Abstract
Bile acid (BA) is de novo synthesized exclusively in the liver and has direct or indirect antimicrobial effects. On the other hand, the composition and size of the BA pool can be altered by intestinal microbiota via the biotransformation of primary BAs to secondary BAs, and subsequently regulate the nuclear farnesoid X receptor (FXR; NR1H4). The BA-activated FXR plays important roles in BA synthesis and metabolism, glucose and lipid metabolism, and even hepatic autophagy. BAs can also play a role in the interplays among intestinal microbes. In this review, we mainly discuss the interactions between BAs and intestinal microbiota and their roles in regulating host metabolism, and probably the autophagic signaling pathway.
Keywords: Bile acid (BA), Farnesoid X receptor (FXR), Intestinal microbiota, Host metabolism, Autophagy
1. Introduction
Bile acid (BA) is biosynthesized de novo in hepatocytes, remained in cholecystis and further converted to secondary BA by intestinal microbiota (Ridlon et al., 2006). BA contributes to the emulsification and solubilization of fats. In addition, it can regulate farnesoid X receptor (FXR; NR1H4) that not only alters the transcription levels of critical genes in BA synthesis, transportation, and metabolism (Makishima et al., 1999), but is also involved in lipid, glucose, and energy metabolism (Zollner et al., 2006; Nguyen and Bouscarel, 2008). Interestingly, FXR is also identified as inhibiting autophagy-related genes (Atgs) in the fed state (Lee et al., 2014; Seok et al., 2014). A large quantity of host and environmental factors can change the composition of intestinal microbes, such as lifestyle, diet, antibiotics, and the host’s environment. In addition, more evidence has revealed that BA is also able to alter gut microbes via direct and indirect antimicrobial effects (Begley et al., 2005), and even contribute to the survival of some BA-tolerant bacteria (Devkota et al., 2012).
Currently, a growing body of research has highlighted that intestinal microbiota disorders can lead to varieties of diseases, for instance, obesity, colon cancer, and Clostridium difficile infection (CDI) (Hill and Artis, 2010; Kau et al., 2011; Tremaroli and Backhed, 2012). In addition, accumulating evidence indicates that intestinal microbiota contributes to the biotransformation of primary BAs to secondary BAs and alters BA composition (Midtvedt, 1974), leading to the activation or inhibition of FXR (Hu et al., 2014) and the subsequent alteration of BA pool size via two FXR-dependent feedback mechanisms of hepatic BA synthesis (Kim et al., 2007). Taken together, BAs can affect the communal structure of intestinal microbes, in turn, intestinal microbes are able to alter BAs. The cross-talk between BAs and intestinal microbes plays essential roles in host metabolism.
Interestingly, BAs play important roles in the interplay among intestinal microbes. For instance, fecal microbiota transplantation (FMT) can restore the intestinal microbiota and fecal BA composition in patients with recurrent CDI (Weingarden et al., 2014). The spore germination of C. difficile can be stimulated by some primary BAs except for chenodeoxycholic acids (CDCAs) (Sorg and Sonenshein, 2008) or inhibited by secondary BAs such as lithocholic acids (LCAs) and ursodeoxycholic acids (UDCAs) (Sorg and Sonenshein, 2010), and FMT can restore gut microbes which can convert primary BAs to secondary BAs, thus inhibiting C. difficile spore germination. In this review, we mainly focus on the metabolism of BA, the interactions between BAs and intestinal microbiota, and the cross-talk between BAs and intestinal microbiota in regulating host metabolism, and probably autophagic signaling pathways.
2. Biosynthesis and enterohepatic circulation (EHC) of BAs
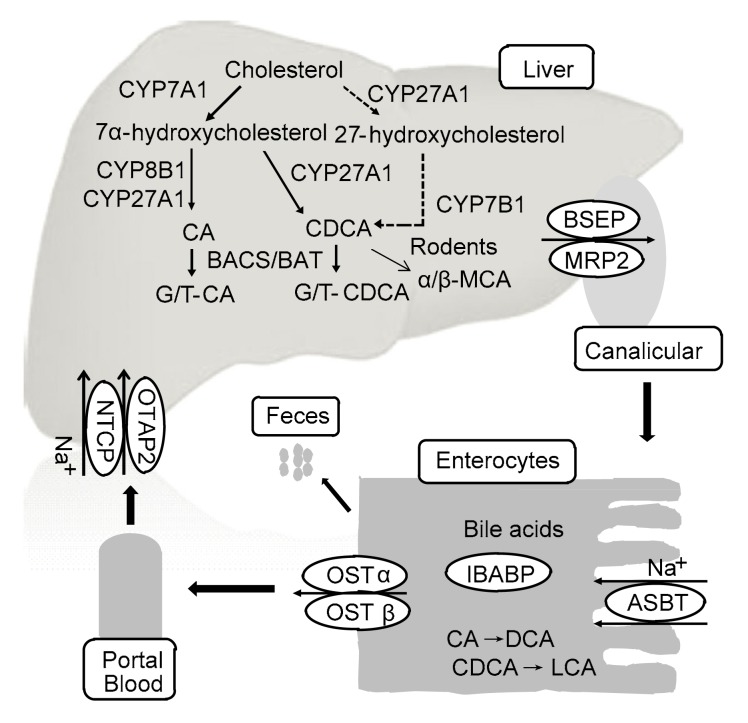
BA is an amphipathic molecule that is synthesized de novo exclusively in hepatocytes (Fig. 1) (Begley et al., 2005). Several previous reviews have summarized in detail the biosynthesis of BA (Chiang, 2004; Lefebvre et al., 2009; de Aguiar Vallim et al., 2013). In brief, two BA biosynthetic pathways are present in the liver. One is the classic or neutral BA synthetic pathway in which cholesterol 7α-hydroxylase (CYP7A1) converts cholesterol into 7α-hydroxycholesterol. This first and rate-limiting hydroxylation step plays a vital role in BA synthesis (Myant and Mitropoulos, 1977). Then, 7α-hydroxycholesterol is catalyzed by sterol 12α-hydroxylase (CYP8B1) and sterol 27-hydroxylase (CYP27A1) to generate cholic acid (CA) and CDCA (Bjorkhem, 1992). In human liver, CA and CDCA are predominant primary BAs. However, in rodent liver, CDCA can be converted into α/β-muricholic acid (α/β-MCA), which is also primary BA (Zhang et al., 2014). The alternative BA synthetic pathway mainly leads to the synthesis of CDCA. The cholesterol side chain is oxidized by CYP27A1 and then the steroid ring is hydroxylated by oxysterol 7α-hydroxylase (CYP7B1). Cholesterol is also converted to 24-, 25-, or 27-hydroxycholesterols in other organs, such as the lung and brain, and these oxysterols are used for hepatic synthesis of BAs.
Fig. 1.
Synthesis and EHC of BAs
This figure is modified from Li and Chiang (2013)
Before being secreted into the bile, primary BAs can conjugate with glycine or taurine under the catalysis of BA coenzyme A synthetase (BACS) and BA amino acid transferase (BAAT) to generate bile salts, which are transported to canaliculi and form bile through the bile salt export protein (BSEP; ABCB11). After a meal, dietary fat stimulates intestinal mucosa to secrete cholecystokinin, which can promote the contraction of the gallbladder, thus facilitating the secretion of bile salts into the small intestine. Most bile salts (95%) are re-absorbed by the intestine via the apical sodium-dependent bile acid transporter (ASBT) and the organic solute transporters (OSTα/β), and then flow into the portal blood (Dawson et al., 2005). The intestinal BA-binding protein (IBABP) contributes to the transportation of BAs across the ileal enterocytes. However, a mini-fraction of CAs, CDCAs, and MCAs are not re-absorbed. They are modified to generate deoxycholic acids (DCAs), LCAs, ursocholic acids (UCAs), and UDCAs in the human, and DCAs, UCAs, UDCAs, murideoxycholic acids (MDCAs), and hyodeoxycholic acids (HDCAs) in mice, which are all secondary BAs, under the catalysis of 7α-dehydroxylase in bacteria (Zhang et al., 2014). Most LCAs are excreted into feces, their loss being compensated by de novo BA biosynthesis. Others return to hepatocytes and are sulfated before excretion to the gallbladder (Wang et al., 2009). Meanwhile, DCAs are re-absorbed in the colon. The re-absorbed primary and secondary BAs return to hepatocytes via the transportation of (Na+)-dependent taurocholic acid co-transporting polypeptide (NTCP) and organic anion transporting polypeptide (OATP) to complete EHC (de Aguiar Vallim et al., 2013).
BAs are potent detergents and contribute to the absorption and transportation of lipids, nutrients, and fat-soluble vitamins (Begley et al., 2005). BAs also are identified to be natural ligands for FXR, which can be regulated by BA and subsequently modulate metabolic pathways (Houten et al., 2006; Eloranta and Kullak-Ublick, 2008).
3. BAs as regulators of intestinal microbiota
The recent literature has suggested that BAs not only have direct antibacterial effects but also affect microbiota indirectly via nuclear receptors (Begley et al., 2005; Inagaki et al., 2006). However, some BA-tolerant microbes can thrive in the presence of BAs (Fig. 2a) (Devkota et al., 2012).
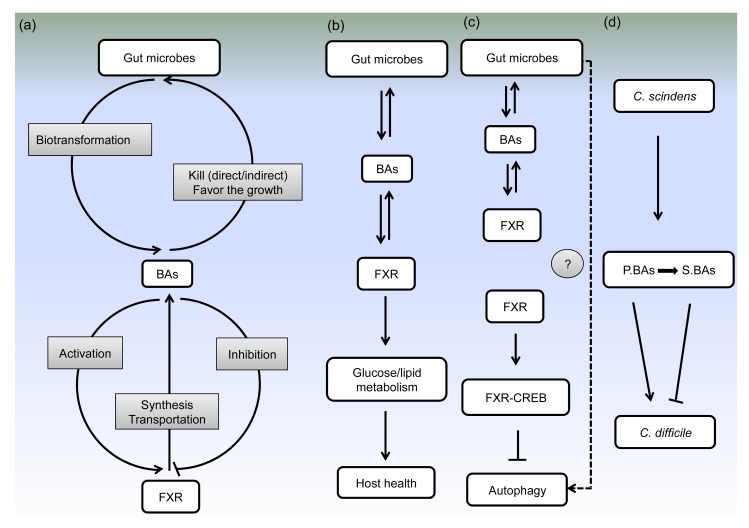
Fig. 2.
Interactions between BAs and gut microbes in host metabolism and probably in autophagy signaling pathways
(a) Interactions among gut microbes, BAs, and FXR. Gut microbes involve in the transformation of primary BAs into second BAs. BAs have direct and indirect antimicrobial effects, and even can favor the growth of BA-tolerant bacteria. BAs can activate or inhibit the expression of FXR, which in turn has profound effects on the synthesis and transportation of BAs. (b) Interactions among gut microbes, BAs, and FXR in host health. (c) Potential relationships between gut microbes and autophagy mediated by BAs. Interactions among gut microbes, BAs and FXR have been elucidated. The inhibition of hepatic autophagy by FXR-CREB complex has also been demonstrated. However, whether gut microbes can regulate autophagy via BAs and FXR pathways needs to be further investigated. (d) BAs as mediators among gut microbes. C. scindens can convert primary BAs (P.BAs) to secondary BAs (S.BAs). Primary BAs can stimulate C. difficile spores germination but secondary BAs can inhibit C. difficile spores germination
3.1. Direct effects of BAs on bacteria
When BA-intolerant bacteria are exposed to BAs, cell membrane integrity can be altered and intracellular enzymes leak out, resulting in the inhibition of bacterial overgrowth (Noh and Gilliland, 1993; de Valdez et al., 1997). High levels of BAs dissolve phospholipids and dissociate intrinsic membranous proteins, leading to the spillage of intracellular materials (Heuman et al., 1996). Low concentrations of BAs can also affect the permeability and fluidity of cell membrane and change membrane characteristics (Noh and Gilliland, 1993). The antimicrobial potency of DCA is 10 times greater than that of CA (Kurdi et al., 2006). Additionally, previous studies have shown that BAs may affect some microbial cells in other ways, such as inducing DNA damage, activating enzymes involved in DNA repair, and altering protein structure (Kandell and Bernstein, 1991; Bernstein et al., 1999). A recent study of Cremers et al. (2014) revealed that bile salts have profound effects on many key proteins in bacteria through protein unfolding, aggregation and disulfide stress. Therefore, BAs are thought to have destructive effects on gut microbes except for some BA-tolerant bacteria.
3.2. Indirect effects of BAs on bacteria
Interestingly, oral BAs can activate FXR and inhibit the bacterial proliferation resulting from biliary obstruction in rodents, and then protect the ileum from microbial infection (Ding et al., 1993; Lorenzo-Zúñiga et al., 2003). Inagaki et al. (2006) showed that BA-activated FXR can induce the expression of genes, such as Ang1, iNos, and Il18, which are involved in enteroprotection. The products of these genes contribute to the inhibition of microbial overgrowth and mucosal damage. Furthermore, in cultured biliary epithelial cells, CDCA and UDCA are able to induce the expression of cathelicidin through FXR and vitamin D receptor. Cathelicidin is an antimicrobial peptide which acts against microbial invasion in the biliary epithelium and this may contribute to biliary tract sterility (D'Aldebert et al., 2009). Taken together, BAs can affect microbiota via nuclear receptors indirectly.
3.3. Growth of BA-tolerant bacteria
However, the proliferation of some BA-tolerant microbes, such as Bilophila wadsworthia, Escherichia coli, and Listeria monocytogenes, can be facilitated by BAs, leading to the suppression of other symbiotic microbes (Hardison, 1978; Laue et al., 1997; David et al., 2014). Furthermore, some gut commensals can also tolerate BAs, such as some Lactobacillus and Bifidobacterium species expressing bile salt hydrolase (Begley et al., 2005). Early studies observed the proliferation of B. wadsworthia in the presence of BAs since sulphite in taurine-conjugated BAs can be used as a terminal electron acceptor (Hardison, 1978; Laue et al., 1997). Another report revealed that sulfur-reducing bacteria (e.g. B. wadsworthia) thrive when interleukin-10-deficient (Il10−/−) mice were fed with a milk-derived fat diet, because milk fat contributes to high levels of taurine-conjugated BAs and provides enough organic sulphur for sulfur-reducing bacteria. Moreover, adding taurocholic acid (T-CA) to a low-fat diet also contributes to the growth of B. wadsworthia in Il10−/− mice (Devkota et al., 2012). In addition, David et al. (2014) showed an increase of BA-tolerant microbes and a decrease of Firmicutes in volunteers taking the animal-based diet.
3.4. Complex relationships among dietary fat, BAs, and intestinal microbes
Research on the human diet has suggested that BA profiles may be changed from glycine to taurine-conjugation after feeding a “Western diet” (i.e. a high-fat diet) for a long time (Hardison, 1978). As described above, BAs are able to act as regulators of intestinal microbes. In this regard, a recent study demonstrated that a “Western diet” is capable of increasing Firmicutes, decreasing Bacteroidetes, and disturbing the ecological balance between the two predominant gut microbes (Murphy et al., 2010). Accordingly, only gut microbes which possess high BA tolerance can survive when a diet supplemented with CA is administered (Islam et al., 2011). A marked reduction in Bacteroidetes and an expansion in Firmicutes after CA intake were shown, resulting in an obvious expansion of BA 7α-dehydroxylating species, which can produce secondary BAs. Therefore, BAs in the intestine have negative effects on Bacteroidetes and Actinobacteria, while they exert beneficial effects on Firmicutes, especially on BA 7α-dehydroxylating species. Taken together, these studies have revealed complex relationships among dietary fat, BAs, and intestinal microbes.
4. Regulation of BA profiles by intestinal microbiota
It has been demonstrated that BA pool size and composition can regulate the structure of intestinal microbiota. By contrast, mechanisms of disturbed intestinal microbiota affecting BA profiles have also been shown (Ridlon et al., 2014). Previously, studies have revealed that intestinal microbiota has profound effects on BA profiles (Sayin et al., 2013). Compared with conventional rats, germ-free (GF) rats fed with a diet supplemented with suitable cholesterol have more BAs in the bile; furthermore, levels of T-CAs are higher (Wostmann, 1973). Consistent with this, a recent study showed that T-CA, especially T-β-MCA, is the major BA in many host tissues (e.g. liver, kidney, and heart) of GF mice (Swann et al., 2011). In addition, Vrieze et al. (2014) revealed that oral vancomycin caused a suppression of Firmicutes (mainly Gram-positive bacteria) and an expansion of Proteobacteria (mainly Gram-negative bacteria), resulting in reduced secondary BAs in feces and increased primary BAs in plasma. In sum, intestinal microbiota can affect BA pool size and composition via regulating the biotransformation, biosynthesis, and transportation of BA.
4.1. Biotransformation of BAs by intestinal microbiota
It has been elucidated that intestinal microbiota profoundly affects the biotransformation of BAs (Fig. 2a) via deconjugation, dehydrogenation, epimerization, and 7α/β-dehydroxylation of primary BAs and generates secondary BAs (Ridlon et al., 2006). Deconjugation is the process of fracturing the C-24 N-acyl amide of conjugated BA which links BA to glycine or taurine and generates unconjugated BA. A wide array of gut bacteria possess bacterial bile salt hydrolases (BSH) and bacterial hydroxysteroid dehydrogenases, which are involved in the deconjugation and dehydrogenation of bile salts, respectively, while only a minor population of gut bacteria in Clostridium clusters XI and XVIa, such as C. sordellii, C. sordelliifell, and C. scindens, which belong to the Firmicutes phylum, possess 7α/β-dehydroxylase and catalyze the dehydrogenation reaction of BA (Midtvedt, 1974; Hayakawa, 1982; Doerner et al., 1997; Ridlon et al., 2006). A recent study revealed a clear elevation of Clostridium clusters XI and XVIa in high-fat-diet mice, and Clostridium cluster XI contained a sole species resembling C. sordellii and accounted for more than 12% of the fecal microbiota (Yoshimoto et al., 2013).
BA profiles can be altered by intestinal microbes owing to their different BA metabolic enzymes. Li et al. (2013) showed that when mice were treated with tempol, an antioxidant which can reduce Lactobacillus and inhibit their BSH activity, a diminution of the Firmicutes:Bacteroidetes ratio, a decrease of secondary BAs, and an increase of T-β-MCA were observed. As noted in the study of Sayin et al. (2013), secondary BAs are potent FXR agonists but T-β-MCA is a FXR antagonist. Thus tempol-treated mice have an enlarged BA pool size attributed to the inhibition of FXR signaling, and the decrease of secondary BAs is mainly caused by the decline of Clostridium clusters XI and XVIa, which are involved in the dehydrogenation of BAs (Li et al., 2013). Notably, high accumulation of secondary BAs in the EHC system of individuals may result in diseases, such as colon cancers and gallstones (Mcgarr et al., 2005).
4.2. Biosynthesis of BAs regulated by intestinal microbiota
BA synthesis is regulated through BA feedback mechanisms which are involved in the expression of BA activated-FXR (Fig. 2a). The regulation of FXR is attributed to the size and composition of the BA pool (Russell, 2003). As summarized by Hu et al. (2014) recently, CA, CDCA, DCA, and LCA are FXR-agonistic, whereas α/β-MCA and UDCA are FXR-antagonistic. They observed an increase in BA synthesis and an enlarged BA pool in Cyp8b1−/− mice, ampicillin (AMP)-treated mice, and GF mice, which was attributed to the expansion of α/β-MCA and UDCA and the reduction of DCA. At present, FXR is identified as modulating the expression of CYP7A1, which is a key enzyme in limiting hepatic BA synthesis through two mechanisms. In hepatocytes, CYP7A1 is inhibited by BA via an FXR/SHP/LRH-1 mechanism. In enterocytes, CYP7A1 is inhibited via the FXR/FGF15/FGFR4 mechanism (Kim et al., 2007). Once activated by BA, hepatic FXR triggers a small heterodimer partner (SHP) to suppress the transcriptional level of liver-relahomolog-1 (LRH-1), thus suppressing CYP7A1 transcription. In addition, SHP can combine with hepatocyte nuclear factor 4α to inhibit CYP7A1 and CYP8B1 transcription (Goodwin et al., 2000). Simultaneously, BA-activated FXR expressed in the gut also contributes to BA homeostasis. Intestinal FXR in mouse can induce fibroblast growth factor (FGF) 15 that can activate fibroblast growth factor receptor 4 (FGFR4) to suppress CYP7A1, thus inhibiting hepatic BA synthesis (Holt et al., 2003).
As mentioned above, gut microbes can modulate the BA pool and composition, which in turn significantly affects the BA profiles. Administration of antibiotic, such as AMP, bacitracin, streptomycin, and neomycin, can elevate hepatic BA biosynthesis in the liver via suppression of FGF15 expression in ileum, indicating that less FXR is activated by BA (Miyata et al., 2009). Previous research has revealed that compared with GF mice, conventionally raised (CONV-R) mice had decreased T-β-MCA and elevated T-CA. Re-derivation of Fxr−/− mice as GF proved that intestinal microbes can induce the expression of SHP and FGF15 in a FXR-dependent mechanism (Sayin et al., 2013). Furthermore, MCAs were identified as natural FXR antagonists in this study. Therefore, they elucidated that intestinal microbes can modulate BA profiles through ameliorating the suppression of FXR in the ileum (Sayin et al., 2013). Another study showed that oral probiotic elevated Firmicutes and Actinobacteria, but decreased Bacteroidetes and Proteobacteria, leading to the elevation of BA excretion in the feces and hepatic BA synthesis (Degirolamo et al., 2014). In this study, Degirolamo et al. (2014) observed an increase of CYP7A1, and CYP8B1, and a decrease of FGF15, with no alteration of SHP or LRH-1 level. Interestingly, they found that probiotic-triggered changes in BA were restored in FXR- and FGF15-deficient mice. All these results demonstrated that probiotics contribute to BA deconjugation in the ileum, BA secretion in the feces, and BA synthesis in the liver through suppressing the FXR/FGF15 negative feedback mechanism of BA synthesis.
4.3. Transportation of BAs regulated by intestinal microbiota
FXR also has profound effects on the transportation of BAs (Fig. 2a) via regulating transporters in the EHC. For instance, BSEP can facilitate bile salts secreted into gallbladder to form bile. A study on human subjects showed that the BSEP promoter possesses a FXR binding site and FXR can trigger BSEP expression (Ananthanarayanan et al., 2001). Previous studies elucidated that FGF15/19 suppresses ASBT which modulates intestinal BA re-absorption in enterocytes and cholangiocytes (Li et al., 2005; Sinha et al., 2008). Moreover, AMP-treated mice have less BA in feces, more BA in portal blood, and elevated levels of ASBT in ileum (Miyata et al., 2009). Interestingly, a decrease in ASBT in ileum was observed when AMP-treated mice were administered T-DCA or CA (Miyata et al., 2011). FXR is also reported to be involved in the activation of IBABP (Hwang et al., 2002), which facilitates bile salts passing through the ileal enterocytes. OSTα/β contributes to the efflux of BA into portal blood and the OSTα/β in mice possesses functional FXR and LRH elements, resulting in the regulation of BA synthesis (Frankenberg et al., 2006). Moreover, NTCP, the major hepatic BA transporter, can be regulated by FXR through inducing SHP (Denson et al., 2001). Recently, it has been shown that two antibiotic combinations clearly elevated the mRNA levels of several EHC transporters, including NTCP, OATP1B2, BSEP, and MRP2, but suppressed the mRNA levels of CYP8B1 (Zhang et al., 2014).
4.4. Role of intestinal microbiota in host metabolism mediated by BAs
As noted above, intestinal microbiota exerts a profound influence on BA pool size and composition. In previous studies, BAs have been identified as regulatory molecules and have an important impact on the modulation of lipid, glucose, and energy metabolism. BA metabolic disturbance may lead to a variety of host metabolic diseases and related complications, such as obesity, type-2 diabetes, cirrhosis, and colon cancer (Zollner et al., 2006; Nguyen and Bouscarel, 2008). Vrieze et al. (2014) speculated that gut microbes are capable of altering glucose metabolism via altering the BA pool and FGF19 signaling in humans. Based on this hypothesis, a decrease of Firmicutes, fecal BAs, and peripheral insulin sensitivity was observed after administration of oral vancomycin. This study demonstrated that gut microbes, especially the Firmicutes, are beneficial for BA and glucose metabolism. Joyce et al. (2014) revealed BSH enzymes expressed in the gut of GF or CONR mice noticeably changed plasma BAs and influenced gene-expression levels in both fat metabolism and metabolic signaling pathways. When CONR mice express higher levels of BSH enzymes, their weight gain, plasma cholesterol, and hepatic triglycerides reduce significantly. In this study, intestinal microbes can regulate host fat metabolism via BSH enzymes. Taken together, intestinal microbiota can alter the BA pool and then profoundly affect host metabolism, which is clearly closely related to host health (Fig. 2b).
4.5. Potential relationships between intestinal microbiota and autophagy mediated by BAs
Autophagy is a highly conserved basic process which can degrade cytoplasmic constituents for cellular survival and the maintenance of cellular homeostasis upon nutritional deprivation (Deretic and Levine, 2009). It is known to be regulated by nutrient-sensing signaling pathways including the mammalian target of the rapamycin (mTOR) pathway. However, two research groups have shown that in the fed state, BA-activated FXR can modulate the autophagy gene network in the liver independent of the mTOR pathway (Lee et al., 2014; Seok et al., 2014). Seok et al. (2014) revealed that after a meal or administration of GW4064 (which is an FXR agonist), FXR is activated and hepatic autophagy is inhibited. Because FXR can generate the FXR-cAMP response element binding protein (CREB) complex and destroy the CREB-CREB regulated transcription coactivator 2 (CRTC2) complex, then suppress the transcriptional levels of Atgs, such as Atg7, Ulk1, and Tfeb. The CREB-CRTC2 complex is able to activate transcription of Atgs under nutritional starvation. Lee et al. (2014) revealed that BA-activated FXR strongly represses hepatic autophagy induced by nutrient deprivation, but in Fxr−/− mice, autophagy is not inhibited. As is well known, autophagy can be induced to counter intestinal pathogenic infection (Joubert et al., 2009), such as infection of Salmonella (Brumell et al., 1999) and Group A Streptococcus (Levine et al., 2011). Interestingly, some gut microbes (e.g. Shigella flexneri, L. monocytogenes, and E. coli) can escape autophagy (Allaoui et al., 1992; Dortet et al., 2011; Baxt et al., 2013). Some gut microbes (e.g. Staphylococcus aureus and Brucella abortus) even can use autophagy for their survival, replication, and intercellular dissemination (Schnaith et al., 2007; Starr et al., 2012). In addition, intestinal microbiota can regulate BA pool size and composition and subsequently alter FXR which can regulate autophagy. Therefore, potential relationships between intestinal microbiota and autophagy mediated by BAs may exist (Fig. 2c) and these relationships may have a profound effect in the therapy of many human diseases caused by autophagy disturbance, for instance, cancers, cardiovascular and cerebrovascular diseases, and neurodegenerative diseases.
5. Role of BAs in the interplay among intestinal microbes
Intestinal microbes can inhibit other intestinal microbes in different ways, such as competing for nutrients or intestinal epithelium receptors, and secreting bacteriocins to kill nearby microorganisms directly (Sekirov and Finlay, 2009). Interestingly, intestinal microbes can regulate BA profiles, which in turn modify intestinal microbes. For example, CDI is the primary stimulus of recurrent ulcerative colitis (Rupnik et al., 2009). To produce toxins, the spore of C. difficile must germinate and then grow. Some primary BAs except for CDCA can efficiently stimulate C. difficile spore germination (Fig. 2d) (Sorg and Sonenshein, 2008; Heeg et al., 2012). The germination-specific protease CspC, which can recognize BAs, has a vital role in C. difficile germination and further C. difficile infection (Francis et al., 2013). However, secondary BAs such as LCAs and UDCAs can suppress C. difficile germination (Sorg and Sonenshein, 2010). Notably, a recent study revealed that intestinal extracts, which can efficiently stimulate C. difficile spore germination in the colon, have a lower content of secondary BAs. This study suggested that secondary BAs may inhibit C. difficile spore germination in vivo (Giel et al., 2010).
FMT, which introduces a fecal enema from a healthy individual into the gut of a patient, can treat recurrent CDI (Borody and Khoruts, 2012). Recently, Weingarden et al. (2014) showed that FMT can restore the intestinal microbiota and fecal BA composition in patients with recurrent CDI. The process in which primary BAs are converted into secondary BAs is disrupted in patients with CDI. However, the normalization of gut microbial composition via FMT can correct BA metabolism, thereby developing an adverse environment for C. difficile spore germination (Weingarden et al., 2014). Importantly, a recent study identified C. scindens, a resistance-associated gut microbe which can efficiently inhibit C. difficile infection. It is BA 7α-dehydroxylating bacteria that can convert primary BAs to secondary BAs. The engraftment of C. scindens can correct the biosynthesis of secondary BAs, thereby inhibiting CDI (Buffie et al., 2015). Taken together, some gut microbes which synthesize 7α-hydroxysteroid dehydrogenase enzymes can indirectly inhibit C. difficile spore germination and growth through converting primary BAs to secondary BAs. Thus, BAs can act as mediators among gut microorganisms and play vital roles in maintaining intestinal homeostasis.
6. Perspectives
Although a great number of studies have demonstrated the complex interactions among intestinal microbes, BAs, host metabolism, and human health, only a few related pieces of research have elucidated that BAs can act as mediators in the regulation of intestinal microbiota and host metabolism. The mechanisms by which BAs affect the interaction between intestinal microbiota and host metabolism are poorly understood. At present, a large fraction of studies on BAs are carried out based on genetically modified mice models. However, the size and composition of BAs and the metabolism of BAs in mice are greatly different from BAs in human; as a consequence, most of these results are not applicable to humans. In this review, a new insight has been provided in the exploration of potential relationships between microbiota and autophagy. The investigation of microbial therapies, such as administering antibiotics, probiotics, and prebiotics, may be combined with the autophagic process. In addition, identification of unknown host factors similar to BA could deepen our understanding of the way in which one gut microbe acts on another. Importantly, these elucidations will provide new insights to alleviate gastrointestinal diseases and maintain intestinal health via altering host factors or supplementing several precise microbes.
Acknowledgments
We thank all Yan laboratory members for discussions and critical reading of the manuscript.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 31322053), the Hubei Province Distinguished Young Scholar (No. 2012FFA015), and the Fundamental Research Funds for the Central Universities (Nos. 2013PY056 and 2013JQ001), China
Compliance with ethics guidelines: Yang-fan NIE, Jun HU, and Xiang-hua YAN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Allaoui A, Mounier J, Prevost MC, et al. icsB: a Shigella flexneri virulence gene necessary for the lysis of protrusions during intercellular spread. Mol Microbiol. 1992;6(12):1605–1616. doi: 10.1111/j.1365-2958.1992.tb00885.x. [DOI] [PubMed] [Google Scholar]
- 2.Ananthanarayanan M, Balasubramanian N, Makishima M, et al. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276(31):28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- 3.Baxt LA, Garza-Mayers AC, Goldberg MB. Bacterial subversion of host innate immune pathways. Science. 2013;340(6133):697–701. doi: 10.1126/science.1235771. [DOI] [PubMed] [Google Scholar]
- 4.Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein H, Payne CM, Bernstein C, et al. Activation of the promoters of genes associated with DNA damage, oxidative stress, ER stress and protein malfolding by the bile salt, deoxycholate. Toxicol Lett. 1999;108(1):37–46. doi: 10.1016/S0378-4274(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 6.Bjorkhem I. Mechanism of degradation of the steroid side chain in the formation of bile acids. J Lipid Res. 1992;33(4):455–471. [PubMed] [Google Scholar]
- 7.Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9(2):88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- 8.Brumell JH, Steele-Mortimer O, Finlay BB. Bacterial invasion: force feeding by salmonella. Curr Biol. 1999;9(8):R277–R280. doi: 10.1016/S0960-9822(99)80178-X. [DOI] [PubMed] [Google Scholar]
- 9.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile . Nature. 2015;517(7533):205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang JY. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J Hepatol. 2004;40(3):539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Cremers CM, Knoefler D, Vitvitsky V, et al. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo . PNAS. 2014;111(16):E1610–E1619. doi: 10.1073/pnas.1401941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Aldebert E, Biyeyeme Bi Mve MJ, Mergey M, et al. Bile salts control the antimicrobial peptide cathelicidin through nuclear receptors in the human biliary epithelium. Gastroenterology. 2009;136(4):1435–1443. doi: 10.1053/j.gastro.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 13.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson PA, Hubbert M, Haywood J, et al. The heteromeric organic solute transporter α-β, Ostα-Ostβ, is an ileal basolateral bile acid transporter. J Biol Chem. 2005;280(8):6960–6968. doi: 10.1074/jbc.M412752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17(5):657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degirolamo C, Rainaldi S, Bovenga F, et al. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 2014;7(1):12–18. doi: 10.1016/j.celrep.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 17.Denson LA, Sturm E, Echevarria W, et al. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 2001;121(1):140–147. doi: 10.1053/gast.2001.25503. [DOI] [PubMed] [Google Scholar]
- 18.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5(6):527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Valdez GF, Martos G, Taranto MP, et al. Influence of bile on β-galactosidase activity and cell viability of Lactobacillus reuteri when subjected to freeze-drying. J Dairy Sci. 1997;80(9):1955–1958. doi: 10.3168/jds.S0022-0302(97)76137-X. [DOI] [PubMed] [Google Scholar]
- 20.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487(7405):104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding JW, Andersson R, Soltesz V, et al. The role of bile and bile acids in bacterial translocation in obstructive jaundice in rats. Eur Surg Res. 1993;25(1):11–19. doi: 10.1159/000129252. [DOI] [PubMed] [Google Scholar]
- 22.Doerner KC, Takamine F, Lavoie CP, et al. Assessment of fecal bacteria with bile acid 7α-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol. 1997;63(3):1185–1188. doi: 10.1128/aem.63.3.1185-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dortet L, Mostowy S, Samba-Louaka A, et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 2011;7(8):e1002168. doi: 10.1371/journal.ppat.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eloranta JJ, Kullak-Ublick GA. The role of FXR in disorders of bile acid homeostasis. Physiology. 2008;23(5):286–295. doi: 10.1152/physiol.00020.2008. [DOI] [PubMed] [Google Scholar]
- 25.Francis MB, Allen CA, Shrestha R, et al. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog. 2013;9(5):e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankenberg T, Rao A, Chen F, et al. Regulation of the mouse organic solute transporter α-β, Ostα-Ostβ, by bile acids. Am J Physiol Gastrointest Liver Physiol. 2006;290(5):G912–G922. doi: 10.1152/ajpgi.00479.2005. [DOI] [PubMed] [Google Scholar]
- 27.Giel JL, Sorg JA, Sonenshein AL, et al. Metabolism of bile salts in mice influences spore germination in Clostridium difficile . PLoS ONE. 2010;5(1):e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodwin B, Jones SA, Price RR, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6(3):517–526. doi: 10.1016/S1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 29.Hardison WG. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology. 1978;75(1):71–75. [PubMed] [Google Scholar]
- 30.Hayakawa S. Microbial transformation of bile acids. A unified scheme for bile acid degradation, and hydroxylation of bile acids. Z Allg Mikrobiol. 1982;22(5):309–326. doi: 10.1002/jobm.19820220505. [DOI] [PubMed] [Google Scholar]
- 31.Heeg D, Burns DA, Cartman ST, et al. Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS ONE. 2012;7(2):e32381. doi: 10.1371/journal.pone.0032381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heuman DM, Bajaj RS, Lin Q. Adsorption of mixtures of bile salt taurine conjugates to lecithin-cholesterol membranes: implications for bile salt toxicity and cytoprotection. J Lipid Res. 1996;37(3):562–573. [PubMed] [Google Scholar]
- 33.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holt JA, Luo G, Billin AN, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17(13):1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25(7):1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu X, Bonde Y, Eggertsen G, et al. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J Intern Med. 2014;275(1):27–38. doi: 10.1111/joim.12140. [DOI] [PubMed] [Google Scholar]
- 37.Hwang ST, Urizar NL, Moore DD, et al. Bile acids regulate the ontogenic expression of ileal bile acid binding protein in the rat via the farnesoid X receptor. Gastroenterology. 2002;122(5):1483–1492. doi: 10.1053/gast.2002.32982. [DOI] [PubMed] [Google Scholar]
- 38.Inagaki T, Moschetta A, Lee YK, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. PNAS. 2006;103(10):3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Islam KB, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141(5):1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 40.Joubert PE, Meiffren G, Gregoire IP, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6(4):354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Joyce SA, Macsharry J, Casey PG, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. PNAS. 2014;111(20):7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kandell RL, Bernstein C. Bile salt/acid induction of DNA damage in bacterial and mammalian cells: implications for colon cancer. Nutr Cancer. 1991;16(3-4):227–238. doi: 10.1080/01635589109514161. [DOI] [PubMed] [Google Scholar]
- 43.Kau AL, Ahern PP, Griffin NW, et al. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim I, Ahn SH, Inagaki T, et al. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48(12):2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Kurdi P, Kawanishi K, Mizutani K, et al. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol. 2006;188(5):1979–1986. doi: 10.1128/JB.188.5.1979-1986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laue H, Denger K, Cook AM. Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl Environ Microbiol. 1997;63(5):2016–2021. doi: 10.1128/aem.63.5.2016-2021.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JM, Wagner M, Xiao R, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516(7529):112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lefebvre P, Cariou B, Lien F, et al. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89(1):147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 49.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F, Jiang C, Krausz KW, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Chen F, Shang Q, et al. FXR-activating ligands inhibit rabbit ASBT expression via FXR-SHP-FTF cascade. Am J Physiol Gastrointest Liver Physiol. 2005;288(1):G60–G66. doi: 10.1152/ajpgi.00170.2004. [DOI] [PubMed] [Google Scholar]
- 52.Li T, Chiang JYL. Nuclear receptors in bile acid metabolism. Drug Metab Rev. 2013;45(1):145–155. doi: 10.3109/03602532.2012.740048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lorenzo-Zúñiga V, Bartoli R, Planas R, et al. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 2003;37(3):551–557. doi: 10.1053/jhep.2003.50116. [DOI] [PubMed] [Google Scholar]
- 54.Makishima M, Okamoto AY, Repa JJ, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284(5418):1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 55.Mcgarr SE, Ridlon JM, Hylemon PB. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J Clin Gastroenterol. 2005;39(2):98–109. [PubMed] [Google Scholar]
- 56.Midtvedt T. Microbial bile acid transformation. Am J Clin Nutr. 1974;27(11):1341–1347. doi: 10.1093/ajcn/27.11.1341. [DOI] [PubMed] [Google Scholar]
- 57.Miyata M, Takamatsu Y, Kuribayashi H, et al. Administration of ampicillin elevates hepatic primary bile acid synthesis through suppression of ileal fibroblast growth factor 15 expression. J Pharmacol Exp Ther. 2009;331(3):1079–1085. doi: 10.1124/jpet.109.160093. [DOI] [PubMed] [Google Scholar]
- 58.Miyata M, Yamakawa H, Hamatsu M, et al. Enterobacteria modulate intestinal bile acid transport and homeostasis through apical sodium-dependent bile acid transporter (SLC10A2) expression. J Pharmacol Exp Ther. 2011;336(1):188–196. doi: 10.1124/jpet.110.171736. [DOI] [PubMed] [Google Scholar]
- 59.Murphy EF, Cotter PD, Healy S, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59(12):1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 60.Myant NB, Mitropoulos KA. Cholesterol 7α-hydroxylase. J Lipid Res. 1977;18(2):135–153. [PubMed] [Google Scholar]
- 61.Nguyen A, Bouscarel B. Bile acids and signal transduction: role in glucose homeostasis. Cell Signal. 2008;20(12):2180–2197. doi: 10.1016/j.cellsig.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 62.Noh DO, Gilliland SE. Influence of bile on cellular integrity and β-galactosidase activity of Lactobacillus acidophilus . J Dairy Sci. 1993;76(5):1253–1259. doi: 10.3168/jds.S0022-0302(93)77454-8. [DOI] [PubMed] [Google Scholar]
- 63.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 64.Ridlon JM, Kang DJ, Hylemon PB, et al. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30(3):332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7(7):526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 66.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 67.Sayin SI, Wahlstrom A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Schnaith A, Kashkar H, Leggio SA, et al. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J Biol Chem. 2007;282(4):2695–2706. doi: 10.1074/jbc.M609784200. [DOI] [PubMed] [Google Scholar]
- 69.Sekirov I, Finlay BB. The role of the intestinal microbiota in enteric infection. J Physiol. 2009;587(17):4159–4167. doi: 10.1113/jphysiol.2009.172742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seok S, Fu T, Choi SE, et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516(7529):108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sinha J, Chen F, Miloh T, et al. β-Klotho and FGF-15/19 inhibit the apical sodium-dependent bile acid transporter in enterocytes and cholangiocytes. Am J Physiol Gastrointest Liver Physiol. 2008;295(5):G996–G1003. doi: 10.1152/ajpgi.90343.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190(7):2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol. 2010;192(19):4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Starr T, Child R, Wehrly TD, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11(1):33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swann JR, Want EJ, Geier FM, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. PNAS. 2011;108(Suppl. 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 77.Vrieze A, Out C, Fuentes S, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60(4):824–831. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 78.Wang DQ, Cohen DE, Carey MC. Biliary lipids and cholesterol gallstone disease. J Lipid Res. 2009;50(Suppl.):S406–S411. doi: 10.1194/jlr.R800075-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weingarden AR, Chen C, Bobr A, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306(4):G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wostmann BS. Intestinal bile acids and cholesterol absorption in the germfree rat. J Nutr. 1973;103(7):982–990. doi: 10.1093/jn/103.7.982. [DOI] [PubMed] [Google Scholar]
- 81.Yoshimoto S, Yoshimoto TM, Atarashi K, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Limaye PB, Renaud HJ, et al. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol Appl Pharmacol. 2014;277(2):138–145. doi: 10.1016/j.taap.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zollner G, Marschall HU, Wagner M, et al. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: pathogenetic and therapeutic considerations. Mol Pharm. 2006;3(3):231–251. doi: 10.1021/mp060010s. [DOI] [PubMed] [Google Scholar]