Abstract
The aim of this study is to investigate the effects of leucine (Leu) and histidine (His) on the expression of both the mammalian target of rapamycin (mTOR) signaling pathway-related proteins and caseins in immortalized bovine mammary epithelial cells (CMEC-H), using a single supplement through Western blotting. The Earle’s balanced salt solution (EBSS) was set as the control group and other treatment groups, based on the EBSS, were added with different concentrations of Leu or His, respectively. The results showed that, compared with the control group, the expression of caseins and the phosphorylation of mTOR (Ser2481), Raptor (Ser792), eIF4E (Ser209), and eEF2 (Thr56) increased with the Leu concentrations ranging from 0.45 to 10.80 mmol/L (P<0.01). The P-4EBP1 (Thr37) at 10.80 mmol/L Leu, and P-RPS6 (Ser235/236) at 5.40 to 10.80 mmol/L Leu all decreased. Similarly, the His supplementation from 0.15 to 9.60 mmol/L increased the expression of αs2-casein, β-casein, κ-casein, P-mTOR (Ser2481), P-Raptor (Ser792), P-S6K1 (Thr389), P-4EBP1 (Thr37), P-eIF4E (Ser209), and P-eEF2 (Thr56) (P<0.01) in CMEC-H, whereas the αs1-casein expression was only reduced at 9.60 mmol/L His, G protein β subunit-like protein (GβL) at 0.15 and 9.60 mmol/L His, and P-RPS6 at 4.80 to 9.60 mmol/L His. Our linear regression model assay suggested that the αs1-casein expression was positively correlated with P-mTOR (P<0.01), P-S6K1 (P<0.01), and P-eEF2 (P<0.01) for the addition of Leu, while the expressions of β-casein (P<0.01) and κ-casein (P<0.01) were positively correlated with P-eEF2 for the addition of His. In conclusion, the milk protein synthesis was up-regulated through activation of the mTOR pathway with the addition of Leu and His in CMEC-H.
Keywords: Bovine mammary epithelial cells, Leucine, Histidine, Western blotting, mTOR, Casein
1. Introduction
Dairy cow nutrient requirement systems regard essential amino acid (EAA) requirements in aggregate as metabolizable protein (MP), and presume a fixed efficiency of MP use for milk protein (Arriola Apelo et al., 2014b). The composition of EAAs in MP vastly affects milk protein synthesis. Since the exact amount of limiting amino acids (LAAs) in dairy cow’s diets was unknown, cows were easily overfed to meet the balance of MP requirements, which leads to a waste of amino acids (AAs) and poor N efficiency (Bionaz et al., 2012; Arriola Apelo et al., 2014a). In current dairy production systems, an average of 25% of dietary N is captured in milk, which results in approximately 75% of the dietary N being excreted in urine and feces (Hristov et al., 2004; Arriola Apelo et al., 2014b). Feeding animals with low-protein diets (Kalscheur et al., 2006) and supplementation of those deficient EAAs (lysine (Lys), methionine, leucine (Leu), isoleucine, valine, and histidine (His)) may maintain production, which can improve the total N efficiency (Kim et al., 2001; Wang et al., 2010; Appuhamy et al., 2011a; 2012), which is in accordance with Liebig’s hypothesis on developing the concept of the order of LAAs. This concept is commonly described by using the analogy of a water barrel with broken staves (Appuhamy et al., 2012). The utilization of AAs is important for mammary protein synthesis in lactating cows (Appuhamy et al., 2012; Arriola Apelo et al., 2014c). Previous research on AA metabolism primarily focused on optimizing balanced diets, which is essential for maintaining and enhancing milk protein synthesis in dairy cattle (Hanigan et al., 2001).
AAs not only serve as a precursor for protein synthesis but also as signaling molecules that regulate the protein synthesis in mammalians (Kimball, 2002; Arriola Apelo et al., 2014c). A great number of reports have shown that AA-induced stimulation of milk protein synthesis is partially mediated by the mammalian target of rapamycin (mTOR), a protein kinase present in the rapamycin-sensitive mTOR complex 1 (mTORC1) (Yang et al., 2006; Prizant and Barash, 2008; Appuhamy et al., 2012; Arriola Apelo et al., 2014c). mTORC1 consists of mTOR, regulatory associated protein of TOR (Raptor), and a G protein β subunit-like protein (GβL) (Kim, 2009). A great deal of evidence indicated that P-mTOR at Ser2448 was an indicator of mTOR pathway activity in immortalized bovine mammary epithelial cells (MAC-T) (Appuhamy et al., 2011b; 2012). Prizant and Barash (2008) showed that P-mTOR in Ser2448 was completely inhibited by His supplementation, but phosphorylation of the ribosomal protein S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E binding protein (4EBP1) could still be detected. AA supplementation may have contributed to other existent phosphorylation sites of mTOR that were not detected, resulting in phosphorylation of downstream effectors. Peterson et al. (2000) and Gerasimovskaya et al. (2005) demonstrated that the P-mTOR at Ser2481 was also a good biomarker for mTOR pathway activation. Cheng et al. (2004) demonstrated that the P-mTOR in Thr2446 acted as a nutrient-regulated phosphorylation site located in the mTOR catalytic domain. We cannot exclude the possibility that phosphorylation of mTOR in Ser2448 was important, yet it was not the only one regulating mTOR activity in bovine mammary epithelial cells (CMECs). Kim et al. (2002) confirmed that P-Raptor is associated with mTOR and modulated by AAs, which regulate the mTOR kinase activity. When activated by AAs, mTORC1 in turn catalyzed S6K1 and 4EBP1 phosphorylation, followed by phosphorylating the ribosomal protein S6 (RPS6) (Wang and Proud, 2006). S6K1 was a target of mTOR activated in Thr389 whose phosphorylation status was a main indicator of the AA-induced mTORC1-activated signaling pathway (Moshel et al., 2006; Prizant and Barash, 2008; Appuhamy et al., 2012; Arriola Apelo et al., 2014c). In addition, phosphorylation of 4EBP1 by mTOR increased the initial reaction rates (Beugnet et al., 2003). When 4EBP1 was activated, the expression of a regulatory protein binding into the mRNA cap-binding protein eukaryotic initiation factor 4E (eIF4E) was also promoted. When 4EBP1 was unphosphorylated, cap-dependent translation was inhibited by competitively blocking the binding of eukaryotic translation initiation factor 4G (eIF4G) to eIF4E (Gingras et al., 1999; Harris and Lawrence, 2003). The elongation step of mRNA translation was enhanced by eukaryotic elongation factor 2 (eEF2) (Wang and Proud, 2006) and was inhibited by its phosphorylation on Thr56, which suggested that eEF2 may be a limiting factor in milk protein synthesis (Christophersen et al., 2002; Appuhamy et al., 2011b).
These studies have shown that Leu and His in addition to Lys and Met are main LAAs in forage diets (Kim et al., 2001; Korhonen et al., 2002). Leu, a branched chain amino acid (BCAA), is currently the most prevalent EAA in the diet (Korhonen et al., 2002; Appuhamy et al., 2012). According to previous reports, Leu was involved in cell growth and differentiation through the mTOR pathway (Proud, 2007; Stipanuk, 2007). Moreover, Leu produced a dose-dependent regulation of mTOR and served as a key intracellular regulatory factor of the mTORC1 pathway in CMECs (Moshel et al., 2006; Prizant and Barash, 2008; Appuhamy et al., 2012; Durán and Hall, 2012).
Kim et al. (2001) found that His was the first limiting factor for milk protein secretion. Some traditional experiments indicated that the perfusion of His promoted milk protein synthesis (Bequette et al., 2000). Likewise, some cell experiments demonstrated that the addition of His activated the mTOR pathway, which up-regulated phosphorylation of the downstream protein and ultimately promoted milk protein synthesis (Appuhamy et al., 2012). Prizant and Barash (2008) also reported that His was a key intracellular regulatory factor of the mTORC1 pathway in L-1 cells, a cloned cell line derived from lactating bovine mammary gland, however, negatively regulated P-S6K1, and decreased the β-casein synthesis rate. So, it is meaningful to explore the mechanism of how His influenced the mTOR pathway.
We hypothesize that AAs affect milk protein synthesis in CMEC-H through phosphorylation of translation factors of the mTOR signaling pathway. Although the effects of EAAs on mTOR cell signaling and milk protein synthesis have been extensively explored (Moshel et al., 2006; Burgos and Cant, 2010; Toerien et al., 2010), effects of different levels of Leu or His on phosphorylation of Raptor on Ser792, mTOR on Ser2481, and GβL expression involved in casein protein synthesis were not clear in bovine mammary cells. Our study investigated the regulatory effects of different concentrations of Leu and His on the mTOR signaling pathway and the relationships between these signaling protein activities with milk protein synthesis.
2. Materials and methods
In the present study, all animal care and experimental procedures were approved and conducted under the established standards of the Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing, China.
Unless otherwise stated, all components were purchased from the Sigma-Aldrich Shanghai Trading Co. Ltd. (Shanghai, China). The Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F12 (diluted 1:1 (v/v), 1439945/1491066, Invitrogen Trading Co. Ltd., China) was used as the basal growth medium, supplemented with 10% fetal bovine serum (FBS) (Invitrogen Trading Co. Ltd., China) and 100 μg/ml penicillin-streptomycin solution (C0222, Beyotime Institute of Biotechnology, China). The differentiation medium contained bovine insulin, progesterone, bovine holo-transferrin, hydrocortisone, bovine epithelial growth factor, and bovine estradiol (Sigma-Aldrich, Cat. Nos. I4434, T1283, P8783, H0888, E4127, and E2758, respectively).
Individual AAs (L-Leu and L-His, Cat. Nos. L8912 and H5659, respectively) were purchased from the Sigma-Aldrich Shanghai Trading Co. Ltd., China. The non-and site-specific phosphorylated antibodies against mTOR (Ser2481, YT2913/YP1134), 4EBP1 (Thr37, YT0018/YP0001), RPS6 (Ser235/236, YT4139/YP0832), and eIF4E (Ser209, YT1516/YP0094) were purchased from Immuno Way. The total and site-specific phosphorylated antibodies against Raptor (Ser792, No. 2280/2083), S6K1 (Thr389, No. 9202/9205), eEF2 (Thr56, No. 2332/2331), and GβL (No. 3274) were purchased from Cell Signaling Technology (Danvers, MA, USA).
The αs1-casein antibody (SAB1401093) and κ-casein antibody (SAB1401094) were purchased from Sigma-Aldrich Shanghai Trading Co., Ltd. China, the αs2-casein antibody (bs-10034R) was purchased from Bioss (Beijing Biosynthesis Biotechnology Co., Ltd., China) and the β-casein antibody (orb18512) was purchased from Biorbyt (Cambridge, UK). β-Actin (ab8226), used as a loading control, was purchased from Abcam Trading (Shanghai) Company Ltd. (China).
2.1. Cell culture and experimental design
The immortalized bovine mammary epithelial cell line (CMEC-H) was established in our previous work (Hu et al., 2014). The cells were cultured at 37 °C with 5% CO2 and digested with 0.25% (2.5 g/L) trypsin-0.02% etilendiaminetetraacetic acid (EDTA) (C0203, Beyotime Institute of Biotechnology, China).
The Earle’s balanced salt solution (EBSS, CC0043, Leagene Biotechnology, China) was used as a substitute of medium without AAs (Table 1), and was set as a negative control in the experimental design. The EBSS was supplemented with 10% FBS and was used as a positive control. The concentrations of Leu and His in the DMEM/F12 medium were 0.45 and 0.15 mmol/L, respectively. EAAs were added according to the concentrations described in Tables 2 and 3. The CMEC-H cells were cultured in 9-cm plates (172958, Thermo Scientific, China) to 80% confluence and induced to be differentiated by the differentiation medium for 24 h. The differentiated CMEC-H cells were incubated in serum-free, complete DMEM/F12 media overnight. Leu or His was supplemented individually for 6 h at the indicated concentrations. Each experiment was triplicated and each of the whole experiments was repeated three times on three different days.
Table 1.
Composition of the Earle’s balanced salt solution (EBSS) medium*
| Component | Concentration (mmol/L) |
| Anhydrous CaCl2 | 1.050 |
| CuSO4∙5H2O | 5.2×10−6 |
| Fe(NO3)3∙9H2O | 1.238×10−4 |
| FeSO4∙7H2O | 0.002 |
| Anhydrous MgCl2 | 0.301 |
| Anhydrous MgSO4 | 0.407 |
| KCl | 4.157 |
| NaHCO3 | 14.286 |
| NaCl | 120.612 |
| Anhydrous Na2HPO4 | 0.500 |
| NaH2PO4∙H2O | 0.453 |
| ZnSO4∙7H2O | 0.002 |
| D-Glucose (dextrose) | 17.506 |
| Phenol red | 0.022 |
Modified from Gao et al. (2015)
Table 2.
Leu levels in the medium for the in vitro culture of CMEC-H from dairy cattle
| Group | EBSS | Leu (mmol/L) | 10% FBS |
| Negative control | + | ||
| Positive control | + | + | |
| Treatment | + | 0.45 | |
| + | 1.35 | ||
| + | 5.40 | ||
| + | 10.80 |
DMEM/F12 base media contained leucine (0.45 mmol/L) at a level sufficient for normal cellular processes. +: add
Table 3.
His levels in the medium for the in vitro culture of CMEC-H from dairy cattle
| Group | EBSS | His (mmol/L) | 10% FBS |
| Negative control | + | ||
| Positive control | + | + | |
| Treatment | + | 0.15 | |
| + | 1.20 | ||
| + | 4.80 | ||
| + | 9.60 |
DMEM/F12 base media contained histidine (0.14999 mmol/L) at a level sufficient for normal cellular processes. +: add
2.2. Protein immunoblot analysis
The cell samples were homogenized in an ice-cold radio immunoprecipitation assay (RIPA) buffer (P0013, Beyotime Institute of Biotechnology, China) containing 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), 1% (v/v) protease, and a phosphatase inhibitor cocktail (78430, Thermo Scientific). Cell lysates were centrifuged at 2000×g for 3 min and the supernatants were transferred to new tubes. The protein concentrations of the samples were determined by bicinchoninic acid (BCA) assay kits (P0010S, Beyotime Institute of Biotechnology, China). The lysates (30 μg of protein) were loaded onto gels, separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and blotted onto polyvinylidene fluoride (PVDF) transfer membranes (0.45 and 0.2 μmol/L immobilon-P, IPVH00010 and ISEQ00010, Millipore). The membranes were blocked for 1 h (blot-PO buffer, WBAVDP001, Millipore) and incubated overnight at 4 °C in primary antibodies (appropriately diluted 1:1000 in phosphate-buffered saline (PBS)) against total and site-specific phosphorylated mTOR (Ser2481), 4EBP1 (Thr37), RPS6 (Ser235/236), eIF4E (Ser209), Raptor (Ser792), S6K1 (Thr389), eEF2 (Thr56), and GβL. The amounts of synthesized milk proteins were determined by antibodies against αs1-casein (1:1000), αs2-casein (1:400), β-casein (1:500), and κ-casein (1:1000). The blots were washed three times with Tris-buffered saline containing 0.02% (v/v) Tween-20 (TBST), and then incubated with a second antibody (anti-rabbit IgG, A9169, Sigma; anti-goat IgG, AP106P, Millipore; anti-Mouse IgG, A9044, Sigma) and diluted at 1:5000 in PBS.
Immunoreactive bands were visualized by Pierce ECL Western Blotting Substrate kits (32106, Thermo Scientific) according to the manufacturer’s instructions and were then exposed to X-ray film. The protein band densities were determined by the Image J2x 2.1.4.7 Analyzer (Rawak Software, Inc., Germany). The relative values from the AA-supplemented cells were related to their AA-deprived controls.
2.3. Statistical analysis
At least three independent experiments were done with identical results and all data were tested using the Duncan’s test for post-hoc multiple comparisons of treatment means by SAS software (SAS 9.2 Inst., Cary, NC, USA). Differences between experimental groups were considered significant at a P-value of <0.05. The random effect in the one-way analysis of variance (ANOVA) model was replicated and the fixed effect was the levels of Leu and His. Meaningful relationships among the phosphorylation state of the signaling proteins and casein protein expression in CMEC-H were quantified with simple linear regression models using the REG procedure of SAS.
3. Results
3.1. Effects of Leu or His supplementation on the expression of caseins
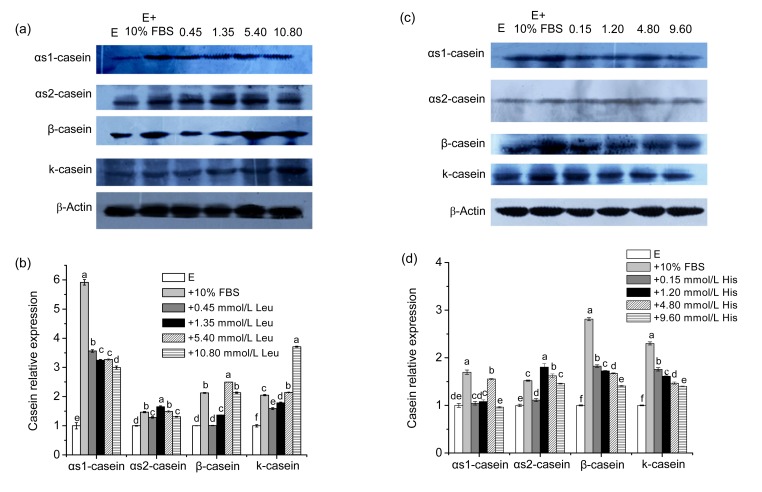
Compared with the negative control group, our data demonstrated that the addition of Leu to EBSS generated dose-responsive effects on the casein expression. Figs. 1a and 1b show that Leu had stimulatory effects on the casein expression in the range of 0.45 to 10.80 mmol/L. Specific to each casein, αs1-casein, αs2-casein, β-casein, and κ-casein reached their highest expression at 0.45, 1.35, 5.40, and 10.80 mmol/L Leu with an increase of 260%, 66%, 150%, and 270%, respectively.
Fig. 1.
Effects of Leu and His on the expression of caseins in CMEC-H
(a) Leu was supplemented for 6 h at the indicated concentrations. Casein levels were determined by immunoblot analysis. Numbers above the lanes refer to the levels (mmol/L) of the supplemented Leu relative to that in the Earle’s balanced salt solution. (b) Densitometric analysis of signals obtained from the casein immunoblot (a). (c) His was supplemented for 6 h at the indicated concentrations. Casein levels were determined by immunoblot analysis. Numbers above the lanes refer to the levels (mmol/L) of the supplemented His relative to that in the Earle’s balanced salt solution. (d) Densitometric analysis of signals obtained from the casein immunoblot (c). β-Actin was assessed as a loading control. A representative blot and quantitation of three independent experiments were shown. In all panels, data represent the mean±SD. E: Earle’s balanced salt solution; E+10% FBS: Earle’s balanced salt solution supplemented with 10% fetal bovine serum. Data in the same concentration marked with different letters represent a significant difference (P<0.05)
Figs. 1c and 1d show the effects of His supplementation on the casein expression. Compared with the negative control, the expression of casein was all promoted by His in the range of 0.15 to 9.60 mmol/L, except that αs1-casein expression was decreased at 9.6 mmol/L His. In addition, each casein, αs1-casein, αs2-casein, β-casein, and κ-casein reached their highest expression at 4.80, 1.20, 0.15, and 0.15 mmol/L His with 1.6-, 1.8-, 1.8-, and 1.8-fold changes, respectively.
3.2. Effects of Leu or His supplementation on the expression of mTORC1
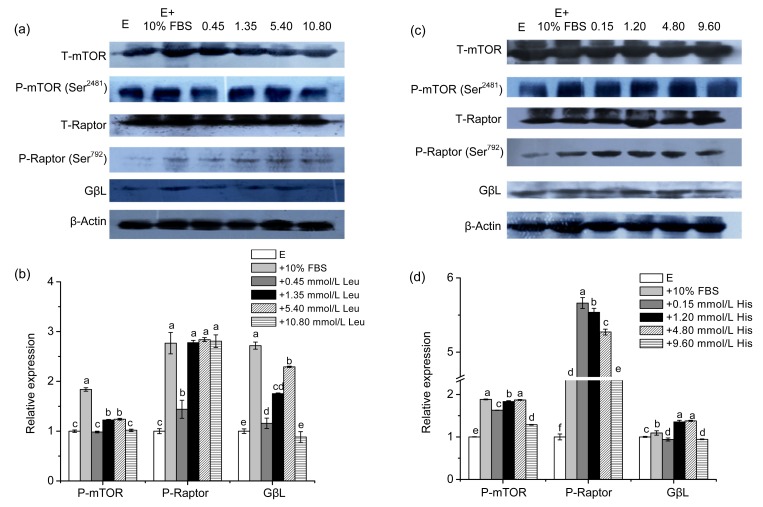
Our results indicated that the Leu supplementation affected the phosphorylation of mTORC1 in a dose-dependent manner. Figs. 2a and 2b show that, compared with the negative control group, only the P-Raptor was markedly increased with the addition of Leu from 0.45 to 10.80 mmol/L. The GβL expression at 10.80 mmol/L Leu and mTOR phosphorylation at 0.45 and 10.80 mmol/L Leu were not significantly changed, and they were significantly enhanced at other concentrations. The changes of mTORC1 all reached their summit at 5.40 mmol/L Leu.
Fig. 2.
Effects of Leu and His on the expression of mTORC1 in CMEC-H
(a) Leu was supplemented for 6 h at the indicated concentrations. mTORC1 levels were determined by immunoblot analysis. Numbers above the lanes refer to the levels (mmol/L) of the supplemented Leu relative to that in the Earle’s balanced salt solution. (b) Densitometric analysis of signals obtained from the mTORC1 immunoblot (a). (c) His was supplemented for 6 h at the indicated concentrations. mTORC1 levels were determined by immunoblot analysis. Numbers above the lanes refer to the levels (mmol/L) of the supplemented His relative to that in the Earle’s balanced salt solution. (d) Densitometric analysis of signals obtained from the mTORC1 immunoblot (c). β-Actin was assessed as a loading control. A representative blot and quantitation of three independent experiments were shown. In all panels, data represent the mean±SD. E: Earle’s balanced salt solution; E+10% FBS: Earle’s balanced salt solution supplemented with 10% fetal bovine serum. Data in the same concentration marked with different letters represent a significant difference (P<0.05)
Figs. 2c and 2d show the influence of His supplementation on mTORC1. P-Raptor was dramatically increased from 0.15 to 9.60 mmol/L His. The P-mTOR and GβL expression were slightly enhanced in the full concentration range, except that the GβL expression was subtly reduced at 0.15 and 9.60 mmol/L His.
3.3. Effects of Leu or His level on the expression of mTOR downstream signaling proteins
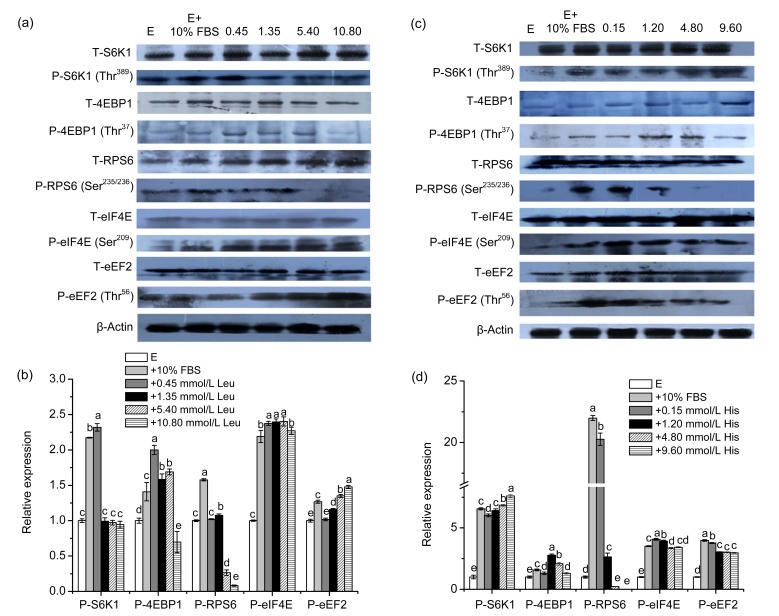
Figs. 3a and 3b show that Leu had obvious stimulatory effects on phosphorylation of eEF2 and eIF4E compared to the negative control group, while S6K1 phosphorylation with the addition of Leu was not significantly changed except at 0.45 mmol/L Leu with 2.2-fold changes. For 4EBP1 phosphorylation, its enhancement was observed from 0.45 to 5.40 mmol/L Leu, while 10.80 mmol/L Leu impaired its phosphorylation. Leu supplementation had a minor enhancing effect on the RPS6 phosphorylation level between 0.45 and 1.35 mmol/L, while the phosphorylation level was inhibited over 1.35 mmol/L Leu.
Fig. 3.
Effects of Leu and His on the expression of mTOR downstream signaling proteins in CMEC-H
(a) Leu was supplemented for 6 h at the indicated concentrations. mTOR downstream signaling protein levels were determined by immunoblot analysis. Numbers above the lanes refer to the levels (mmol/L) of the supplemented Leu relative to that in the Earle’s balanced salt solution. (b) Densitometric analysis of signals obtained from the mTOR downstream signaling protein immunoblot (a). (c) His was supplemented for 6 h at the indicated concentrations. mTOR downstream signaling protein levels were determined by immunoblot analysis. Numbers above the lanes refer to the levels (mmol/L) of the supplemented His relative to that in the Earle’s balanced salt solution. (d) Densitometric analysis of signals obtained from the mTOR downstream signaling protein immunoblot (c). β-Actin was assessed as a loading control. A representative blot and quantitation of three independent experiments were shown. In all panels, data represent the mean±SD. E: Earle’s balanced salt solution; E+10% FBS: Earle’s balanced salt solution supplemented with 10% fetal bovine serum. Data in the same concentration marked with different letters represent a significant difference (P<0.05)
Figs. 3c and 3d suggest that His supplementation up-regulated all of the studied mTOR downstream proteins in the full concentration range except P-RPS6 at 9.60 mmol/L His and S6K1 phosphorylation which increased in a dose-dependent manner. In addition, at 9.60 mmol/L His, P-S6K1 was nearly 7 times greater than that of the control group. The phosphorylation of 4EBP1 reached its peak (P<0.01) at 1.20 mmol/L His. It was noteworthy that P-RPS6 at 0.15 mmol/L His was 20 times greater than that of the control group.
3.4. Relationships between casein expression and mTOR pathway-related protein changes with Leu supplementation
A simple linear regression model assay demonstrated that αs1-casein expression was positively associated with phosphorylation of mTOR, GβL, Raptor, S6K1, 4EBP1, eEF2, RPS6, and eIF4E (P<0.01), respectively (Table 4). With the addition of Leu, a 1-fold increase in αs1-casein expression was related to 4.85-, 2.04-, 1.15-, 2.48-, 2.44-, 8.14-, 1.41-, and 2.12-fold changes in mTOR, GβL, Raptor, S6K1, 4EBP1, eEF2, RPS6, and eIF4E phosphorylation, respectively. A notably positive relationship (R 2=0.7820, 0.5815, 0.7881, 0.7835, and 0.6164; P<0.01) was found between αs1-casein expression and mTOR, GβL, S6K1, eEF2 and eIF4E phosphorylation, respectively. No phosphorylation of other mTOR signaling proteins explained the further variation in αs1-casein protein expression in CMEC-H (Table 4).
Table 4.
Regression analyses for relationships among phosphorylation ratios of signaling proteins and αs1-casein, αs2-casein, β-casein, κ-casein in bovine mammary epithelial cells cultured in media with Leu for 6 h
| Dependent variable | Independent variable | P-value | Regression coefficient | R 2 |
| αs1-casein | mTOR | 0.0001 | 4.84738 | 0.7820 |
| GβL | 0.0001 | 2.03558 | 0.5815 | |
| Raptor | 0.0052 | 1.15177 | 0.3572 | |
| S6K1 | 0.0001 | 2.48016 | 0.7881 | |
| 4EBP1 | 0.0010 | 2.44002 | 0.4710 | |
| eEF2 | 0.0001 | 8.14314 | 0.7835 | |
| RPS6 | 0.0087 | 1.40988 | 0.3179 | |
| eIF4E | 0.0001 | 2.11775 | 0.6164 | |
| αs2-casein | mTOR | 0.0285 | 0.41805 | 0.2200 |
| GβL | 0.0046 | 0.24768 | 0.3663 | |
| Raptor | 0.0001 | 0.21865 | 0.6179 | |
| S6K1 | 0.0008 | 0.29594 | 0.4823 | |
| 4EBP1 | 0.0172 | 0.28417 | 0.2630 | |
| eEF2 | 0.0341 | 0.68184 | 0.2043 | |
| RPS6 | 0.3133 | 0.08850 | 0.0049 | |
| eIF4E | 0.0021 | 0.26688 | 0.4225 | |
| β-casein | mTOR | 0.0170 | 1.20726 | 0.2638 |
| GβL | 0.0013 | 0.73002 | 0.4534 | |
| Raptor | 0.0001 | 0.60842 | 0.6670 | |
| S6K1 | 0.0068 | 0.68103 | 0.3370 | |
| 4EBP1 | 0.4660 | 0.25321 | −0.0267 | |
| eEF2 | 0.0188 | 2.00066 | 0.2556 | |
| RPS6 | 0.4630 | −0.17444 | −0.0262 | |
| eIF4E | 0.0013 | 0.73944 | 0.4531 | |
| κ-casein | mTOR | 0.2831 | 0.82785 | 0.0136 |
| GβL | 0.6733 | 0.15885 | −0.0504 | |
| Raptor | 0.0018 | 0.71268 | 0.4338 | |
| S6K1 | 0.0475 | 0.74591 | 0.1751 | |
| 4EBP1 | 0.2856 | −0.52169 | 0.0128 | |
| eEF2 | 0.2485 | 1.48963 | 0.0249 | |
| RPS6 | 0.0626 | −0.60010 | 0.1503 | |
| eIF4E | 0.0017 | 1.03420 | 0.4375 |
The αs2-casein expression was positively correlated with phosphorylation of mTOR, GβL, Raptor, S6K1, 4EBP1, eEF2, and eIF4E (P<0.05), respectively (Table 4). A 1-fold increase in αs2-casein protein expression was related to 0.42-, 0.25-, 0.22-, 0.30-, 0.28-, 0.68-, and 0.27-fold changes in mTOR, GβL, Raptor, S6K1, 4EBP1, eEF2, and eIF4E phosphorylation, respectively. However, Raptor phosphorylation appeared to be the primary driver when computed in a multivariate model, which explained 61.79% of the observed variation (Table 4).
The β-casein expression was positively associated with phosphorylation of mTOR, GβL, Raptor, S6K1, eEF2, and eIF4E (P<0.05), respectively (Table 4). A 1-fold increase in the β-casein protein expression was related to 1.21-, 0.73-, 0.61-, 0.68-, 2.00-, and 0.74-fold changes in mTOR, GβL, Raptor, S6K1, eEF2, and eIF4E phosphorylation, respectively. A notably positive relationship (R 2=0.6670, P<0.01) was found between the β-casein expression and P-Raptor, while no correlations were found between the expression of β-casein and phosphorylation of 4EBP1 (P=0.4660) or RPS6 (P=0.4630) in CMEC-H (Table 4).
The κ-casein expression was positively associated with phosphorylation of Raptor, S6K1, and eIF4E (P<0.05), respectively (Table 4). No correlations were found between the κ-casein expression and phosphorylation of mTOR (P=0.2831), GβL (P=0.6733), 4EBP1 (P=0.2856), eEF2 (P=0.2485), or RPS6 (P=0.0626) in CMEC-H (Table 4).
3.5. Relationships between casein expression and mTOR pathway-related protein changes with His supplementation
The expression of αs1-casein was positively correlated with phosphorylation of mTOR, GβL, and eEF2 (P<0.05), respectively (Table 5). A 1-fold increase in the αs1-casein expression was related to 0.59-, 0.60-, and 0.14-fold changes in mTOR, GβL, and eEF2 phosphorylation, respectively. A notable positive relationship (R 2=0.5133, P<0.01) was found between the αs1-casein expression and eEF2 phosphorylation.
Table 5.
Regression analyses for relationships among phosphorylation ratios of signaling proteins and αs1-casein, αs2-casein, β-casein, κ-casein in bovine mammary epithelial cells cultured in media with His for 6 h
| Dependent variable | Independent variable | P-value | Regression coefficient | R 2 |
| αs1-casein | mTOR | 0.0023 | 0.58689 | 0.4149 |
| GβL | 0.0234 | 0.60173 | 0.2370 | |
| Raptor | 0.5879 | 0.02210 | −0.0424 | |
| S6K1 | 0.1926 | 0.10670 | 0.0476 | |
| 4EBP1 | 0.3503 | 0.05559 | −0.0044 | |
| eEF2 | 0.0005 | 0.13542 | 0.5133 | |
| RPS6 | 0.3110 | 0.01383 | 0.0055 | |
| eIF4E | 0.1374 | 0.08256 | 0.0784 | |
| αs2-casein | mTOR | 0.0007 | 0.60885 | 0.4954 |
| GβL | 0.0018 | 0.74109 | 0.4307 | |
| Raptor | 0.0598 | 0.07033 | 0.1544 | |
| S6K1 | 0.0020 | 0.21580 | 0.4267 | |
| 4EBP1 | 0.0001 | 0.17724 | 0.5790 | |
| eEF2 | 0.1536 | 0.06191 | 0.0682 | |
| RPS6 | 0.9829 | 0.00029 | −0.0625 | |
| eIF4E | 0.0029 | 0.14346 | 0.3995 | |
| β-casein | mTOR | 0.0003 | 1.22687 | 0.5341 |
| GβL | 0.0263 | 1.10642 | 0.2271 | |
| Raptor | 0.3081 | 0.07720 | 0.0063 | |
| S6K1 | 0.0628 | 0.27713 | 0.1500 | |
| 4EBP1 | 0.2005 | 0.14074 | 0.0440 | |
| eEF2 | 0.0001 | 0.33806 | 0.9638 | |
| RPS6 | 0.0003 | 0.07675 | 0.5368 | |
| eIF4E | 0.0049 | 0.26810 | 0.3624 | |
| κ-casein | mTOR | 0.0006 | 0.85113 | 0.5050 |
| GβL | 0.0544 | 0.69444 | 0.1629 | |
| raptor | 0.1936 | 0.06931 | 0.0472 | |
| S6K1 | 0.0501 | 0.20637 | 0.1704 | |
| 4EBP1 | 0.2484 | 0.09071 | 0.0250 | |
| eEF2 | 0.0001 | 0.23346 | 0.9048 | |
| RPS6 | 0.0001 | 0.05838 | 0.6228 | |
| eIF4E | 0.0015 | 0.20877 | 0.4464 |
The αs2-casein expression was positively associated with phosphorylation of mTOR, GβL, S6K1, 4EBP1, and eIF4E (P<0.01), respectively (Table 5). A 1-fold increase in the αs2-casein expression was related to 0.61-, 0.74-, 0.22-, 0.18-, and 0.14-fold changes in mTOR, GβL, S6K1, 4EBP1, and eIF4E phosphorylation, respectively. A positive relationship (R 2=0.5790, P<0.01) was found between the αs2-casein expression and P-4EBP1.
The β-casein expression was positively associated with phosphorylation of mTOR, GβL, eEF2, RPS6, and eIF4E (P<0.05), respectively (Table 5). A 1-fold increase in the β-casein expression was related to 1.23-, 1.11-, 0.34-, and 0.27-fold changes in mTOR, GβL, eEF2, and eIF4E phosphorylation, respectively. A notable positive relationship (R 2=0.5341, 0.9638, and 0.5368, P<0.01) was found between the β-casein expression and mTOR, eEF2 and RPS6 phosphorylation, respectively.
The κ-casein expression was positively associated with phosphorylation of mTOR, eEF2, RPS6, and eIF4E (P<0.01), respectively (Table 5). A 1-fold increase in the κ-casein expression was related to 0.85-, 0.23-, and 0.21-fold changes in mTOR, eEF2, and eIF4E phosphorylation, respectively. A notable positive relationship (R 2=0.5050, 0.9048, and 0.6228, P<0.01) was found between the κ-casein expression and mTOR, eEF2 and RPS6 phosphorylation, respectively.
4. Discussion
In this paper, the functions and potential molecular mechanisms of Leu and His effects on the mTOR signaling pathway in the regulation of milk protein synthesis were studied. In order to eliminate the impact of the other AAs on CMEC-H, all AAs were removed from EBSS and Leu or His was then individually added to it.
In our study, all four forms of caseins were up-regulated by Leu or His supplementation with the exception of 9.60 mmol/L His on αs1-casein, which supported the notion that both Leu and His are key limiting factors for milk protein synthesis.
It is widely accepted that mTOR is a key regulator of milk protein synthesis and most reports were concerned with the role of AAs in the regulation of P-mTOR on Ser2448 in milk protein synthesis (Prizant and Barash, 2008; Appuhamy et al., 2012; Arriola Apelo et al., 2014c). Here we identify another mTOR phosphorylation site on Ser2481 as an indicator of activated mTOR pathway in CMEC-H, which was increased by the addition of Leu or His in the full concentration range. An exception to this was the supplementation of Leu at 0.45 mmol/L with no obvious change. This data indicated that P-mTOR on Ser2481 could act as an alternative switch to integrate signals from Leu and His to regulate protein translation. The effects of Leu and His supplementation on two other mTORC1 components, Raptor and GβL, were also studied. The association between Raptor and mTOR was manipulated by AAs. Raptor acts as a scaffold protein regulating the assembly, localization, and substrate binding of mTORC1 (Laplante and Sabatini, 2012), and it is also used for the mTOR-catalyzed phosphorylation of 4EBP1 and mediates TOR action in vivo (Hara et al., 2002). With AA deprivation, the mTOR kinase activity is inactive (Kim et al., 2013). The addition of Leu and His activates mTOR kinase to increase the Raptor expression (Kim et al., 2002). GβL also plays a positive role in mTOR activation by AA supplementation. The binding of GβL to the mTOR kinase domain stabilized the interaction between the Raptor and mTOR (Kim et al., 2003). Our results indicated that Raptor notably increased (1.8 and 4.7 times) in comparison with the control group, when added Leu and His, respectively. GβL expression increased in the range of 0.45 and 5.40 mmol/L of Leu with a 2.5-fold change in comparison with the control, whereas His supplementation slightly enhanced GβL expression in the same range. Both GβL and P-mTOR demonstrated the highest changes at 5.40 mmol/L Leu or 4.80 mmol/L His. Previous studies indicated that GβL positively correlated with P-mTOR on Ser2448 (Kim et al., 2003), and that there was a strong relationship between GβL and P-mTOR on Ser2481.
S6K1, a major downstream target of mTOR, serves as an indicator of activation of the mTORC1 pathway (Yang et al., 2006; Prizant and Barash, 2008; Appuhamy et al., 2012; Arriola Apelo et al., 2014c). As most reports showed, 0.45 mmol/L Leu dramatically increased S6K1 phosphorylation.
The effect of Leu concentrations greater than 0.45 mmol/L on S6K1 phosphorylation was monitored, and there was no obvious change in comparison with the control group. P-S6K1 increased along with His supplementation in the full concentration range. This was in contrast to the negative effects of His on P-S6K1 in CMECs reported by Prizant and Barash (2008). However, as a whole, P-S6K1 has a dose-dependent effect from His concentration. However, our results support the traditional perfused experiment in which His was positively associated with P-S6K1 in dairy cows (Toerien et al., 2010). The difference between those two studies was potentially due to the different time response of the His addition. In the study of Prizant and Barash (2008), CMECs were incubated in His for 10 min; however, in our study 6 h was used for incubation. We thought after starvation, the CMECs were in a stress period responding to the His addition in a short time, because they needed a period to repair the damages resulting from the devoid of AAs. So the addition of His may first maintain the cell survival instead of proving the milk synthesis.
4EBP1 is another major downstream target of mTOR (Li et al., 2005). Activated mTOR regulates protein translation by directly inducing P-4EBP1. Our results indicated that the addition of Leu and His to the EBSS increased P-4EBP1 in CMEC-H. Furthermore, only the highest concentrations of Leu inhibited its phosphorylation, which further validated the notion that 4EBP1 was up-regulated by mTOR.
RPS6 is phosphorylated by S6K1. In our study, Leu supplementation slightly improved RPS6 phosphorylation levels between 0.45 and 1.35 mmol/L, while the phosphorylation level was inhibited over 1.35 mmol/L. As a substrate of S6K1, RPS6 was positively correlated with S6K1 phosphorylation between 0.45 and 1.35 mmol/L Leu, which was consistent with the results of Appuhamy et al. (2012) that focused on 0.45 mmol/L Leu. On the other hand, His supplementation vastly enhanced the RPS6 phosphorylation level at 0.15 mmol/L, followed by a sharply decreased phosphorylation level between 1.20 and 9.60 mmol/L. Our study revealed that there may have been a negative feedback between RPS6 and S6K1 beyond 1.35 mmol/L Leu or 0.15 mmol/L His supplementation.
The eIF4E is involved in directing ribosomes to the cap structure of mRNAs (Stipanuk, 2007). The mTORC1 phosphorylates 4EBP1 to release eIF4E from the 4EBP1-eIF4E complex, thus promoting eIF4E-dependent translation initiation. The stimulatory effects of His and Leu supplementation on the phosphorylation of eIF4E in CMECs were notably enhanced, which was in accordance with P-4EBP1 changes.
The eEF2 was known to catalyze the mRNA translocation step in the elongation process during protein synthesis (Kaul et al., 2011). Compared with initiation, eEF2 may be an important factor and is likely to be a rate-limiting enzyme for milk protein synthesis under AA-deprived conditions in the mammary gland of cows only in late-lactation because it consumes more energy in elongation (Merrick, 1992; Wilde et al., 1997; Christophersen et al., 2002). Direct spatially-controlled inhibition of eEF2 phosphorylation induces local translational activation (Li et al., 2005; Kaul et al., 2011). Our study demonstrated that both Leu and His (P<0.01) enhanced eEF2 phosphorylation levels in comparison with a negative control. However, Leu supplementation had enhanced eEF2 phosphorylation by no more than a 20% increase.
A linear regression model was employed to analyze the relationships among EAA, milk protein and mTOR pathway-related proteins. Appuhamy et al. (2014) reported that intracellular EAA tended (P=0.09) to positively affect mTOR phosphorylation, and the phosphorylation of mTOR (P<0.01) was positively associated with fractional rates of mammary protein synthesis (FSR) in bovine mammary tissue slices. Consistently, in the current work, a 1-fold increase in mTOR phosphorylation and GβL expression was associated with a remarkable increase in αs1-casein, αs2-casein, and β-casein expression (P<0.05), provision of extracellular Leu and His in CMEC-H. However, Leu supplementation on four casein expressions explained 115%, 22%, 60%, and 71% (P<0.01), respectively, of the variability in Raptor phosphorylation. Alternatively, the addition of His to casein protein expression was insignificantly associated with Raptor phosphorylation (P=0.59, 0.06, 0.31, and 0.19, respectively). Both Leu and His significantly increased mTOR phosphorylation. Leu significantly activated phosphorylation of S6K1, which enhanced four casein expressions in CMEC-H (P<0.05). However, when the addition of His was analyzed with the same regression model, the S6K1 phosphorylation had a notable association with αs2-casein and κ-casein (P<0.05).
With the addition of Leu or His, αs1-casein was positively related to eEF2 phosphorylation (P<0.01) in CMEC-H. In previous experiments, P-eEF2 was associated with P-mTOR, which tended (P=0.10) to impose a negative effect on casein synthesis rates, independent of mTOR phosphorylation (Appuhamy et al., 2014). Therefore, the inconsistent results of the effects of Leu and His on eEF2 phosphorylation could be due to different substitutes of the medium. In our study, we did not contain AAs and insulin, while insulin and nonessential amino acids (NEAAs) were present in the medium of the study of Appuhamy et al. (2012). It is possible that the potential presence of greater nutrients in substituting in the medium may have partly masked the single AA effects on the cellular status in CMECs. We believe that the interaction of AAs and insulin is more significant than the influence of adding a single AA on mTOR pathways.
5. Conclusions
In summary, the results of this study demonstrated that Leu or His supplementation stimulated αs-casein, β-casein, and κ-casein expression. Casein expression, P-mTOR (Ser2481), P-Raptor (Ser792), P-eIF4E (Ser209), and P-eEF2 (Thr56) increased with Leu concentration in the range of 0.45 to 10.80 mmol/L. Leu supplementation suppressed P-4EBP1 (Thr37) at 10.80 mmol/L, and inhibited P-RPS6 (Ser235/236) over 5.40 mmol/L. Supplementing His from 0.15 to 9.60 mmol/L increased the αs2-casein, β-casein, and κ-casein expression and phosphorylation of S6K1 (Thr389), 4EBP1 (Thr37), eIF4E (Ser209), and eEF2 (Thr56) in CMEC-H cells, at 0.15 and 9.60 mmol/L reduced the GβL expression, and over 4.80 mmol/L inhibited RPS6 (Ser235/236). When adding Leu, the αs1-casein was positively correlated with P-mTOR (Ser2481) (R 2=0.7820, P<0.01) and P-S6K1 (Thr389) (R 2=0.7881, P<0.01), respectively. With the addition of His, eEF2 was positively correlated with β-casein (R 2=0.9638, P<0.01) and κ-casein (R 2=0.9048, P<0.01), respectively. Our results provide basic information for further study to clarify the regulation mechanism of Leu and His on casein expression through the mTOR pathway in the dairy cattle mammary gland.
Footnotes
Project supported by the National Basic Research Program (973) of China (No. 2011CB100805), the Modern Agro-Industry Technology Research System of China (No. nycytx-04-01), and the Agricultural Science and Technology Innovation Program (No. ASTIP-IAS12), China
Compliance with ethics guidelines: Hai-na GAO, Han HU, Nan ZHENG, and Jia-qi WANG declare that they have no conflict of interest.
All institutional and national guidelines for the care and use of laboratory animals were followed.
References
- 1.Appuhamy JA, Knapp JR, Becvar O, et al. Effects of jugular-infused lysine, methionine, and branched-chain amino acids on milk protein synthesis in high-producing dairy cows. J Dairy Sci. 2011;94(4):1952–1960. doi: 10.3168/jds.2010-3442. [DOI] [PubMed] [Google Scholar]
- 2.Appuhamy JA, Bell AL, Nayananjalie WA, et al. Essential amino acids regulate both initiation and elongation of mRNA translation independent of insulin in MAC-T cells and bovine mammary tissue slices. J Nutr. 2011;141(6):1209–1215. doi: 10.3945/jn.110.136143. [DOI] [PubMed] [Google Scholar]
- 3.Appuhamy JA, Knoebel NA, Nayananjalie WA, et al. Isoleucine and leucine independently regulate mTOR signaling and protein synthesis in MAC-T cells and bovine mammary tissue slices. J Nutr. 2012;142(3):484–491. doi: 10.3945/jn.111.152595. [DOI] [PubMed] [Google Scholar]
- 4.Appuhamy J, Nayananjalie WA, England EM, et al. Effects of AMP-activated protein kinase (AMPK) signaling and essential amino acids on mammalian target of rapamycin (mTOR) signaling and protein synthesis rates in mammary cells. J Dairy Sci. 2014;97(1):419–429. doi: 10.3168/jds.2013-7189. [DOI] [PubMed] [Google Scholar]
- 5.Arriola Apelo SI, Bell AL, Estes K, et al. Effects of reduced dietary protein and supplemental rumen-protected essential amino acids on the nitrogen efficiency of dairy cows. J Dairy Sci. 2014;97(9):5688–5699. doi: 10.3168/jds.2013-7833. [DOI] [PubMed] [Google Scholar]
- 6.Arriola Apelo SI, Knapp JR, Hanigan MD. Invited review: current representation and future trends of predicting amino acid utilization in the lactating dairy cow. J Dairy Sci. 2014;97(7):4000–4017. doi: 10.3168/jds.2013-7392. [DOI] [PubMed] [Google Scholar]
- 7.Arriola Apelo SI, Singer LM, Lin XY, et al. Isoleucine, leucine, methionine, and threonine effects on mammalian target of rapamycin signaling in mammary tissue. J Dairy Sci. 2014;97(2):1047–1056. doi: 10.3168/jds.2013-7348. [DOI] [PubMed] [Google Scholar]
- 8.Bequette BJ, Hanigan MD, Calder AG, et al. Amino acid exchange by the mammary gland of lactating goats when histidine limits milk production. J Dairy Sci. 2000;83(4):765–775. doi: 10.3168/jds.S0022-0302(00)74939-3. [DOI] [PubMed] [Google Scholar]
- 9.Beugnet A, Tee AR, Taylor PM, et al. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J. 2003;372(Pt 2):555–566. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bionaz M, Hurley W, Loor J. Milk protein synthesis in the lactating mammary gland: insights from transcriptomics analyses. In: Hurley W, editor. Milk Protein. InTech, Chapter 11. 2012. pp. 285–324. [DOI] [Google Scholar]
- 11.Burgos SA, Cant JP. IGF-1 stimulates protein synthesis by enhanced signaling through mTORC1 in bovine mammary epithelial cells. Domest Anim Endocrinol. 2010;38(4):211–221. doi: 10.1016/j.domaniend.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Cheng SW, Fryer LG, Carling D. Thr2446 is a novel mammalian target of rapamycin (mTOR) phosphorylation site regulated by nutrient status. J Biol Chem. 2004;279(16):15719–15722. doi: 10.1074/jbc.C300534200. [DOI] [PubMed] [Google Scholar]
- 13.Christophersen CT, Karlsen J, Nielsen MO, et al. Eukaryotic elongation factor-2 (eEF-2) activity in bovine mammary tissue. J Dairy Res. 2002;69(2):205–212. doi: 10.1017/S0022029902005526. [DOI] [PubMed] [Google Scholar]
- 14.Durán RV, Hall MN. Leucyl-tRNA synthetase: double duty in amino acid sensing. Cell Res. 2012;22(8):1207–1209. doi: 10.1038/cr.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao HN, Hu H, Wang JQ, et al. Effects of leucine and histidine on milk protein synthesis via mammalian target of rapamycin signaling pathway in the bovine mammary epithelial cells. Chin J Anim Nutr. 2015;27(4):1124–1134. doi: 10.3969/j.issn.1006-267x.2015.04.016. (in Chinese) [DOI] [Google Scholar]
- 16.Gerasimovskaya EV, Tucker DA, Stenmark KR. Activation of phosphatidylinositol 3-kinase, Akt, and mammalian target of rapamycin is necessary for hypoxia-induced pulmonary artery adventitial fibroblast proliferation. J Appl Physiol. 2005;98(2):722–731. doi: 10.1152/japplphysiol.00715.2004. [DOI] [PubMed] [Google Scholar]
- 17.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68(1):913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 18.Hanigan MD, Crompton LA, Metcalf JA, et al. Modelling mammary metabolism in the dairy cow to predict milk constituent yield, with emphasis on amino acid metabolism and milk protein production: model evaluation. J Theor Biol. 2001;213(2):223–239. doi: 10.1006/jtbi.2001.2417. [DOI] [PubMed] [Google Scholar]
- 19.Hara K, Maruki Y, Long X, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 20.Harris TE, Lawrence JC., Jr TOR signaling. Sci STKE. 2003;212:re15. doi: 10.1126/stke.2122003re15. [DOI] [PubMed] [Google Scholar]
- 21.Hristov AN, Price WJ, Shafii B. A meta-analysis examining the relationship among dietary factors, dry matter intake, and milk and milk protein yield in dairy cows. J Dairy Sci. 2004;87(7):2184–2196. doi: 10.3168/jds.S0022-0302(04)70039-9. [DOI] [PubMed] [Google Scholar]
- 22.Hu H, Zheng N, Dai WT. Immortalization of a primary bovine mammary epithelial cell line by the SV40 large T-antigen gene. Kansas City, MO: The American Dairy Science Association; 2014. [DOI] [Google Scholar]
- 23.Kalscheur KF, Baldwin VI RL, Glenn BP, et al. Milk production of dairy cows fed differing concentrations of rumen-degraded protein. J Dairy Sci. 2006;89(1):249–259. doi: 10.3168/jds.S0022-0302(06)72089-6. [DOI] [PubMed] [Google Scholar]
- 24.Kaul G, Pattan G, Rafeequi T. Eukaryotic elongation factor-2 (eEF2): its regulation and peptide chain elongation. Cell Biochem Funct. 2011;29(3):227–234. doi: 10.1002/cbf.1740. [DOI] [PubMed] [Google Scholar]
- 25.Kim CH, Choung JJ, Chamberlain DG. Estimates of the efficiency of transfer of L-histidine from blood to milk when it is the first-limiting amino acid for secretion of milk protein in the dairy cow. J Sci Food Agric. 2001;81(12):1150–1155. doi: 10.1002/jsfa.923. [DOI] [Google Scholar]
- 26.Kim DH, Sarbassov DD, Ali SM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 27.Kim DH, Sarbassov DD, Ali SM, et al. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11(4):895–904. doi: 10.1016/S1097-2765(03)00114-X. [DOI] [PubMed] [Google Scholar]
- 28.Kim E. Mechanisms of amino acid sensing in mTOR signaling pathway. Nutr Res Pract. 2009;3(1):64–71. doi: 10.4162/nrp.2009.3.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SG, Buel GR, Blenis J. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol Cells. 2013;35(6):463–473. doi: 10.1007/s10059-013-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimball SR. Regulation of global and specific mRNA translation by amino acids. J Nutr. 2002;132(5):883–886. doi: 10.1093/jn/132.5.883. [DOI] [PubMed] [Google Scholar]
- 31.Korhonen M, Vanhatalo A, Huhtanen P. Evaluation of isoleucine, leucine, and valine as a second-limiting amino acid for milk production in dairy cows fed grass silage diet. J Dairy Sci. 2002;85(6):1533–1545. doi: 10.3168/jds.S0022-0302(02)74223-9. [DOI] [PubMed] [Google Scholar]
- 32.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Alafuzoff I, Soininen H, et al. Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer’s disease brain. FEBS J. 2005;272(16):4211–4220. doi: 10.1111/j.1742-4658.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- 34.Merrick WC. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56(2):291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moshel Y, Rhoads RE, Barash I. Role of amino acids in translational mechanisms governing milk protein synthesis in murine and ruminant mammary epithelial cells. J Cell Biochem. 2006;98(3):685–700. doi: 10.1002/jcb.20825. [DOI] [PubMed] [Google Scholar]
- 36.Peterson RT, Beal PA, Comb MJ, et al. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J Biol Chem. 2000;275(10):7416–7423. doi: 10.1074/jbc.275.10.7416. [DOI] [PubMed] [Google Scholar]
- 37.Prizant RL, Barash I. Negative effects of the amino acids Lys, His, and Thr on S6K1 phosphorylation in mammary epithelial cells. J Cell Biochem. 2008;105(4):1038–1047. doi: 10.1002/jcb.21904. [DOI] [PubMed] [Google Scholar]
- 38.Proud CG. Amino acids and mTOR signalling in anabolic function. Biochem Soc Trans. 2007;35(5):1187–1190. doi: 10.1042/BST0351187. [DOI] [PubMed] [Google Scholar]
- 39.Stipanuk MH. Leucine and protein synthesis: mTOR and beyond. Nutr Rev. 2007;65(3):122–129. doi: 10.1111/j.1753-4887.2007.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 40.Toerien CA, Trout DR, Cant JP. Nutritional stimulation of milk protein yield of cows is associated with changes in phosphorylation of mammary eukaryotic initiation factor 2 and ribosomal S6 kinase 1. J Nutr. 2010;140(2):285–292. doi: 10.3945/jn.109.114033. [DOI] [PubMed] [Google Scholar]
- 41.Wang C, Liu HY, Wang YM, et al. Effects of dietary supplementation of methionine and lysine on milk production and nitrogen utilization in dairy cows. J Dairy Sci. 2010;93(8):3661–3670. doi: 10.3168/jds.2009-2750. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 43.Wilde CJ, Quarrie LH, Tonner E, et al. Mammary apoptosis. Livest Prod Sci. 1997;50(1-2):29–37. doi: 10.1016/S0301-6226(97)00070-5. [DOI] [Google Scholar]
- 44.Yang Q, Inoki K, Kim E, et al. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. PNAS. 2006;103(18):6811–6816. doi: 10.1073/pnas.0602282103. [DOI] [PMC free article] [PubMed] [Google Scholar]