Abstract
Condensin plays fundamental roles in chromosome dynamics. In this study, we determined the binding sites of condensin on fission yeast (Schizosaccharomyces pombe) chromosomes at the level of nucleotide sequences using chromatin immunoprecipitation (ChIP) and ChIP sequencing (ChIP-seq). We found that condensin binds to RNA polymerase I-, II- and III-transcribed genes during both mitosis and interphase, and we focused on pol II constitutive and inducible genes. Accumulation sites for condensin are distinct from those of cohesin and DNA topoisomerase II. Using cell cycle stage and heat-shock-inducible genes, we show that pol II-mediated transcripts cause condensin accumulation. First, condensin's enrichment on mitotically activated genes was abolished by deleting the sep1+ gene that encodes an M-phase-specific forkhead transcription factor. Second, by raising the temperature, condensin accumulation was rapidly induced at heat-shock protein genes in interphase and even during mid-mitosis. In interphase, condensin accumulates preferentially during the postreplicative phase. Pol II-mediated transcription was neither repressed nor activated by condensin, as levels of transcripts per se did not change when mutant condensin failed to associate with chromosomal DNA. However, massive chromosome missegregation occurred, suggesting that abundant pol II transcription may require active condensin before proper chromosome segregation.
Introduction
In eukaryotic cells, condensin, a principal regulator of chromosomal dynamics, consists of heteropentameric subunits containing two structural maintenance of chromosomes (SMC) subunits and three non-SMC subunits (Koshland & Strunnikov 1996; Hudson et al. 2009; Yanagida 2009; Wood et al. 2010; Cuylen & Haering 2011; Hirano 2012). SMC was initially identified in budding yeast as SMC1, as its mutation smc1-1 caused an increased rate of minichromosome nondisjunction (Strunnikov et al. 1993). SMC2/Cut14 and SMC4/Cut3 were identified as condensin SMC subunits in fission yeast, Schizosaccharomyces pombe, by analyzing temperature-sensitive mutants defective in mitotic chromosome condensation, as well as segregation (Saka et al. 1994). Budding yeast SMC2 and SMC4 were also identified as condensin subunits (Strunnikov et al. 1995), whereas SMC1 and SMC3 are cohesin subunits (Michaelis et al. 1997). Second only to DNA topoisomerase II, vertebrate condensin subunit ScII/SMC2 was found to be the most abundant protein in mitotic chromosome scaffolds (Adolphs et al. 1977; Saitoh et al. 1994). Hirano & Mitchison (1994) discovered that Xenopus XCAP-E is a frog egg condensin SMC protein, which is essential for promoting mitotic chromosome condensation in vitro (Hirano & Mitchison 1994). There are two different types of eukaryotic condensin complexes: condensin I and II (Ono et al. 2003; Hirano 2012). In budding and fission yeast, condensin II has not yet been identified, suggesting that only condensin I is actually present.
In S. pombe, mitotic chromosome condensation in this eukaryotic microbe is visible with light microscopy; hence, mutants defective in condensation can be screened. Two mutants, cut3-477 and cut14-208, which show severe defects in mitotic chromosome arm compaction, were isolated, and gene cloning indicated that the gene products, Cut3 and Cut14, belong to SMC family proteins (Saka et al. 1994). Three non-SMC condensin subunits (Cnd2/Barren and HEAT repeat-containing Cnd1 and Cnd3) are bound to the ATPase-containing globular domains of the SMC heterodimer (Sutani et al. 1999; Yoshimura et al. 2002). Heteropentameric holocondensin and SMC2/4 dimers possess DNA reannealing activity that promotes conversion from complementary single-stranded DNA (ssDNA) to double-stranded DNA (dsDNA), but this activity is not found in cohesin, suggesting that DNA reannealing is required for condensin function (Sutani & Yanagida 1997; Sakai et al. 2003).
In S. pombe, condensin disperses to both the nucleus and the cytoplasm during interphase, but moves to the nucleus upon entry into mitosis. The N-terminal T19 residue of the SMC4/Cut3 subunit is phosphorylated by Cdc2 (Sutani et al. 1999). Then, the condensin holocomplex is transferred into the nucleus by Cut15, one of two importin-α proteins (Matsusaka et al. 1998). Condensin binding at mitotic chromosomes requires not only its nuclear entry, but also phosphorylation of non-SMC subunit Cnd2/Barren by Aurora B-like kinase Ark1 (Nakazawa et al. 2011; Tada et al. 2011). Phosphorylation of the Cnd2 subunit promotes physical interaction between condensin and histone H2A (Tada et al. 2011). Condensin phosphorylated by Ark1 is continuously required for chromosome segregation throughout mitosis, as is DNA topoisomerase II (Top2) (Nakazawa et al. 2011).
Genomewide ChIP (Chromatin immunoprecipitation)-chip analyses that employ microarrays for detecting DNA fragments bound to condensin have been previously carried out in budding yeast (Wang et al. 2005; D'Ambrosio et al. 2008). These analyses showed that condensin associates with pericentromeric DNA, subtelomeres, RNA polymerase I (pol I)-transcribed ribosomal DNA (rDNA) repeats, RNA polymerase III (pol III)-transcribed tRNA genes, and other genes bound by the pol III transcription machinery TFIIIC. In S. pombe, condensin accumulates at centromeres and pol I-transcribed rDNAs, depending upon the presence of several kinetochore- and rDNA-associating proteins (Nakazawa et al. 2008). S. pombe condensin also associates with pol III-transcribed genes and retrotransposons, and mediates their clustering at centromeres (D'Ambrosio et al. 2008; Iwasaki et al. 2010; Tanaka et al. 2012). In contrast, condensin association with pol II-transcribed genes is not well understood in either budding or fission yeast. Recently, ChIP coupled with high-throughput DNA sequencing (ChIP-seq) and chromatin affinity purification sequencing (ChAP-seq) showed that chicken condensin I, worm condensins, and mouse condensin II bind predominantly to promoter or regulatory sequences of actively transcribed loci (Dowen et al. 2013; Kim et al. 2013; Kranz et al. 2013). Association sites of condensin thus appear to be closely correlated with transcriptional activation. However, it has not been clearly shown that transcriptional activation actually promotes condensin association at loci.
ChIP-seq allows the identification of binding sites of chromosome-interacting proteins with high resolution, compared to ChIP-chip followed by microarray analysis (Schmidt et al. 2009b). To determine condensin-enriched sites along S. pombe's three chromosomes, we carried out ChIP-seq analysis of FLAG-tagged Cut14/SMC2 protein in both mitotically arrested and asynchronously cultured cells. We present evidence that condensin accumulates at transcription sites with abundant RNA pol II, during mitosis and interphase, and we show that condensin-binding sites are distinct from those for cohesin Rad21 and DNA topoisomerase II (Top2). Furthermore, we examined condensin association at two types of inducible genes, Sep1-dependent mitotically up-regulated genes and heat-shock protein (Hsp) genes. ChIP-quantitative PCR (qPCR) assays at these inducible genes show that pol II-mediated transcripts cause condensin accumulation in postreplicative G2 phase and even in mid-mitosis.
Results
ChIP-seq analysis of Cut14/SMC2 showed condensin accumulation at RNA polymerase II transcription sites
For chromatin immunoprecipitation (ChIP) of condensin SMC2 subunit Cut14, we employed an S. pombe strain designated as Cut14-FLAG, which contains a chromosomally integrated FLAG-tagged cut14+ gene under the native promoter (Nakazawa et al. 2008). Holocondensin was obtained by immunoprecipitation of Cut14-FLAG proteins using anti-FLAG antibody (Nakazawa et al. 2011). To determine the relative degree of condensin association with mitotic and interphase chromosomes, ChIP-seq analysis that combines ChIP with whole-genome sequencing was carried out according to published procedures (Schmidt et al. 2009b) for the S. pombe genome (consisting of three chromosomes). Cross-linked and sonicated chromatin was prepared from two cell extracts: wild-type (WT) asynchronously grown culture (70–80% of cells are in interphase) and β-tubulin cold-sensitive (cs) mutant nda3-KM311, synchronously mitotically arrested at a restrictive temperature (20 °C) (Hiraoka et al. 1984). DNA samples coprecipitated with anti-FLAG antibody were prepared along with input DNA (whole cell extract [WCE] DNA). Then, both DNA samples were sequenced using an Illumina GA IIx (Experimental procedures).
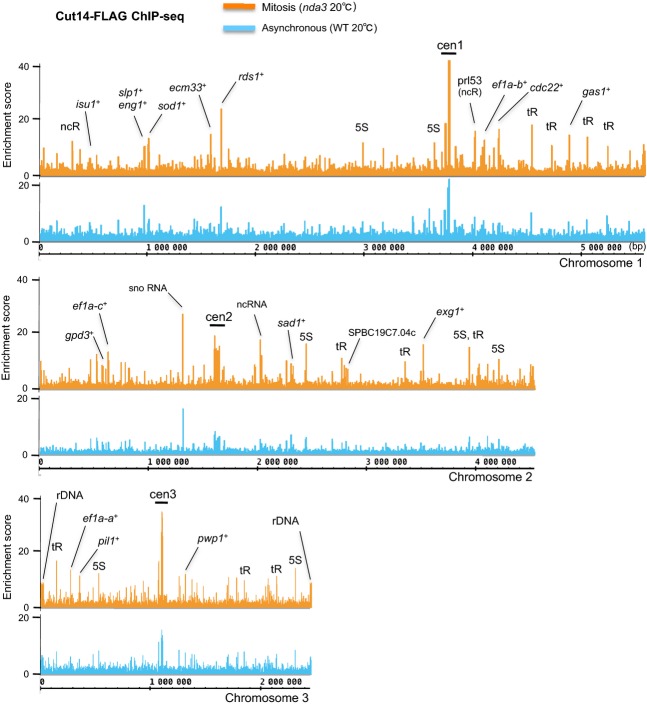
High enrichment of Cut14-FLAG was obtained for a number of chromosomal DNA sites at three chromosomes in mitosis (nda3, orange) and asynchronous (WT, blue) cells (Fig.1). Scores of condensin-enriched peaks were generally higher in nda3 samples than in WT samples. Enrichment scores for centromeres (cen1, cen2, cen3) were also high in mitosis. Most Cut14-enriched sites coincided with loci actively transcribed by pol II and pol I- or pol III-transcribed RNA genes. Some representative pol II-transcribed genes (see also Tables1 and 2, explained below) and prominent RNA genes were marked (tR, tRNA; 5S, 5S rRNA; ncR, noncoding RNA; snoRNA, small nucleolar RNA; cen1, cen2 and cen3, centromeric DNA; rDNA, ribosomal DNA). The S. pombe genome has ∽150 repeats of 10.4-kb rDNA units clustered at both ends of chromosome 3 (Schaak et al. 1982; Toda et al. 1984). Only 2 and 1 rDNA repeats were represented on the left and right ends of chromosome 3, respectively (Fig.1). These results suggest that condensin accumulates heavily at pol II-transcribed genes, in addition to previously reported condensin association sites (centromeric DNAs, pol I-transcribed rDNA repeats, pol III-transcribed tRNA and 5S rRNA genes; D'Ambrosio et al. 2008; Nakazawa et al. 2008; Iwasaki et al. 2010). We validated condensin enrichment at pol II-transcribed genes using ChIP followed by quantitative, real-time PCR (ChIP-qPCR) (Fig. S1 in Supporting Information). FLAG-tagged non-SMC subunit, Cnd1, also accumulates at pol II-transcribed genes to about the same extent as Cut14, suggesting that condensin holocomplex binds to these loci. Our ChIP-seq results for pol II-transcribed genes are in good agreement with those obtained by Drs. Takashi Sutani and Katsuhiko Shirahige (University of Tokyo, personal communication).
Figure 1.
ChIP-seq analysis of condensin Cut14/SMC2 throughout the whole Schizosaccharomyces pombe genome in mitotic and asynchronous cells. FLAG-tagged S. pombe condensin subunit SMC2 (Cut14-FLAG) enrichment profiles along S. pombe chromosomes 1, 2, and 3 were determined using ChIP-seq. Specimens were obtained from mitotically arrested, cold-sensitive (cs) beta-tubulin mutant nda3-KM311 at the restrictive temperature 20 °C for 8 h (Mitosis, shown in orange) and asynchronously grown wild type (WT) at 20 °C (shown in blue). Ratios of precipitated DNA (IP) to input DNA are shown as the enrichment score of Cut14-FLAG-bound DNA. Representative genes are marked: cen, centromere; tR, tRNA gene; 5S, 5S rRNA gene; ncR, noncoding RNA gene; snoRNA, small nucleolar RNA; rDNA, ribosomal DNA repeats. Only 2 and 1 rDNA repeats are shown on the left and right ends of chromosome 3, respectively, because of its repetitive sequences (see text). Condensin binds to various pol II-transcribed protein-coding genes, as well as to pol I- or III-transcribed RNA genes and centromeric regions.
Table 1.
The top 50 protein-coding genes associated with condensin (Cut14) in mitotically arrested cells
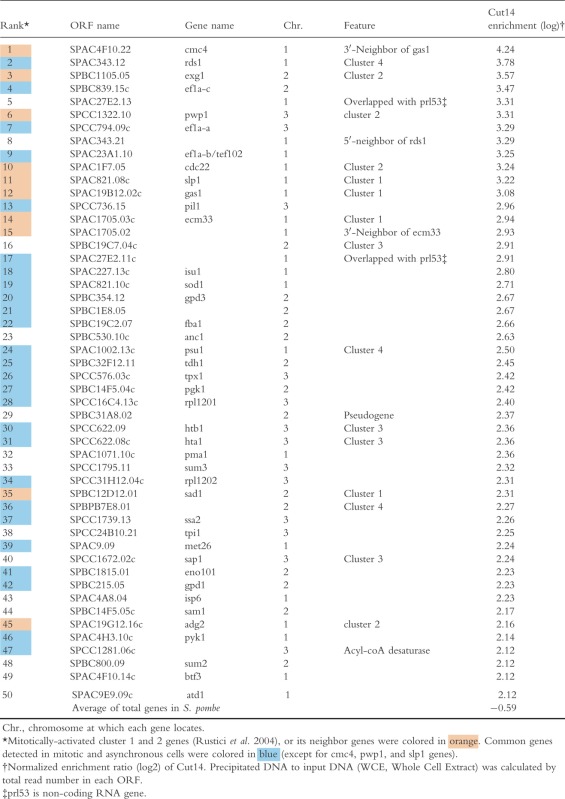 |
Table 2.
The top 50 protein-coding genes associated with condensin (Cut14) in asynchronously dividing cells
 |
For comparison, we conducted genomewide ChIP-seq analyses for cohesin and DNA topoisomerase II (Top2), using FLAG-tagged Rad21 (cohesin non-SMC subunit; Fig. S2A in Supporting Information) and Top2 (Fig. S3A in Supporting Information). Mitotic enrichment of Rad21- and Top2-FLAG was commonly found at centromeres and some RNA genes, but was considerably different from that of condensin Cut14-FLAG. Rad21 was significantly enriched at subtelomeric regions at both ends of chromosomes 1 and 2. Unlike Cut14, Rad21 and Top2 accumulated at some intergenic regions (Figs S2B and S3B Supporting Information, enlarged ChIP-seq profiles of Figs S2A and S3A in Supporting Information, respectively). Accumulation of cohesin and Top2 at these sites is consistent with ChIP-chip data previously reported in fission yeast (Schmidt et al. 2009a; Durand-Dubief et al. 2010; Norman-Axelsson et al. 2013).
Condensin is enriched at mitotically up-regulated genes
In S. pombe, groups of genes, so-called clusters 1–4 (designated C1-C4, hereafter), which oscillate transcriptionally during the cell division cycle, peaked even in mitosis (Rustici et al. 2004). C1 and C2 genes, respectively, were periodically expressed in M and M-G1 phases. ChIP-seq of Cut14-FLAG showed that condensin was enriched at many C1 and C2 genes during mitosis. Tables1 and 2 present the 50 most condensin-enriched protein-coding genes in nda3-arrested mitotic and asynchronous (mostly interphase) wild-type cells, respectively. Among the top 15 genes in mitotic cells, eight belonged to clusters C1 and C2 or their neighbors (Table1, orange colored). Considering that only ∽3% of all protein-coding genes are assigned to C1 or C2 in the S. pombe genome (165 of ∽5000 genes), Cut14 enrichment at these genes is quite significant. Figure2A shows the enlarged ChIP-seq profiles from Fig.1 around ∽10-kb regions containing C1 genes, ecm33+ and gas1+, encoding cell wall materials. Condensin Cut14 was more abundant at these C1 genes in mitosis than in asynchronous cells. Cut14 appeared to bind preferentially to the 3′-ends of these genes. These results suggest that condensin preferentially binds to mitotically activated genes. Rad21 and Top2 were much less abundant at these genes, regardless of cell cycle stage.
Figure 2.

Condensin enriched at mitotically up-regulated genes and constitutively expressed genes. (A, B) Condensin enrichment at mitotically up-regulated ecm33+ and gas1+ genes (A), and constitutively expressed ef1a-a+ and ef1a-b+ genes (B) in mitosis (orange) and asynchronous culture (blue). These are obtained as enlarged profiles from Fig.1 around ∽10-kb regions of ecm33+, gas1+, ef1a-a+, and ef1a-b+ genes. Rad21- and Top2-FLAG profiles are also presented for comparison (enlarged profiles of Figs S2A and S3A in Supporting Information). Profiles are presented as in Fig.1.
Condensin associates with genes that are actively being transcribed
Twenty-six condensin-enriched, protein-coding genes are shared between mitotic and asynchronous cells (Tables1 and 2, blue colored). These common genes include a large number of constitutively activated housekeeping genes, encoding translation elongation factors (ef1a-a+, ef1a-b+, and ef1a-c+), enzymes for glycolysis or glycerol biosynthesis (gpd1+, gpd3+, fba1+, tdh1+, pgk1+, and eno101+), and histones (hta1+ and htb1+). To determine pol II occupancy at condensin-enriched, protein-coding genes, we retrieved previously published ChIP-seq data for pol II association in asynchronous cultured cells (Carlsten et al. 2012). The pol II enrichment ratio of precipitated DNA to input DNA is given in Table2, along with Cut14 enrichment. Among the top 50 Cut14-enriched genes in asynchronous cells, 44 show 2× and 28 genes show 4× pol II enrichment compared to the average for all pol II-transcribed genes. Hence, condensin-enriched genes in asynchronous cells were highly occupied by pol II. As more than half of the top 50 condensin-enriched genes are common to both mitotic and asynchronous cells (Tables1 and 2), condensin appears to associate with genes constitutively expressing abundant transcripts. Figure2B shows an example of Cut14 enrichment at constitutively activated genes, ef1a-a+ and ef1a-b+ that encode translation elongation factors. Rad21 association was not prominent, but Top2 was relatively abundant at the constitutive ef1a-a+ and ef1a-b+ loci.
Sep1-dependent accumulation of condensin at mitotically up-regulated genes
A forkhead transcription factor, Sep1, is essential for M-phase-specific transcription in fission yeast (Buck et al. 2004; Rustici et al. 2004). Using reverse transcription-PCR (RT-PCR), we confirmed that the deletion of sep1 gene abolishes mitotic transcription of C1 (ecm33+, gas1+, and slp1+) and C2 (exg1+) genes in mitotically arrested nda3 mutant cells (Fig.3A). Mitotic expression of these four genes was not significantly affected in a deletion mutant of the atf1 gene encoding the other transcription factor (Takeda et al. 1995). To examine whether condensin enrichment at mitotically activated genes depends on their transcripts, ChIP-qPCR assay was carried out using DNA probes corresponding to four C1 and C2 genes (Fig.3B). Cut14-FLAG proteins were immunoprecipitated with anti-FLAG antibodies in nda3 mitotic cells. Then, coprecipitated and WCE DNAs were amplified with real-time qPCR. The levels of precipitated DNAs were drastically diminished in Δsep1 mutant extracts, but not in Δatf1, showing their dependency on Sep1. In contrast, condensin enrichment at centromere DNA (Cen) did not decrease in Δsep1, suggesting that centromeric accumulation of condensin does not depend upon Sep1.
Figure 3.

Loss of mitotically activated transcription in the Δsep1 mutant results in diminished association of condensin. (A) Expression of mitotically activated genes was greatly reduced in the Δsep1 mutant (Buck et al. 2004), but not in Δatf1. Transcript levels of four mitotically up-regulated ecm33+, gas1+, slp1+, and exg1+ genes were determined in mitotically arrested nda3 single, nda3 Δsep1, and nda3 Δatf1 double mutant cells by quantitative reverse transcription (RT)-PCR. Total extracted RNA was reverse-transcribed to cDNA, and resulting cDNA levels were quantified with error bars showing the standard deviation (n = 3). Relative RNA levels are shown as the ratio between the target gene and act1+ gene. (B) Enrichment of condensin Cut14-FLAG at ecm33+, gas1+, slp1+, and exg1+ genes. Cells were grown as shown in (A). ChIP-qPCR analysis of Cut14-FLAG was carried out in the nda3 single, nda3 Δsep1, and nda3 Δatf1 (control) double mutant background along with a no-tag strain. PCR primers at 6 regions (ecm33+, gas1+, slp1+, exg1+, act1+, and centromeric loci) were used. Relative enrichment was calculated as the ratio of IP to WCE with error bars showing standard deviations (n = 3). Condensin enrichment at these four mitotically transcribed genes requires the presence of Sep1.
Condensin accumulates at inducible heat-shock protein (Hsp) genes following a temperature-shift
We then examined whether condensin was mobilized to transcriptionally inducible genes. To this end, we carried out ChIP-seq of Cut14-FLAG in asynchronously cultured wild-type cells at 20 °C and cells that were heat-shocked for 9 min with a temperature-shift (T-shift) to 36 °C. ChIP-seq of Rad21 and Top2 was also carried out under the same conditions. Five heat-shock protein (Hsp) genes, ssa1+, hsp90+, hsp16+, hsp9+, and SPBC16D10.08c/hsp104+, and the SPBC660.16 gene can be readily induced after a T-shift to 36 °C in asynchronously cultured cells (Chen et al. 2003). Condensin Cut14 association greatly increased at coding and adjacent regions of the six genes 9 min after the T-shift from 20 °C (Fig.4). Except for hsp9+, condensin accumulation appeared to be enhanced in the 3′-region. Association of Top2, but not Rad21, also increased at coding regions after the T-shift. Thus, condensin and Top2, but not Rad21, showed strong affinity for transcription-induced Hsp genes.
Figure 4.

Condensin association at heat-shock protein (Hsp) genes following a temperature-shift. ChIP-seq profiles of Cut14-, Rad21-, and Top2-FLAG are shown at five heat-shock protein (Hsp) genes (ssa1+, hsp90+, hsp16+, hsp9+, and SPBC16D10.08c/hsp104+) and SPBC660.16 genes. Wild-type strains expressing FLAG-tagged proteins were cultured asynchronously at 20 °C (blue) and shifted to 36 °C for 9 min (orange, T-shift) in order to activate Hsp gene transcription. Although SPBC660.16 encodes phosphogluconate dehydrogenase, its transcription is reported to be strikingly up-regulated following a temperature-shift to 36 °C (Chen et al. 2003). Quantified profiles were obtained as described in the legend of Fig.1. Condensin and Top2, but not Rad21, are rapidly mobilized to transcriptionally inducible Hsp genes.
Hsp genes in postreplicative cells preferentially associate with condensin
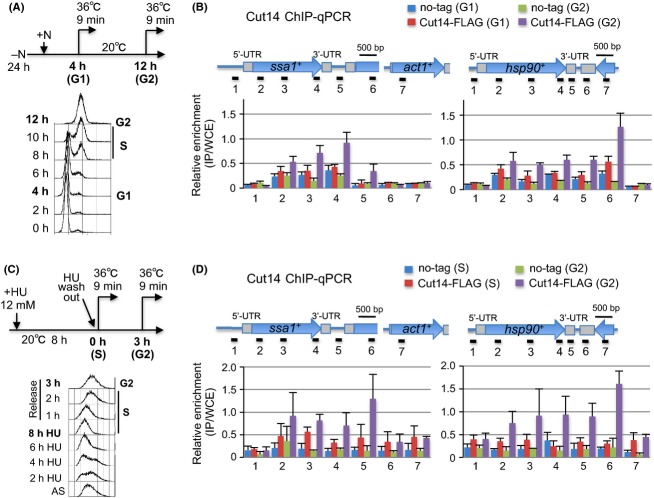
To compare the levels of condensin association with Hsp genes between pre- and postreplicative DNA states, ChIP-qPCR assays were carried out in two moderately synchronous cultivations. First, nitrogen-starved G1 cells from Cut14-FLAG and no-tag strains were shifted to complete medium at 20 °C (Fig.5A). FACS analysis showed that entry into S phase occurred 8–10 h after the shift. At 4 h (G1) and 12 h (G2) in complete medium, cells were shifted to 36 °C for 9 min to activate Hsp genes. ChIP-qPCR assay showed that Cut14-FLAG was precipitated with ssa1+ and hsp90+ genes in G2, whereas precipitated DNA in G1 phase was two- or threefold less, like that in nontagged strains (Fig.5B). Second, by using the hydroxyurea (HU) block and release technique, cells were synchronously cultured in S and G2 phase and then shifted to 36 °C for 9 min (Fig.5C). Hydroxyurea is a DNA replication inhibitor that allows cells to arrest reversibly at S phase. Cut14 associated with ssa1+ and hsp90+ genes during G2, but the association was comparable to the no-tag control during S phase (Fig.5D). In both nitrogen starvation and HU block experiments, accumulations of Cut14 tended to be enhanced in the 3′-region of Hsp genes. These results strongly suggested that condensin binds primarily to postreplicative DNA, at least for heat-shock ssa1+ and hsp90+ genes induced during interphase.
Figure 5.
Condensin preferentially associates with Hsp genes in postreplicative cells. (A) (top) Cells expressing FLAG-tagged or nontagged Cut14 were first arrested at the prereplicative G0 phase in nitrogen-deficient medium (EMM2-N) at 26 °C for 24 h, and shifted to a nitrogen-replenished medium (YPD) at 20 °C for 4 h or 12 h (Sajiki et al. 2009). Cells were briefly incubated at 36 °C for 9 min to activate Hsp genes and harvested for ChIP. (bottom) FACS analysis in the Cut14-FLAG strain showed S phase occurring at 8-10 h. Hence, cells taken at 4 and 12 h, respectively, were derived from prereplicative G1 and postreplicative G2 phases. (B) ChIP-qPCR analysis of Cut14-FLAG was carried out in G1 and G2 cells along with nontagged controls. Positions of PCR primers (horizontal lines with numbers) in the ssa1+ and hsp90+ genes are indicated. 5′- and 3′-UTRs are shown as gray boxes. Relative enrichment was obtained as described above (Fig.3B). (C) Cut14-FLAG and nontagged control cells were arrested in S phase at 20 °C for 8 h with 12 mM hydroxyurea (HU) and then released from the arrest upon washout of HU. At 0 h (S phase) and 3 h after release (G2 phase), cells were incubated at 36 °C for 9 min and harvested for ChIP. (D) ChIP-qPCR analysis of Cut14-FLAG was carried out in S and G2 cells as shown in (B).
Condensin accumulates at Hsp genes even in mid-mitosis, whereas mutant condensin fails to accumulate, resulting in a chromosome segregation defect
To examine whether condensin associates with transcriptionally induced Hsp genes even during mid-mitosis if cells were exposed to high temperature (36 °C), we carried out ChIP-qPCR analysis of Cut14 using nda3 block (at 20 °C) and release (to 36 °C, as nda3 mutant is cs, cold-sensitive) experiments (Hiraoka et al. 1984; Nakazawa et al. 2011) as follows. We used the cs nda3 mutant strain expressing Cut14 wild-type protein (Cut14 WT) or Cut14-208 TS mutant protein (Cut14 TS) tagged with FLAG (Fig.6A). Cut14-208 TS mutant protein, which contains the mutation S861P (Sutani & Yanagida 1997) in the coiled-coil region, remains functional at permissive temperatures (26–30 °C), but is quickly inactivated at the restrictive temperature (36 °C). Both nda3 Cut14 WT and nda3 Cut14 TS strains were first arrested in prometaphase-like stage at 20 °C for 8 h and then shifted to 36 °C. This T-shift enables rapid induction of Hsp gene expression during mitosis, and simultaneously releases arrested cells to anaphase (Nakazawa et al. 2011). Importantly, the T-shift also immediately inactivates Cut14-208 TS mutant protein. After 15 min (corresponding to late anaphase or telophase), with DAPI (4,6-diamino-2-phenylindole) staining, the Cut14 TS strain displayed highly streaked or extended chromosomal DNA, whereas normal segregation was observed for the Cut14 WT cells (Fig6B, C, right) (Nakazawa et al. 2011).
Figure 6.
Condensin accumulates at Hsp genes even in mid-mitosis, whereas the levels of Hsp gene transcripts are not affected by condensin inactivation. (A) Experimental scheme. A temperature-shift was carried out using the cs nda3 β-tubulin mutant expressing Cut14 wild-type protein (strain Cut14 WT) or Cut14-208 TS mutant protein (strain Cut14 TS) tagged with FLAG. At 20 °C for 8 h, both strains were arrested at the prometaphase-like stage (because of the inactivation of β-tubulin). Cells were then transferred to 36 °C, inactivating condensin and simultaneously reactivating β-tubulin. This temperature-shift also induces Hsp gene expression. Culture aliquots were then obtained at 0, 3, 9, 15, and 30 min after the shift. (B) Time-course changes (in %) of different cell types for the two strains (left panel, Cut14 WT; and right panel, Cut14-208 TS). Blue; condensed, red; partially separated, green; two nuclei (without septum), purple; two nuclei (with septum), light blue; streaked or ‘cut’ phenotype, which appeared in Cut14-208 TS strain. (C) ChIP-qPCR analysis of Cut14 WT-FLAG (blue) and Cut14-208 TS-FLAG (red) for four regions (ssa1+, hsp90+, act1+ loci and centromere 3 DNA). Cells were prepared and harvested as shown in (A) and (B), along with nontagged control nda3 mutant cells (green). Positions of PCR primers (horizontal lines with numbers) are indicated. 5′- and 3′-UTRs are shown as gray boxes. Relative enrichment was calculated as the ratio of IP to WCE with error bars showing standard deviation (n = 3). Condensin rapidly accumulates at induced Hsp genes even during mitosis, whereas association of mutant condensin diminished. Micrographs (right) show the phenotypes of FLAG-tagged Cut14 WT and Cut14-208 TS mutant strains stained with DAPI at 15 min after T-shift to 36 °C. Aberrantly streaked chromosomes were observed in Cut14-208 TS mutant. Scale bar, 10 μm. (D) mRNA levels of ssa1+ and hsp90+ genes were determined by reverse transcription (RT)-PCR. The above two strains were grown as shown in (C). Relative RNA levels are shown as the ratio between the target gene and the act1+ gene. Primer position 4 for ssa1+, hsp90+ and act1+ in (C) was used in each gene. Transcription of Hsp genes does not appear to be affected by condensin.
We measured the levels of precipitated DNA at Hsp genes, ssa1+ and hsp90+, by Cut14 ChIP-qPCR in nda3 block and release experiments (Fig.6C). At 0 min (20 °C) in prometaphase-like cells, neither Cut14-FLAG nor Cut14-208 TS-FLAG coprecipitated with ssa1+ and hsp90+ DNAs, because of noninduction of Hsp genes. Contrarily, both Cut14- and Cut14 TS-FLAG were abundantly bound to inner centromere cnt3 DNAs at permissive temperatures for the cut14-208 ts mutant. Then, by shifting cells to 36 °C for 3 min, mutant nda3cs β-tubulin immediately became functional and rapidly produced the spindle, and the cells entered the metaphase. At 3 min, both wild-type Cut14-FLAG and mutant Cut14-208 TS-FLAG could associate with the 3′-UTR of Hsp genes. At 9 min (early anaphase) and 15 min (late anaphase or telophase), WT Cut14-FLAG association increased and accumulated intensely around the 3′-UTR regions, whereas mutant Cut14 TS protein association did not increase. The timing of Cut14-FLAG accumulation measured by ChIP-qPCR correlates well with that of Hsp gene activation (see below). Association of both WT Cut14 and Cut14-208 TS proteins was diminished after 30 min in G1/S phase. Taken together, these results indicate that only functional condensin rapidly accumulated at Hsp genes upon a T-shift.
We then examined whether the rapid induction of Hsp gene transcription was affected by inactivation of condensin. To this end, the levels of ssa1+ and hsp90+ transcripts were measured by RT-PCR in nda3 block and release experiments. At 30 min after T-shift (20–36 °C), ssa1+ and hsp90+ transcripts increased approximately 15- and 5-fold (compared to 0 min), respectively, in both WT Cut14- and mutant Cut14-208 TS protein-expressing nda3 strains (Fig.6D). These results suggest that transcription of Hsp genes was neither repressed nor activated by condensin. The failure to accumulate Cut14-208 TS mutant protein at Hsp loci did not greatly affect transcriptional activation of the genes after T-shift. Instead, condensin mutant cells produced a severe defect in chromosome segregation shortly after the initiation of mitosis (Fig.6B,C, right).
Discussion
In this study, using genomewide ChIP-seq analysis, we identified representative RNA pol II-transcribed genes as condensin-binding sites in both mitosis and interphase. Using ChIP experiments at two types of regulatable genes, we present evidence that pol II-mediated transcripts cause condensin accumulation at genes. Condensin accumulated at induced Hsp genes in interphase and even in mid-mitosis. In two synchronous cultivations, we showed that condensin preferentially binds to postreplicative rather than to prereplicative DNA. These findings establish that abundant pol II transcripts form condensin-binding sites on fission yeast chromosomes during mitosis and postreplicative interphase.
Our ChIP-seq results with the SMC2/Cut14 subunit confirmed the association of S. pombe condensin at centromeric DNA, pol I-transcribed rDNA repeats, and pol III-transcribed tRNA and 5S rRNA genes (D'Ambrosio et al. 2008; Nakazawa et al. 2008; Iwasaki et al. 2010). We found that condensin also associates with pol II-enriched active genes on both interphase and mitotic chromosomes, suggesting that all three RNA polymerases facilitate condensin enrichment. These results raise the possibility that chromosomal loci with abundant polymerases and/or transcribed RNA may form condensin-bound sites. Genomewide analyses showed that condensin I in chicken DT40 cells and worm condensins I and II bind predominantly to promoter or regulatory sequences of pol II-transcribed active genes (Kim et al. 2013; Kranz et al. 2013). Our study showed that S. pombe condensin binds to ORF regions and relatively 3′-ends of pol II-transcribed active genes. This distribution of S. pombe condensin was distinct from that of chicken and worm condensins. In metazoans, the promoter-proximal pausing of pol II near the transcription start site (TSS) is a major cellular mechanism to regulate gene expression (Adelman & Lis 2012). In contrast, paused pol II at TSS is not obvious in budding and fission yeast. S. pombe pol II is reportedly enriched at open reading frames (ORF) and 3′-ends, rather than in promoter regions (Carlsten et al. 2012; Castel et al. 2014). Thus, differences in the condensin association pattern within pol II-transcribed genes may be closely related to pol II distribution.
Condensin enrichment at mitotically up-regulated genes was drastically diminished in a sep1 deletion mutant, suggesting that Sep1-dependent pol II transcription is a major recruiter of condensin at the loci. During mitosis, it is generally understood that global silencing of gene expression occurs, and transcription is thought to be unsuited for chromosome condensation and segregation (Taylor 1960; Clemente-Blanco et al. 2009). In fission yeast, however, a substantial number of genes are periodically up-regulated even during mitosis, by specific transcription factors, including Sep1 (Buck et al. 2004; Rustici et al. 2004). Some of these genes encode products necessary for cytokinesis and cell division immediately after chromosome segregation. The sep1 deletion mutant shows defects in cytokinesis and cell morphology, but not in chromosome segregation (Ribar et al. 1999). Therefore, condensin accumulation at Sep1-dependent genes might not be necessary if transcription of these genes is silent. In other words, we speculate that condensin may be required at the loci only when pol II-mediated transcription is active during mitosis.
Condensin localizes to central centromeric DNAs (cnt and imr) where CENP-A chromatin assembles for kinetochore formation (Aono et al. 2002; Nakazawa et al. 2008). In this study, condensin enrichment at centromeric DNA (cnt) did not decrease in Δsep1 mutant cells. This result suggests that Sep1-promoted pol II transcription is not relevant to condensin association at central centromeres. It has been reported that the central centromeric region also encodes a noncoding RNA transcribed by pol II (Choi et al. 2011). Further study is required to examine whether condensin association at centromeres depends on pol II transcription, and if so, which transcription factor(s) are involved in that process.
We showed that condensin rapidly associates with transcriptionally induced Hsp genes 9 min after T-shift in interphase and even in mid-mitosis. This result strongly suggests that transcriptional activation immediately promotes condensin association at Hsp genes. Considering that the timing of Cut14 accumulation correlated well with that of increasing transcript abundance during mitosis, condensin might be directly or indirectly recruited by the accumulated RNA. We further showed that Cut14-208 TS mutant protein failed to accumulate at Hsp genes after a T-shift in mitosis. In this mutant background, a severe defect of chromosome segregation was frequently observed in anaphase. As DNA reannealing activity is lost in cut14-208 ts mutant at restrictive temperature (Sutani & Yanagida 1997), functional condensin is assumed to be required for its accumulation at activated genes. Cut14 association with Hsp genes diminished after 30 min in G1/S phase, even when transcripts were abundant. It is possible that condensin accumulation is promoted only in the short period (∽15 min) after transcriptional activation. Alternatively, condensin might accumulate at postreplicative transcribed DNAs before chromosome segregation. Indeed, Cut14 ChIP in two kinds of synchronized cultivations suggested that condensin's preferential binding is at postreplicative DNA. It remains to be seen whether condensin is capable of distinguishing postreplicative from prereplicative DNA.
No association of cohesin Rad21 and Top2 was evident at Sep1-dependent ecm33+ and gas1+ genes, but they were weakly associated with constitutively activated genes, ef1a-a+ and ef1a-b+. Thus, cohesin and Top2 may also bind to pol II-transcribed genes in interphase and presumably remain associated in mitosis. After heat-shock treatment in asynchronously cultured cells, Top2 associates significantly with Hsp genes, at levels comparable to those of condensin Cut14, suggesting that Top2 is more like condensin than cohesin, regarding its affinity to pol II-transcribed genes.
In mitosis, Cut14 enrichment is enhanced even at ef1a-a+ and ef1a-b+ genes that are not classified as mitotically up-regulated genes (Tables1 and 2, Fig.2B). S. pombe condensin is highly enriched in mitotic nuclei, whereas it is located throughout cells during interphase (Sutani et al. 1999). Condensin binding to mitotic chromosomes is also promoted by mitosis-specific kinases (Nakazawa et al. 2008; St-Pierre et al. 2009; Tada et al. 2011). Hence, condensin accumulation at actively transcribed genes in interphase might be further increased in mitosis, even if pol II-dependent transcription is not further activated at the gene. Unlike Cut14, association of Rad21 and Top2 at ef1a-a+ and ef1a-b+ genes is not enhanced in mitosis. This is consistent with the notion that cohesin and Top2 are enriched in nuclei throughout the cell cycle (Shiozaki & Yanagida 1992; Birkenbihl & Subramani 1995; Tomonaga et al. 2000).
In S. pombe mitosis, condensin is cytologically enriched in the nucleus from prophase to telophase, with two intense signals for kinetochores and rDNAs clearly seen until metaphase (Nakazawa et al. 2008). Signal intensity of GFP-tagged condensin protein in mitotic nuclei (expressed from a chromosomally integrated gene under the native promoter) is approximately 20-fold compared to that in nuclei of G2 phase cells. However, our ChIP-seq results showed that enrichment scores of condensin on overall mitotic chromosomes are only three- to fourfold higher, relative to that on chromosomes in asynchronous cells (70–80% cells are in G2 phase). This difference raises the possibility that not all GFP condensin signals in mitotic nuclei necessarily represent chromosome-bound condensin. After nuclear transport of condensin, mediated by Cdc2 and Cut15/importin α, its binding to mitotic chromosomes requires additional steps that include phosphorylation of Cnd2 (non-SMC subunits) by Ark1/Aurora B (Nakazawa et al. 2011; Tada et al. 2011). Therefore, in mitotic nuclei, a large fraction of condensin molecules might not be tightly associated with chromosomes, but rather may be mobile between chromosomes and nucleoplasm. Alternatively, the amount of chromosome-bound condensin in interphase may be higher than expected from the GFP-tagged condensin signal in interphase nucleus. This idea is consistent with the previous result that G2 phase cells treated with detergent, which removed proteins unbound to chromatin, showed significant nuclear signals of Cnd2- and Cut3/SMC4-GFP (Aono et al. 2002).
The mechanism by which condensin accumulates at pol II-transcribed active genes remains enigmatic. We speculate that condensin's ability to reanneal ssDNA in order to eliminate bound proteins and/or RNAs may be relevant to its accumulation at highly transcribed genes (Yanagida 2009; Akai et al. 2011). In condensin mutants, chromatin DNA is hypersensitive to ssDNA-specific nuclease (Sutani & Yanagida 1997), consistent with the notion that condensin acts on the unwound region of chromosomal DNA. However, we also suppose that pol II-dependent transcription does not sufficiently explain condensin accumulation at all enriched sites. In a reverse approach to the analysis presented in Tables1 and 2, Cut14 occupancy at pol II-enriched top 50 protein-coding genes is shown in Table S1 (Supporting Information). Among the top 50 genes, 37 had more than twice as much condensin enrichment as the average for all pol II-transcribed genes in asynchronous cells. However, pol II abundance does not necessarily cause condensin accumulation at all 50 genes. This result suggests that gene-specific loading factor(s) may also be involved in condensin accumulation. In budding yeast, cohesin- (and condensin-) loader Scc2/Scc4 complex preferentially associates with highly expressed genes by cooperating with the RSC chromatin remodeling complex (Lopez-Serra et al. 2014). Similarly, accumulation sites for condensin are presumably determined by a combination of pol II-mediated transcription and condensin recruiters at its binding sites.
In this study, condensin inactivation caused severe defects in chromosome segregation, but not in transcriptional activation or repression of Hsp genes. Moreover, our results indicate that condensin preferentially associates with postreplicative DNA at transcribed Hsp genes. Therefore, we hypothesize that condensin may have to interact with abundant pol II transcripts before faithful segregation of postreplicative chromosomes. Further study is definitely needed to understand how condensin acts on pol II-transcribed genes and/or transcription apparatus carrying transcribed RNAs to effect successful chromosome segregation and to play diverse roles in mitosis and interphase.
Experimental procedures
Strains and media
The S. pombe haploid, wild-type strain 972h− and its derivative mutant strains, including the temperature-sensitive (ts) cut14-208 (Saka et al. 1994) and cold-sensitive (cs) nda3-KM311 (Hiraoka et al. 1984), were used. Strains with chromosomally integrated 3FLAG-tagged Cut14 or Rad21 have been described previously (Adachi et al. 2008; Nakazawa et al. 2008). The Top2-3FLAG, Cnd1-3FLAG, and Cut14-208 TS-3FLAG strains were also made by chromosomal integration under the native promoter in the same manner. Deletion mutants of the sep1+ and atf1+ genes were kind gifts from Dr. C. J. McInerny (University of Glasgow) and Dr. T. Toda (Cancer Research UK), respectively. Strains used are shown in Table S2 (Supporting Information). S. pombe was cultured in YPD and MEA sporulation media (Saka et al. 1994; Forsburg & Rhind 2006). Cells were counted using a Sysmex CDA-500 hematology analyzer.
Synchronous culture and temperature-shift experiments
Temperature-shift experiments were carried out as previously reported (Nakazawa et al. 2011), using single nda3-KM311 or double nda3-KM311 cut14-208 mutants. Cs or cs ts mutant cells were grown in YPD at 30 °C (a permissive temperature for both strains). When cell concentrations reached 4×106 cells/mL, cultures were transferred to 20 °C and incubated for 8 h to (reversibly) block spindle formation and to arrest cells at a prometaphase-like stage (Hiraoka et al. 1984). For release experiments, cells were shifted to 36 °C (a permissive temperature for nda3 mutants) to allow spindle formation and cell cycle progression. After this transfer, cell aliquots were obtained at 0, 3, 9, 15, and 30 min and fixed with 2.5% glutaraldehyde to check anaphase progression. Asynchronously cultured cells were also harvested at 20 °C for 8 h in the absence of nda3-KM311. For temperature-shift (T-shift) conditions in asynchronously cultured cells, cells incubated at 20 °C for 8 h were shifted to 36 °C for 9 min. Prereplicative G1 cells were obtained by published procedures through nitrogen starvation and release into YPD medium (Sajiki et al. 2009). EMM2-N, lacking NH4Cl, was used for yeast starvation (2 × 106 cells/mL for 24 h at 26 °C). For HU block and release experiments, 12 mm HU was added to YPD medium at 20 °C for 8 h. Cells were then washed extensively in distilled water and released into YPD without HU. FACScan analysis was carried out as described previously (Akai et al. 2011).
Chromatin Immunoprecipitation (ChIP)
ChIP was carried out as described (Nakazawa et al. 2008) with minor modifications. Cells were disrupted with glass beads, using a Multi-Beads Shocker (Yasui Kikai), and DNA was quantified by real-time PCR (ExiCycler; Bioneer) with SYBR Premix Ex Taq II solution (TaKaRa). PCR primers used are shown in Table S3 (Supporting Information).
ChIP sequencing (ChIP-seq)
For ChIP sequencing, coprecipitated DNA with antibodies and WCE DNA were prepared as described above, except that a fourfold volume of cell culture and antibodies were used. Samples for ChIP-seq were prepared as described previously (Schmidt et al. 2009b). Prepared DNA libraries were processed by single-end sequencing (36 bp) on the Illumina Genome Analyzer GA IIx, according to the manufacturer's instructions.
Data analysis of ChIP-seq
After each completed sequencing run, raw image files from the Illumina sequencer were computationally processed to obtain nucleotide-base calls. The standard analytical pipeline provided by Illumina (CASAVA 1.8.2) was used for base-calling and demultiplexing of samples with an index sequence that was attached to the template during sample preparation. Subsequent to base-calling and demultiplexing, sequencing reads were aligned to the S. pombe genome sequence (downloaded from Ensemble genomes [ftp://ftp.ensemblgenomes.org/pub/fungi/current/fasta/schizosaccharomyces_pombe/dna/], Schizosaccharomyces_pombe.ASM294v1.16.dna.chromosome I, II, III & MT FASTA format) using Bowtie alignment software (bowtie-bio.sourceforge.net) with default parameters. To avoid artifacts associated with spuriously high read numbers, the 10× average read number was set as maximum at each location. To compare precipitated DNA (IP) and input DNA (WCE) data, the total read number of each sample was normalized to 10 million. The whole genome was then scanned using a sliding window to smooth the data (window = 500 bp, step = 10 bp). Before calculating a ratio, to avoid dividing by a small number caused by low WCE coverage, we added a pseudocount of 10 to all windows of each sample. ChIP enrichment was calculated as the ratio of IP to WCE for each window. This ratio was used for visualization of the ChIP-seq profile along three chromosomes using Integrated Genome Browser (IGB, ver. 7.0.3, Nicol et al. 2009). To calculate Cut14 enrichment at protein-coding genes, the same preprocesses were carried out to remove artifacts associated with spuriously high read numbers and normalize the data. Read numbers in each protein-coding gene were counted, and Cut14 enrichment was calculated as the log2 ratio of IP to WCE. Pol II enrichment was also calculated using published data (Carlsten et al. 2012).
RNA extraction and reverse transcription-PCR
Total RNA from S. pombe was extracted using the hot-phenol method. Purified RNA was reverse-transcribed using PrimeScript RT reagent (TaKaRa) with oligo dT primers according to the manufacturer's instructions. The genomic DNA eraser supplied with reverse transcription reagent was used to remove contaminated genomic DNA in the RNA sample. cDNA was quantified using real-time PCR with SYBR Premix Ex Taq II solution (TaKaRa).
Accession number
ChIP sequencing data have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE65956.
Acknowledgments
We are indebted to Drs Christopher. J. McInerny and Takashi Toda for the S. pombe strains. We thank Dr Takashi Sutani and Professor Katsuhiko Shirahige for generously sharing unpublished results with us, and Dr Steven D. Aird for editing the manuscript. This study was partly supported by the CREST Research Fund from the Japan Science and Technology Corporation when M.Y. was affiliated with Kyoto University. N.N. was supported by a Grant-in-Aid for Young Scientists (B) 25840013 from the Japan Society for the Promotion of Science (JSPS). X.X. was supported by a Grant-in-Aid for JSPS Fellows from JSPS. We are also grateful for the generous support of OIST Graduate University.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web site:
Figure S1ChIP-qPCR analyses of two condensin subunits.
Figure S2 ChIP-seq profiles of cohesin subunit Rad21.
Figure S3 ChIP-seq profiles of DNA topoisomerase II (Top2).
Table S1 Cut14 occupancy at pol II-enriched top 50 protein-coding genes in asynchronously dividing cells
Table S2 List of strains used in this study
Table S3 List of PCR primers used in this study
References
- Adachi Y, Kokubu A, Ebe M, Nagao K. Yanagida M. Cut1/separase-dependent roles of multiple phosphorylation of fission yeast cohesion subunit Rad21 in post-replicative damage repair and mitosis. Cell Cycle. 2008;7:765–776. doi: 10.4161/cc.7.6.5530. [DOI] [PubMed] [Google Scholar]
- Adelman K. Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolphs KW, Cheng SM, Paulson JR. Laemmli UK. Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc. Natl Acad. Sci. USA. 1977;74:4937–4941. doi: 10.1073/pnas.74.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akai Y, Kurokawa Y, Nakazawa N, Tonami-Murakami Y, Suzuki Y, Yoshimura SH, Iwasaki H, Shiroiwa Y, Nakamura T, Shibata E. Yanagida M. Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open Biol. 2011;1:110023. doi: 10.1098/rsob.110023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono N, Sutani T, Tomonaga T, Mochida S. Yanagida M. Cnd2 has dual roles in mitotic condensation and interphase. Nature. 2002;417:197–202. doi: 10.1038/417197a. [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP. Subramani S. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J. Biol. Chem. 1995;270:7703–7711. doi: 10.1074/jbc.270.13.7703. [DOI] [PubMed] [Google Scholar]
- Buck V, Ng SS, Ruiz-Garcia AB, Papadopoulou K, Bhatti S, Samuel JM, Anderson M, Millar JB. McInerny CJ. Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J. Cell Sci. 2004;117:5623–5632. doi: 10.1242/jcs.01473. [DOI] [PubMed] [Google Scholar]
- Carlsten JO, Szilagyi Z, Liu B, Lopez MD, Szaszi E, Djupedal I, Nystrom T, Ekwall K, Gustafsson CM. Zhu X. Mediator promotes CENP-a incorporation at fission yeast centromeres. Mol. Cell. Biol. 2012;32:4035–4043. doi: 10.1128/MCB.00374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Ren J, Bhattacharjee S, Chang AY, Sanchez M, Valbuena A, Antequera F. Martienssen RA. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell. 2014;159:572–583. doi: 10.1016/j.cell.2014.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N. Bahler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi ES, Stralfors A, Castillo AG, Durand-Dubief M, Ekwall K. Allshire RC. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J. Biol. Chem. 2011;286:23600–23607. doi: 10.1074/jbc.M111.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Blanco A, Mayan-Santos M, Schneider DA, Machin F, Jarmuz A, Tschochner H. Aragon L. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature. 2009;458:219–222. doi: 10.1038/nature07652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S. Haering CH. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 2011;21:552–559. doi: 10.1016/j.tcb.2011.06.003. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K. Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Bilodeau S, Orlando DA, Hubner MR, Abraham BJ, Spector DL. Young RA. Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem Cell Rep. 2013;1:371–378. doi: 10.1016/j.stemcr.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Persson J, Norman U, Hartsuiker E. Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010;29:2126–2134. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T. Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Hudson DF, Marshall KM. Earnshaw WC. Condensin: architect of mitotic chromosomes. Chromosome Res. 2009;17:131–144. doi: 10.1007/s10577-008-9009-7. [DOI] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI. Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell. 2010;21:254–265. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Zhang T, Wong NC, Davidson N, Maksimovic J, Oshlack A, Earnshaw WC, Kalitsis P. Hudson DF. Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat. Commun. 2013;4:2537. doi: 10.1038/ncomms3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. Strunnikov A. Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Kranz AL, Jiao CY, Winterkorn LH, Albritton SE, Kramer M. Ercan S. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol. 2013;14:R112. doi: 10.1186/gb-2013-14-10-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Serra L, Kelly G, Patel H, Stewart A. Uhlmann F. The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat. Genet. 2014;46:1147–1151. doi: 10.1038/ng.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusaka T, Imamoto N, Yoneda Y. Yanagida M. Mutations in fission yeast Cut15, an importin alpha homolog, lead to mitotic progression without chromosome condensation. Curr. Biol. 1998;8:1031–1034. doi: 10.1016/s0960-9822(07)00425-3. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R. Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Nakazawa N, Mehrotra R, Ebe M. Yanagida M. Condensin phosphorylated by the Aurora-B-like kinase Ark1 is continuously required until telophase in a mode distinct from Top2. J. Cell Sci. 2011;124:1795–1807. doi: 10.1242/jcs.078733. [DOI] [PubMed] [Google Scholar]
- Nakazawa N, Nakamura T, Kokubu A, Ebe M, Nagao K. Yanagida M. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J. Cell Biol. 2008;180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG, Jr, Raja A. Loraine AE. The integrated genome browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman-Axelsson U, Durand-Dubief M, Prasad P. Ekwall K. DNA topoisomerase III localizes to centromeres and affects centromeric CENP-A levels in fission yeast. PLoS Genet. 2013;9:e1003371. doi: 10.1371/journal.pgen.1003371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF. Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Ribar B, Grallert A, Olah E. Szallasi Z. Deletion of the sep1(+) forkhead transcription factor homologue is not lethal but causes hyphal growth in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 1999;263:465–474. doi: 10.1006/bbrc.1999.1333. [DOI] [PubMed] [Google Scholar]
- Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P. Bahler J. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Goldberg IG, Wood ER. Earnshaw WC. ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J. Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajiki K, Hatanaka M, Nakamura T, Takeda K, Shimanuki M, Yoshida T, Hanyu Y, Hayashi T, Nakaseko Y. Yanagida M. Genetic control of cellular quiescence in S. pombe. J. Cell Sci. 2009;122:1418–1429. doi: 10.1242/jcs.046466. [DOI] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y. Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Hizume K, Sutani T, Takeyasu K. Yanagida M. Condensin but not cohesin SMC heterodimer induces DNA reannealing through protein-protein assembly. EMBO J. 2003;22:2764–2775. doi: 10.1093/emboj/cdg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaak J, Mao J. Soll D. The 5.8S RNA gene sequence and the ribosomal repeat of Schizosaccharomyces pombe. Nucleic Acids Res. 1982;10:2851–2864. doi: 10.1093/nar/10.9.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CK, Brookes N. Uhlmann F. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 2009a;10:R52. doi: 10.1186/gb-2009-10-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J. Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009b;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K. Yanagida M. Functional dissection of the phosphorylated termini of fission yeast DNA topoisomerase II. J. Cell Biol. 1992;119:1023–1036. doi: 10.1083/jcb.119.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Douziech M, Bazile F, Pascariu M, Bonneil E, Sauve V, Ratsima H. D'Amours D. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol. Cell. 2009;34:416–426. doi: 10.1016/j.molcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E. Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Larionov VL. Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T. Yanagida M. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature. 1997;388:798–801. doi: 10.1038/42062. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K. Yanagida M. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K, Susumu H, Sakuno T. Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M. Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Tanizawa H, Sriswasdi S, Iwasaki O, Chatterjee AG, Speicher DW, Levin HL, Noguchi E. Noma KI. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol. Cell. 2012;48:532–546. doi: 10.1016/j.molcel.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JH. Nucleic acid synthesis in relation to the cell division cycle. Ann. NY Acad. Sci. 1960;90:409–421. doi: 10.1111/j.1749-6632.1960.tb23259.x. [DOI] [PubMed] [Google Scholar]
- Toda T, Nakaseko Y, Niwa O. Yanagida M. Mapping of rRNA genes by integration of hybrid plasmids in Schizosaccharomyces pombe. Curr. Genet. 1984;8:93–97. doi: 10.1007/BF00420224. [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, Nasmyth K. Yanagida M. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BD, Eyre D, Basrai M, Lichten M. Strunnikov A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 2005;25:7216–7225. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF. Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M. Clearing the way for mitosis: is cohesin a target? Nat. Rev. Mol. Cell Biol. 2009;10:489–496. doi: 10.1038/nrm2712. [DOI] [PubMed] [Google Scholar]
- Yoshimura SH, Hizume K, Murakami A, Sutani T, Takeyasu K. Yanagida M. Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr. Biol. 2002;12:508–513. doi: 10.1016/s0960-9822(02)00719-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1ChIP-qPCR analyses of two condensin subunits.
Figure S2 ChIP-seq profiles of cohesin subunit Rad21.
Figure S3 ChIP-seq profiles of DNA topoisomerase II (Top2).
Table S1 Cut14 occupancy at pol II-enriched top 50 protein-coding genes in asynchronously dividing cells
Table S2 List of strains used in this study
Table S3 List of PCR primers used in this study