamakura, Japan). The annotation and oligonucleotide sequences of the probes conformed to the miRBase microRNA database (http://www.mirbase.org/). Following stringent washes, fluorescent signals were scanned with the ScanArray Express Scanner (PerkinElmer, Waltham, MA, USA) and analyzed using GenePix Pro version 5.0 (Molecular Devices, Sunnyvale, CA, USA). The raw data for each spot were normalized by substitution with a mean intensity of the background signal, which was determined by the signal intensities of all the blank spots with 95% confidence intervals. Measurements of the duplicate spots with signal intensities greater than two standard deviation (SD) of the background signal intensity were considered valid. The relative expression level of miRNA was calculated by comparing the signal intensities of the averaged valid spots with their mean value throughout the microarray experiments following normalization by their equivalently adjusted median values.
Cell proliferation assay
Cells were uniformly seeded (3×103/well) in triplicate into 96-well dishes (day 0). Cells were counted using the Cell Counting kit-8 (Dojindo, Kumamoto, Japan) on days 1–3. Plate absorbance was measured in a microplate reader at 450 nm.
Growth inhibition assay
The growth inhibition assay was performed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (Sigma-Aldrich Co., St. Louis, MO, USA) assay, as described previously (23). Briefly, cells were incubated for 72 h in several concentrations of 5-FU and CDDP. After re-incubation for 4 h with MTT solution, an acid-isopropanol mixture was added to dissolve the resulting formazan crystals. Plate absorbance was measured in a microplate reader at a wavelength of 550 nm with a 650 nm reference and the results were expressed as a percentage of absorbance relative to that of the untreated controls.
Invasion assay
The invasion assay was performed using transwell culture chambers (BD Biosciences, Bedford, MA, USA) according to the manufacturer's instructions. The upper chamber was loaded with cell suspension and the lower chamber was loaded with 10% FBS. After a 24-h incubation, cells that had invaded the undersurface of the membrane were counted under a microscope. Three microscopic fields were randomly selected for cell counting.
Statistical analysis
Data are presented as mean ± SD. Between-group differences were assessed using the χ2 test and continuous variables were compared using Student's t-test. Statistical analysis was performed using JMP software version 9.0.2 (SAS Institute Inc., Cary, NC, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-615-3p expression is higher in the recurrence group compared to the non-recurrence group
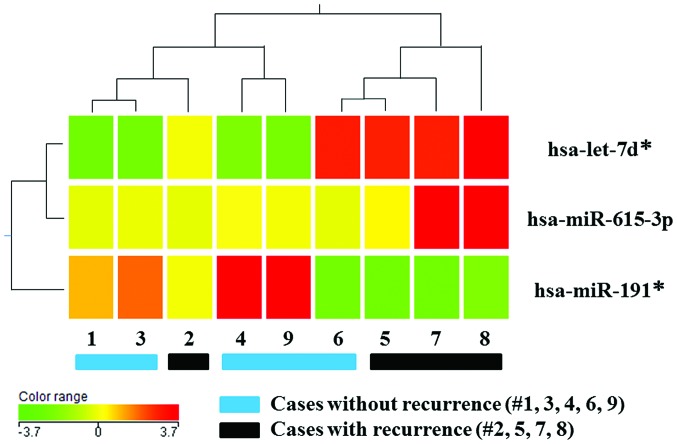
Previously, we reported that postoperative HCC metastasis is represented by recurrence within a 2-year postoperative period (24). During 2 years of observation after surgery, postoperative HCC recurrence was identified in four of nine patients (the recurrence group); the five patients without recurrence were termed the non-recurrence group. To investigate the miRNA expression profiles in DTCs and their regulation of postoperative HCC recurrence, the miRNA from DTCs isolated preoperatively from the BM of the recurrence group was compared against the non-recurrence group. Cluster analysis revealed distinct miRNA expression profiles between the two groups; the expression levels of three out of 941 miRNAs (0.3%) exhibited a mean change of >1.5-fold in the recurrence group compared to the non-recurrence group (Fig. 1). When including adequate expression quantities and excluding miRNA*s, miR-615-3p was identified as a candidate miRNA in DTCs regulating HCC recurrence. Subsequently, further in vitro investigations were performed focusing on the effect of miR-615-3p on malignant characteristics of HCC cells.
Figure 1.
Heatmap of the microRNA (miRNA) profile in disseminated tumor cells from hepatocellular carcinoma patients with and without recurrence. The miRNA levels were determined using miRNA microarray analysis. Cluster analysis showed three miRNAs (hsa-let-7d*, hsa-miR-615-3p and hsa-miR-191*) that were significantly differentially expressed between the two groups, with a >1.5-fold change. The heatmap summarizes the expression levels of these three miRNAs, which range from low (green) to high (red). Patients 2, 5, 7 and 8 had recurrence and patients 1, 3, 4, 6 and 9 had no recurrence.
miR-615-3p alters proliferative activity
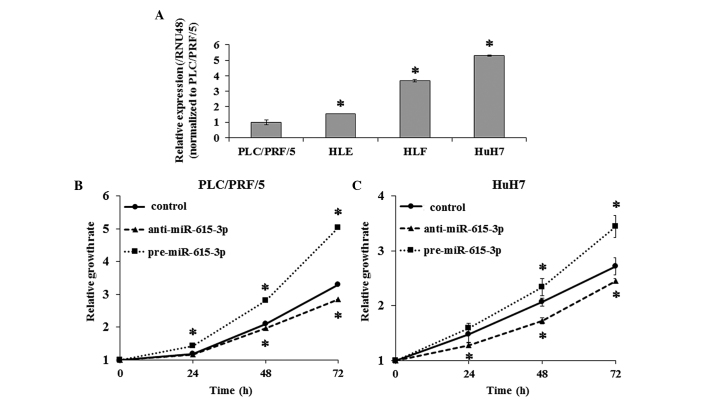
To evaluate the effect of miR-615-3p on cell proliferation, pre-miR-615-3p and anti-miR-615-3p were transfected into PLC/PRF/5 and HuH7 cells, which exhibited the lowest and highest expression of miR-615-3p among the four HCC cell lines, respectively (Fig. 2A). RT-qPCR confirmed that miR-615-3p expression was significantly increased and decreased in cells transfected with pre-miR-615-3p and anti-miR-615-3p, respectively. The cell proliferation assay demonstrated that PLC/PRF/5 and HuH7 cells overexpressing miR-615-3p were significantly more proliferative compared to the controls and that miR-615-3p-suppressed cells exhibited decreased cell proliferation compared to the controls (Fig. 2B and C).
Figure 2.
Expression level of miR-615-3p in hepatocellular carcinoma (HCC) cell lines and its effect on cell proliferation. The expression level of miR-615-3p was examined by reverse transcription-quantitative polymerase chain reaction in four HCC cell lines: PLC/PRF/5, HLE, HLF and HuH7. (A) PLC/PRF/5 and HuH7 exhibited the lowest and highest expression levels of miR-615-3p among the four cell lines, respectively. The effect of miR-615-3p on cell proliferation was investigated using a cell proliferation assay in (B) PLC/PRF/5 and (C) HuH7 cells. Cell proliferative capacity is enhanced by transfection with pre-miR-615-3p and suppressed by transfection with anti-miR-615-3p. Experiments were performed in triplicate and data are expressed as the mean ± standard deviation. *P<0.05 compared to (A) PLC/PRF/5 and (B and C) each control.
miR-615-3p induces resistance to chemotherapeutic agents
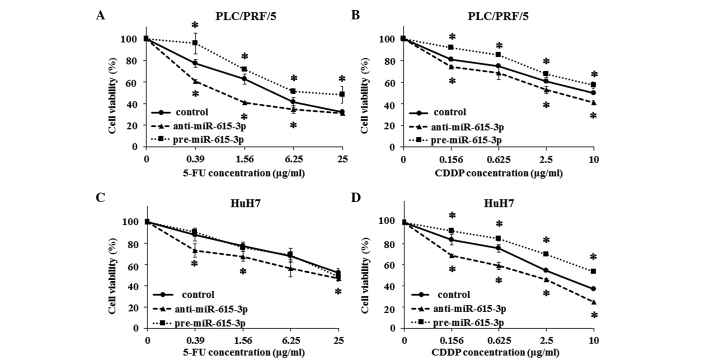
The growth inhibition assay was also used to examine the resistance to chemotherapeutic agents, such as 5-FU and CDDP. PLC/PRF/5 cells transfected with pre-miR-615-3p and anti-miR-615-3p were significantly more and less resistant to the two drugs, respectively, compared to the control cells (Fig. 3A and B). HuH7 cells exhibited a pattern similar to that of PLC/PRF/5 cells, although miR-615-3p-overexpressing HuH7 cells did not show more resistance compared to the control cells (Fig. 3C and D).
Figure 3.
Evaluation of hepatocellular carcinoma cell resistance to chemotherapeutic agents following modulation of miR-615-3p expression. Resistance to (A and C) 5-fluorouracil (5-FU) and (B and D) cisplatin (CDDP) was investigated by the growth inhibition assay in (A and B) PLC/PRF/5 and (C and D) HuH7 cells following transfection with pre-miR-615-3p and anti-miR-615-3p. Resistance of PLC/PRF/5 cells to 5-FU and CDDP was enhanced by transfection with pre-miR-615-3p and suppressed by transfection with anti-miR-615-3p. In HuH7 cells, pre-miR-615-3p transfection induced resistance to CDDP only, while anti-miR-615-3p transfection attenuated resistance to the two drugs. Experiments were performed in triplicate and data are expressed as the mean ± standard deviation. *P<0.05 compared to each control.
Expression level of miR-615-3p is associated with epithelial-mesenchymal transition (EMT)
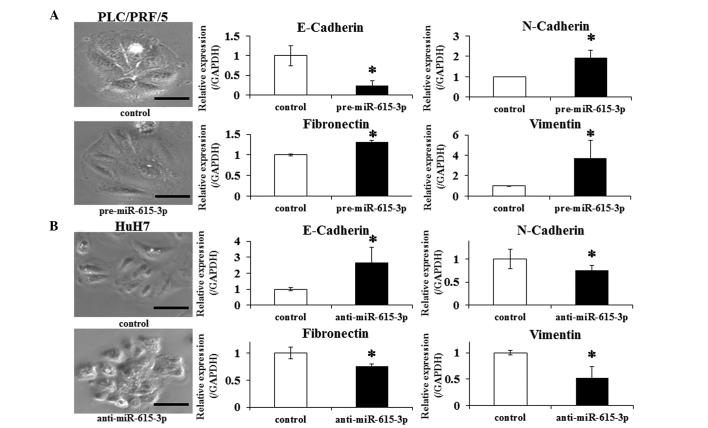
The EMT, a highly conserved developmental program activated during mesoderm formation and neural crest development, is indicated in promoting the dissemination of malignant cells from primary tumors (25). In this context, whether changing the miR-615-3p expression affects EMT in HCC cells was investigated. PLC/PRF/5 cells were transfected with pre-miR-615-3p and HuH7 cells with anti-miR-615-3p. Control PLC/PRF/5 cells exhibited a well-defined epithelial polygonal shape, while pre-miR-615-3p-transfected PLC/PRF/5 cells exhibited a clear morphological change characterized by a spindle-like fibroblastoid appearance (Fig. 4A). Consistent with this finding, a significantly greater expression of N-cadherin, fibronectin and vimentin, and lower expression of E-cadherin was observed in the transfected cells compared to the control cells (Fig. 4B). The changes of the morphological appearance and expression of the above genes were also examined in HuH7 cells transfected with anti-miR-615-3p. The transfected HuH7 cells exhibited a pattern of the changes that was opposite to the pattern observed in PLC/PRF/5 cells (Fig. 4B). These observations indicated that the miR-615-3p-overexpressing cells promote EMT-like phenotypes in HCC cell lines.
Figure 4.
Effect of miR-615-3p on epithelial-mesenchymal transition (EMT) in hepatocellular carcinoma cells. Morphological findings were observed in (A) PLC/PRF/5 following pre-miR-615-3p transfection and (B) HuH7 following anti-miR-615-3p transfection. Expression levels of genes associated with EMT (such as E-cadherin, N-cadherin, fibronectin and vimentin) were also examined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in (A) PLC/PRF/5 following pre-miR-615-3p transfection and in (B) HuH7 following anti-miR-615-3p transfection. The images (left) are representative photographs of the morphological appearance and the bar charts (right) show comparisons of the expression levels of the EMT-associated genes examined by RT-qPCR. Experiments were performed in triplicate and data are expressed as the mean ± standard deviation. *P<0.05 compared to each control. Scale bar, 50 µm.
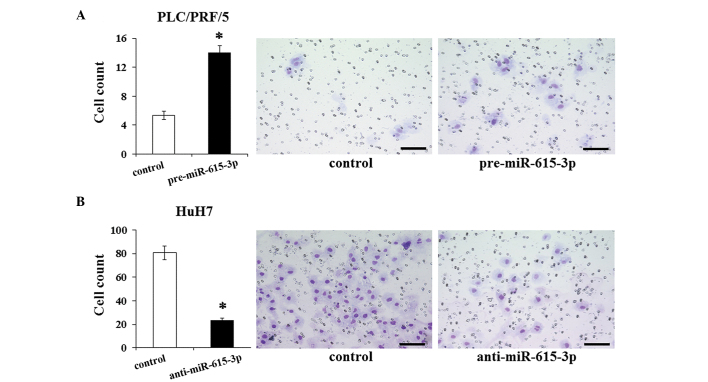
miR-615-3p promotes invasive capacity
The association of miR-615-3p with EMT led us to consider the effect of miR-615-3p on the invasive capacity of cells. The invasive capacity was examined in HCC cells following the modulation of their miR-615-3p expression level. The invasion assay demonstrated that the invasive capacity of PLC/PRF/5 cells transfected with pre-miR-615-3p is significantly greater than that of the control cells (Fig. 5A). The miR-615-3p-suppressed HuH7 cells exhibited a significantly less invasive capacity compared to the control cells (Fig. 5B). These results suggested that miR-615-3p influences the invasive capacity, which was consistent with the results of the EMT features induced by miR-615-3p.
Figure 5.
Evaluation of the invasive capacity in hepatocellular carcinoma cells following modulation of the miR-615-3p expression. Invasive capacity was investigated by the invasion assay in (A) PLC/PRF/5 cells following pre-miR-615-3p transfection and in (B) HuH7 cells following anti-miR-615-3p transfection. The images (right) show representative photographs of each sample and the bar charts (left) depict the numbers of invaded cells following modulation of the miR-615-3p expression level compared to the controls. Experiments were performed in triplicate and data are expressed as the mean ± standard deviation. *P<0.05 compared to each control. Scale bar, 100 µm.
Discussion
In the present study, microarray data were used to compare miRNA expression profiles of DTCs in BM collected preoperatively from HCC patients with and without postoperative recurrence. DTCs of the HCC patients with recurrence exhibited distinctly different miRNA expression profiles compared to the patients without recurrence. In particular, miR-615-3p expression was significantly increased in the BM of patients with recurrence compared to patients without recurrence. Additional in vitro experiments revealed that the expression level of miR-615-3p is significantly associated with the malignant characteristics of HCC cells, such as proliferative capacity, chemotherapeutic resistance and acquisition of EMT-like features. These results suggested the possibility that DTCs from different individuals may have varying characteristics that affect their capacity to develop recurrence/metastasis, which supported the concept that characteristics of existing DTCs, rather than the presence or absence of DTCs, may determine cancer recurrence. Notably, miR-615-3p induced chemoresistance and acquisition of EMT, which are representative phenotypes of cancer stem cells (CSCs) (26,27). This finding indicates that DTCs from patients with recurrence may have features of CSCs, suggesting consistency with previous studies that CSC features are frequently observed in isolated tumor cells in cancer patients with metastatic lesions (28,29).
Previously, we investigated the prognostic impact of α-fetoprotein (AFP) mRNA in BM samples from HCC patients and identified that survival rates did not differ significantly between patients with AFP mRNA-positive BM verses AFP mRNA-negative BM (17). However, during the same period, another study demonstrated that AFP mRNA expression in the BM of HCC patients is of significant prognostic value (30). The controversy appears to be unexpected when considering the possibility that characteristics of DTCs play a role in postoperative tumor recurrence. These previous studies subjected the entire mononuclear cell fraction to analysis; by contrast, the present study analyzed a sorted fraction of viable epithelial tumor cells. Considering the range of cell characteristics among fractions, differences in the fraction analyzed may be a source of the controversy regarding the prognostic significance of AFP mRNA in the whole mononuclear cell fraction of BM.
miR-615-3p, a member of the miR-615 family, reportedly promotes the phagocytic capacity of splenic macrophages through upregulation of peroxisome proliferator-activated receptor γ (31). Regarding the correlation of miR-615-3p with malignant diseases, there has been only one study, which reported that miR-615-3p is associated with survival in mantle cell lymphoma (32). However, the mechanism by which miR-615-3p influences the prognosis has not been investigated. To validate the results of the present study, it is also necessary to investigate whether modulating the level of miR-615-3p expression in an in vivo HCC model would cause a change in metastatic capacity. In this context, the present study will help clarify the function of miR-615-3p in malignant disease. In addition, considering that no studies have focused on the target genes of miR-615-3p, identifying which of its target genes regulate cancer recurrence is required.
The results of the present study potentially lead to several clinical applications in the field of HCC. One is the development of a novel clinical model for predicting the postoperative prognosis in HCC patients by examining the miR-615-3p expression level in DTCs in BM. When considering the sample origin for this potential prediction model, peripheral blood may also be used, as circulating tumor cells (CTCs) may be an alternative to DTCs. CTCs are also reportedly associated with hematogenous dissemination and poor prognosis in cancer patients (5,33). When the upregulation of miR-615-3p is confirmed in the DTCs and the CTCs of HCC patients with recurrence, prognosis may be more easily predicted by analyzing CTCs instead of DTCs in BM. However, several studies comparing the detection rate of the tumor cells (and their prognostic significance) between DTCs and CTCs have reported that the two cell populations do not always correlate and further investigation is required before CTCs are analyzed as a prognostic alternative to DTCs (34,35). Another possible application is a potential miRNA-targeted therapeutic option against HCC. New therapeutic options targeting specific miRNAs have recently been developed (36,37). The results of the present study may lead to a novel, effective therapeutic option that targets miR-615-3p against HCC recurrence; inhibition of miR-615-3p may lead to the prevention of recurrence in HCC patients by suppressing DTCs/CTCs with high malignant potential. However, several challenges must be overcome in order to realize such a new therapy, including the drug delivery system, off-target effects and possible toxicities (38).
In conclusion, the results of the present study demonstrated that DTCs in the BM of HCC patients with and without postoperative recurrence exhibit distinct miRNA expression profiles. This finding suggests that the characteristics of DTCs defined by miRNA expression profiles may act in postoperative tumor recurrence. Furthermore, among the miRNAs with altered expression, miR-615-3p was significantly upregulated in the DTCs of patients with recurrence and its expression level correlated significantly with malignant characteristics of HCC cells in vitro. These results may indicate that the expression level of miR-615-3p in DTCs in the BM is one of the factors that determine postoperative tumor recurrence and suggest that miR-615-3p is a potential target molecule for regulating postoperative HCC recurrence.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Schutte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma – epidemiological trends and risk factors. Dig Dis. 2009;27:80–92. doi: 10.1159/000218339. [DOI] [PubMed] [Google Scholar]
- 3.Zhu AX. Systemic therapy of advanced hepatocellular carcinoma: how hopeful should we be? Oncologist. 2006;11:790–800. doi: 10.1634/theoncologist.11-7-790. [DOI] [PubMed] [Google Scholar]
- 4.Braun S, Vogl FD, Naume B, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353:793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 5.Pantel K, Izbicki J, Passlick B, et al. Frequency and prognostic significance of isolated tumour cells in bone marrow of patients with non-small-cell lung cancer without overt metastases. Lancet. 1996;347:649–653. doi: 10.1016/S0140-6736(96)91203-9. [DOI] [PubMed] [Google Scholar]
- 6.Natsugoe S, Nakashima S, Nakajo A, et al. Bone marrow micrometastasis detected by RT-PCR in esophageal squamous cell carcinoma. Oncol Rep. 2003;10:1879–1883. doi: 10.3892/or.10.6.1879. [DOI] [PubMed] [Google Scholar]
- 7.Oki E, Kakeji Y, Baba H, et al. Clinical significance of cytokeratin positive cells in bone marrow of gastric cancer patients. J Cancer Res Clin Oncol. 2007;133:995–1000. doi: 10.1007/s00432-007-0258-1. [DOI] [PubMed] [Google Scholar]
- 8.Mimori K, Fukagawa T, Kosaka Y, et al. Hematogenous metastasis in gastric cancer requires isolated tumor cells and expression of vascular endothelial growth factor receptor-1. Clin Cancer Res. 2008;14:2609–2616. doi: 10.1158/1078-0432.CCR-07-4354. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 11.Hoshino I, Matsubara H. MicroRNAs in cancer diagnosis and therapy: from bench to bedside. Surg Today. 2013;43:467–478. doi: 10.1007/s00595-012-0392-5. [DOI] [PubMed] [Google Scholar]
- 12.Yang P, Li QJ, Feng Y, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomokuni A, Eguchi H, Tomimaru Y, et al. miR-146a suppresses the sensitivity to interferon-α in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2011;414:675–680. doi: 10.1016/j.bbrc.2011.09.124. [DOI] [PubMed] [Google Scholar]
- 14.Tomimaru Y, Eguchi H, Nagano H, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167–175. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Tomimaru Y, Eguchi H, Nagano H, et al. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-α/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer. 2010;103:1617–1626. doi: 10.1038/sj.bjc.6605958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeyama H, Yamamoto H, Yamashita S, et al. Decreased miR-340 expression in bone marrow is associated with liver metastasis of colorectal cancer. Mol Cancer Ther. 2014;13:976–985. doi: 10.1158/1535-7163.MCT-13-0571. [DOI] [PubMed] [Google Scholar]
- 17.Morimoto O, Nagano H, Miyamoto A, et al. Association between recurrence of hepatocellular carcinoma and α-fetoprotein messenger RNA levels in peripheral blood. Surg Today. 2005;35:1033–1041. doi: 10.1007/s00595-005-3077-5. [DOI] [PubMed] [Google Scholar]
- 18.Akiyoshi S, Fukagawa T, Ueo H, et al. Clinical significance of miR-144-ZFX axis in disseminated tumour cells in bone marrow in gastric cancer cases. Br J Cancer. 2012;107:1345–1353. doi: 10.1038/bjc.2012.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang MH, Chen CL, Chau GY, et al. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology. 2009;50:1464–1474. doi: 10.1002/hep.23221. [DOI] [PubMed] [Google Scholar]
- 20.Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori M, Mimori K, Inoue H, et al. Detection of cancer micrometastases in lymph nodes by reverse transcriptase-polymerase chain reaction. Cancer Res. 1995;55:3417–3420. [PubMed] [Google Scholar]
- 22.Yamamoto T, Nagano H, Sakon M, et al. Partial contribution of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)/TRAIL receptor pathway to antitumor effects of interferon-α/5-fluorouracil against Hepatocellular Carcinoma. Clin Cancer Res. 2004;10:7884–7895. doi: 10.1158/1078-0432.CCR-04-0794. [DOI] [PubMed] [Google Scholar]
- 23.Eguchi H, Nagano H, Yamamoto H, et al. Augmentation of antitumor activity of 5-fluorouracil by interferon α is associated with up-regulation of p27Kip1 in human hepatocellular carcinoma cells. Clin Cancer Res. 2000;6:2881–2890. [PubMed] [Google Scholar]
- 24.Sakon M, Umeshita K, Nagano H, et al. Clinical significance of hepatic resection in hepatocellular carcinoma: analysis by disease-free survival curves. Arch Surg. 2000;135:1456–1459. doi: 10.1001/archsurg.135.12.1456. [DOI] [PubMed] [Google Scholar]
- 25.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 27.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raimondi C, Gradilone A, Naso G, et al. Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res Treat. 2011;130:449–455. doi: 10.1007/s10549-011-1373-x. [DOI] [PubMed] [Google Scholar]
- 30.Kamiyama T, Takahashi M, Nakagawa T, et al. AFP mRNA detected in bone marrow by real-time quantitative RT-PCR analysis predicts survival and recurrence after curative hepatectomy for hepatocellular carcinoma. Ann Surg. 2006;244:451–463. doi: 10.1097/01.sla.0000234840.74526.2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang A, Zhang S, Li Z, et al. miR-615-3p promotes the phagocytic capacity of splenic macrophages by targeting ligand-dependent nuclear receptor corepressor in cirrhosis-related portal hypertension. Exp Biol Med (Maywood) 2011;236:672–680. doi: 10.1258/ebm.2011.010349. [DOI] [PubMed] [Google Scholar]
- 32.Goswami RS, Atenafu EG, Xuan Y, et al. MicroRNA signature obtained from the comparison of aggressive with indolent non-Hodgkin lymphomas: potential prognostic value in mantle-cell lymphoma. J Clin Oncol. 2013;31:2903–2911. doi: 10.1200/JCO.2012.45.3050. [DOI] [PubMed] [Google Scholar]
- 33.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 34.Fehm T, Muller V, Alix-Panabieres C, Pantel K. Micrometastatic spread in breast cancer: detection, molecular characterization and clinical relevance. Breast Cancer Res. 2008;10(Suppl 1):S1. doi: 10.1186/bcr1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ignatiadis M, Georgoulias V, Mavroudis D. Micrometastatic disease in breast cancer: clinical implications. Eur J Cancer. 2008;44:2726–2736. doi: 10.1016/j.ejca.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 36.Kota J, Chivukula RR, O'Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]