Abstract
Pancreatic ductal adenocarcinoma (PDAC) is among the most fatal of malignancies with an extremely poor prognosis. The objectives of this study were to provide a detailed understanding of PDAC pathophysiology in view of the host immune response. We examined the PDAC tissues, sera, and peripheral blood cells of PDAC patients using immunohistochemical staining, the measurement of cytokine/chemokine concentrations, gene expression analysis, and flow cytometry. The PDAC tissues were infiltrated by macrophages, especially CD33+CD163+ M2 macrophages and CD4+ T cells that concomitantly express programmed cell death-1 (PD-1). Concentrations of interleukin (IL)-6, IL-7, IL-15, monocyte chemotactic protein-1, and interferon-γ-inducible protein-1 in the sera of PDAC patients were significantly elevated. The gene expression profile of CD14+ monocytes and CD4+ T cells was discernible between PDAC patients and healthy volunteers, and the differentially expressed genes were related to activated inflammation. Intriguingly, PD-1 was significantly upregulated in the peripheral blood CD4+ T cells of PDAC patients. Correspondingly, the frequency of CD4+PD-1+ T cells was increased in the peripheral blood cells of PDAC patients, and this increase correlated to chemotherapy resistance. In conclusion, inflammatory conditions in both PDAC tissue and peripheral blood cells in PDAC patients were prominent, highlighting monocytes/macrophages as well as CD4+ T cells with influence of the clinical prognosis.
We examined the inflammatory features of PDAC patients using the PDAC tissues, sera, and peripheral blood by immunohistochemical staining, measurement of cytokines/chemokines, gene expression analysis, and flow cytometry. We foundg that monocyte/macrophage cells and CD4+ T cells were highlighted immune-mediating cells in local cancer tissue as well as in peripheral blood of PDAC patients, among which the important subfraction with clinical impact influencing PDAC prognosis by chemotherapy was involved.
Keywords: CD4+ T cells, macrophages, monocytes, pancreatic ductal adenocarcinoma, programmed cell death-1
Pancreatic ductal adenocarcinoma (PDAC) is one of the most fatal solid malignancies with a 5-year survival rate of <5% and a median survival of 4–6 months in Japan as well as in other countries.1,2 Surgical resection is the only method for achieving radical treatment; however, only 15–20% of patients are diagnosed in the operable early stage.3 For unresectable PDAC, gemcitabine-based chemotherapeutic regimens are efficacious; however, improvements in survival are limited to only several months, and no complete remission is fundamentally expected.4 Therefore, it is extremely important to understand the pathophysiology of PDAC in order to establish novel diagnostic methods for early detection as well as to develop novel effective therapies.
Cancer is frequently associated with inflammation induced by the host immune system,5 involving a variety of immune-mediating cells, including the Th1 helper T cells and cytotoxic T lymphocytes,6 which inhibit cancer progression, and the myeloid-derived suppressor cells7 and regulatory T cells,8 which are cancer-promoting inflammatory cells. These anticancer and cancer-promoting immune-mediating cells have a complex involvement in the persistent inflammation associated with cancer that influences the patient's prognosis. Some inflammatory cells such as regulatory T cells, myeloid-derived suppressor cells, as well as humoral mediator interleukin (IL)-6, are reported to be involved in PDAC;9 however, details of the systemic inflammatory condition of PDAC has not been sufficiently studied. Accordingly, the purpose of this study was to elucidate the systemic inflammatory state of PDAC by investigating the inflammatory markers in the local cancer tissue, serum, and peripheral blood.
Materials and Methods
Patients and pancreatic ductal adenocarcinoma tissues
Twenty specimens that were surgically removed from PDAC patients (Table1) were used for pathological analysis. Both PDAC patients and healthy volunteers were enrolled after providing informed consent prior to the serum concentration analysis of cytokines and chemokines, gene expression analysis of peripheral blood cells, and flow cytometry analysis. The clinical characteristics of the study participants are provided in Tables S1–S3. The clinical stages were evaluated in accordance with the TNM staging system for pancreatic carcinoma issued by the Union of International Cancer Control (7th edition). The therapeutic effect of chemotherapy was assessed in terms of partial responsiveness, stable disease, and progressive disease in accordance with the Response Evaluation Criteria in Solid Tumors. The study was approved by the institutional review board and was carried out in accordance with the Declaration of Helsinki.
Table 1.
Inflammatory features of pancreatic ductal adenocarcinoma tissues and clinical characteristics of patients
| Patient no. | Age, years | Sex | Stage | Tumor size, mm | Degree of inflammation | Infiltrating inflammatory cells† | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4 | T-bet | FoxP3 | PD-1 | CD33 | CD14 | CD163 | ||||||
| 1 | 71 | F | II | 25 | Mild | >100 | <5 | 24 | 6 | >100 | 78 | >100 |
| 2 | 57 | M | III | 25 | Moderate | 58 | <5 | 35 | 14 | 33 | 73 | >100 |
| 3 | 61 | F | II | 15 | Severe | 48 | <5 | 39 | 26 | >100 | >100 | >100 |
| 4 | 54 | F | III | 55 | Mild | >100 | <5 | 27 | <5 | >100 | 12 | 56 |
| 5 | 70 | F | IV | 33 | Mild | 46 | <5 | 12 | 45 | >100 | 38 | >100 |
| 6 | 66 | M | II | 25 | Moderate | 26 | <5 | 16 | 15 | >100 | 85 | >100 |
| 7 | 60 | F | III | 18 | Moderate | >100 | <5 | 37 | 6 | >100 | 62 | >100 |
| 8 | 78 | M | III | 37 | Moderate | >100 | <5 | 48 | 8 | >100 | 28 | 65 |
| 9 | 77 | M | II | 30 | Moderate | 55 | 39 | 26 | 18 | >100 | 54 | >100 |
| 10 | 57 | M | III | 55 | Moderate | 71 | <5 | 13 | 57 | >100 | <10 | 98 |
| 11 | 65 | M | III | 43 | Moderate | >100 | 12 | 51 | 6 | >100 | >100 | >100 |
| 12 | 68 | F | III | 25 | Severe | >100 | <5 | 22 | 12 | >100 | >100 | >100 |
| 13 | 62 | M | II | 22 | Mild | >100 | <5 | <10 | 7 | >100 | 19 | 59 |
| 14 | 65 | M | II | 25 | Moderate | >100 | 5 | <10 | 32 | >100 | 23 | 73 |
| 15 | 59 | F | II | 18 | Mild | >100 | <5 | <10 | <5 | >100 | 43 | 92 |
| 16 | 66 | M | I | 10 | Moderate | >100 | 13 | 74 | 58 | >100 | <10 | <10 |
| 17 | 70 | M | III | 5 | Moderate | >100 | <5 | 14 | <5 | >100 | >100 | 41 |
| 18 | 57 | F | II | 18 | Moderate | >100 | <5 | 15 | <5 | >100 | 24 | >100 |
| 19 | 63 | M | II | 25 | Mild | >100 | <5 | 23 | <5 | >100 | 27 | 83 |
| 20 | 64 | F | II | 21 | Moderate | >100 | 23 | 51 | 42 | >100 | >100 | >100 |
The number of each inflammatory cells was assessed per high power field. F, female; M, male.
Isolation of peripheral blood mononuclear cells
Peripheral blood was obtained prior to any treatments for PDAC. Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood using Ficoll–Hypaque density gradient centrifugation (Sigma-Aldrich, St. Louis, MO, USA), as previously described.10 The obtained fraction of PBMCs was incubated with bead-labeled anti-CD4, anti-CD8, anti-CD14, or anti-CD15 antibodies (Miltenyi, Cologne, Germany), followed by isolation using a magnet.
Additional procedures
Additional materials and methods are presented in Data S1.
Results
Features of local immune-mediating cells in PDAC tissues
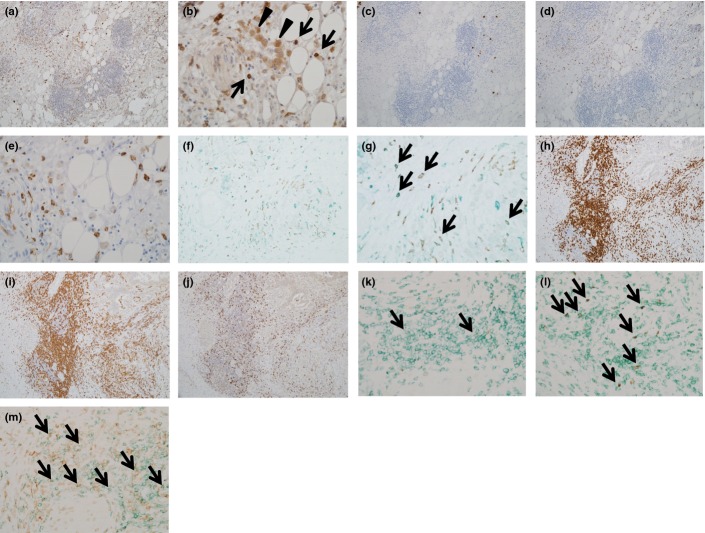
To elucidate local inflammatory conditions of PDAC, we immunohistochemically analyzed the surgically resected PDAC tissues. We found that CD33+ myeloid cells markedly infiltrated PDAC tissues (Fig.1a,b, Table1). Neutrophil elastase-positive cells were rarely observed among CD33+ cells (Fig.1c), suggesting that CD33+ myeloid-lineage cells were likely monocytes/macrophages. There was a fraction of CD14+ cells (Fig.1d,e, Table1) and a more prominent fraction of CD163+ monocytes/macrophages among CD33+ cells (Fig.1f,g, Table1), indicating infiltration of M2 suppressive macrophages. Lymphoid follicles were observed adjacent to PDAC tissues, where most of the infiltrating lymphocytes were CD3+ T cells (Fig.1h). Among infiltrating CD3+ T cells, CD4+ T cells were predominant compared to the CD8+ T cells (Fig.1i,j). T-bet+ cells were not frequently observed (Fig.1k), whereas FoxP3+ cells as well as programmed cell death-1 (PD-1)+ cells were more frequently observed (Fig.1l,m, Table1), suggesting that regulatory or activated CD4+ T cells had infiltrated the PDAC tissues.
Figure 1.
Immunohistochemical analysis of pancreatic ductal adenocarcinoma (PDAC) tissues. Surgically resected PDAC tissues were immunohistochemically stained. (a, b) CD33: several positive cells were scattered in the fibroadipose area around PDAC, which were mainly composed of a monocyte (arrows in b) and macrophage (arrowheads in b) morphology. (c) Neutrophil elastase: a few positive cells were observed. (d, e) CD14: several positive cells highlighting the monocyte morphology. (f, g) Double immunostaining of CD163 (brown) and CD33 (green). Most of the CD163+ cells were also positive for CD33. (h) CD3: lymphoid aggregation around PDAC was mainly composed of CD3+ T cells. (i) CD4: the majority of lymphoid aggregation was CD4+ T cells. (j) CD8: the number of CD8+ cells was small compared to that of CD4+ cells shown in (i). (k) Double immunostaining of T-bet (brown, nuclear expression) and CD4 (green). Double-positive cells were observed (arrows), but the number was very small. (l) Double immunostaining of FoxP3 (brown, nuclear expression) and CD4 (green). Several double-positive cells were scattered (arrows). (m) PD-1 (brown) and CD4 (green): several double-positive cells were found (arrows). Magnification: a, c, d, f, h, i, and j, ×40; b, e, g, k, l, and m, ×100.
Serum cytokine and chemokine concentration in PDAC patients
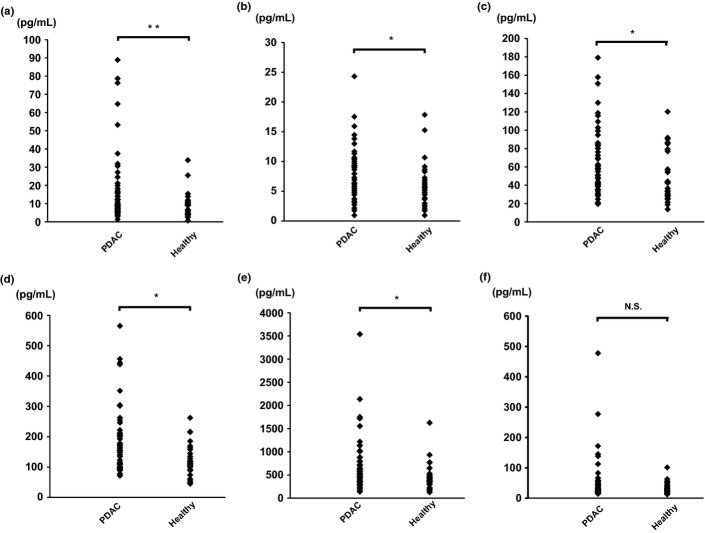
We next assessed concentration level of panels of cytokines and chemokines in sera of PDAC patients (Table S1). Serum concentrations of the cytokines IL-6, IL-7, and IL-15 were significantly elevated in PDAC patients compared to healthy volunteers (Fig.2). Among chemokines, monocyte chemotactic protein-1 (MCP-1) and interferon-γ-inducible protein-10 (IP-10) were significantly elevated, and IL-8 was relatively high in PDAC patients, although this was not statistically significant (P = 0.06; Fig.2). We used real time detection-PCR (RTD-PCR) to measure the expression levels of mRNAs encoding these cytokines and chemokines in CD14+ monocytes/macrophages and CD4+ T cells of PDAC patients. We found that IL-15 expression by CD14+ monocytes/macrophages and IL-6 and IL-7 expression by CD4+ T cells of PDAC patients were significantly upregulated compared to those of healthy volunteers (Fig. S1), suggesting that peripheral CD14+ monocytes/macrophages and CD4+ T cells contribute to the elevations in cytokine levels evident in the sera of PDAC patients. Such cells are the local macrophages and CD4+ T cells associated with inflammation of PDAC tissues.
Figure 2.
Concentration of cytokines and chemokines in sera obtained from pancreatic ductal adenocarcinoma patients (n = 50) prior to treatment and from healthy volunteers (n = 27). The serum concentration of cytokines and chemokines was measured using a multiplex bead immunoassay system. (a) Interleukin (IL)-6, (b) IL-7, (c) monocyte chemotactic protein-1, (d) IL-15, (e) interferon-γ-inducible protein-1, and (f) IL-8. *P < 0.05, **P < 0.01.
Distinct gene expression profile of CD14+ monocytes and CD4+ T cells in PBMCs of patients with PDAC
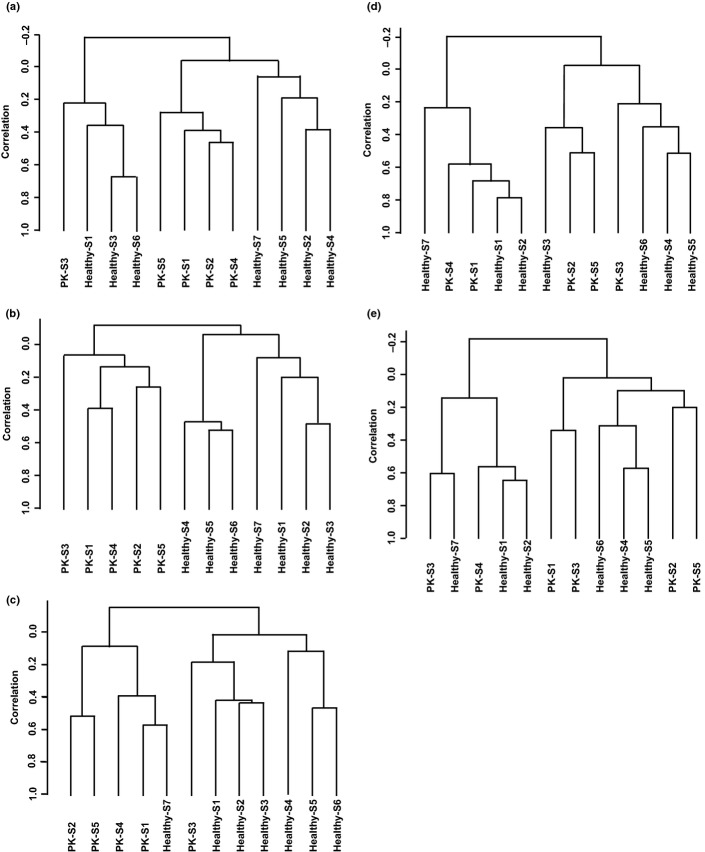
Local infiltrating inflammatory cells and serum cytokine/chemokine elevation highlighted macrophages and CD4+ T cells in the context of PDAC inflammation. We further examined whether peripheral blood cells were affected using gene expression analysis with DNA microarray. Unsupervised clustering analysis of gene expression showed a clear gene expression pattern for all blood cells (Fig.3a), which was consistent with our previous report.11 The analysis of peripheral blood cell subfractions in PDAC patients showed a discernible gene expression profile of the CD14+ monocyte and CD4+T cell fractions (Fig.3b,c), whereas CD8+ and CD15+ cell fractions did not (Fig.3d,e).
Figure 3.
Unsupervised clustering analysis of gene expression profiles of subfractions of peripheral blood cells from patients with pancreatic ductal adenocarcinoma (PK; n = 7) and healthy volunteers (n = 5). RNA was isolated from entire blood cells or each subfraction of peripheral blood cells, followed by gene expression analysis using DNA microarray. Genes that passed the quality check control were used in each clustering analysis. (a) Entire blood cells, 7039 genes; (b) CD14+ cells, 6602 genes; (c) CD4+ cells, 6770 genes; (d) CD8+ cells, 7621 genes; and (e) CD15+ cells, 9728 genes.
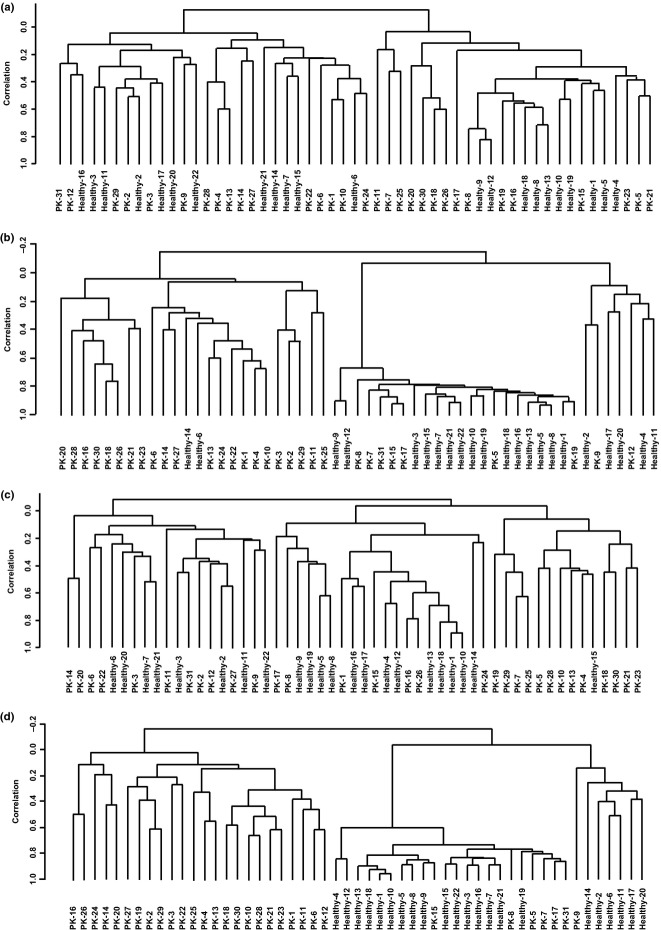
Unsupervised analysis of the gene expression profile in isolated CD14+ monocytes and CD4+ T cells in a larger cohort (Table S3) also showed relatively discernible clusters for PDAC patients and healthy volunteers (Fig.4a,c). Significantly altered gene expression by ≥1.5-fold in peripheral CD14+ monocytes of PDAC patients compared to healthy volunteers was observed in 261, 126, and 85 genes at P-values of <0.05, <0.01, and <0.005, respectively. Most of these genes were upregulated (177/261, 87/126, and 61/85, respectively). The numbers of significantly altered genes by ≥1.5-fold in CD4+ cells were 690, 496, and 419 with P-values of <0.05, <0.01, and <0.005, respectively. Most of these were also upregulated (459/690, 349/496, and 298/419, respectively). Unsupervised analysis of the gene expression profile using the 261 significantly altered genes from CD14+ monocytes and 496 genes from CD4+ T cells showed distinct clusters for PDAC patients and healthy volunteers (Fig.4b,d).
Figure 4.
Unsupervised clustering analysis of the gene expression profile of CD14+ monocytes and CD4+ T cells in the peripheral blood of pancreatic ductal adenocarcinoma (PDAC) patients (PK) and healthy volunteers. RNA was isolated from all blood cells or each subfraction of peripheral blood cells from 31 PDAC patients and 22 healthy volunteers, followed by gene expression analysis using DNA microarray. (a, b) Hierarchical analysis of gene expression for isolated CD4+ cells in peripheral blood using all 10 868 filtered genes (a) or 266 genes whose expression was significantly altered between PDAC patients and healthy volunteers ≥1.5-fold with P < 0.001 (b). (c, d) Hierarchical analysis of gene expression for isolated CD14+ cells in peripheral blood using all 11 947 filtered genes (c) or 126 genes whose expression was significantly altered between PDAC patients and healthy volunteers ≥1.5-fold at P < 0.01 (d).
The biological process networks related to the 261 genes, whose expression was significantly altered ≥1.5-fold in CD14+ monocytes/macrophages of PDAC patients, included the cell cycle, inflammation, blood coagulation, cell adhesion, and development (Table2). We randomly selected 17 genes from the list of those 50 most significantly upregulated upon microarray analysis (Table3), and measured transcriptional expression levels by RTD-PCR. We found that most of these genes were indeed upregulated, including the adhesion-related gene CD226 and the cell cycle-related gene CDK6 (Table S4). Biological process networks related to the 496 genes whose expression was significantly altered ≥1.5-fold in CD4+ T cells of PDAC patients mostly included the cell cycle and inflammation as well as DNA damage and apoptosis (Table4). We randomly selected 18 genes from the list of those 50 most significantly upregulated, as revealed by microarray analysis (Table5), and measured transcriptional expression levels using RTD-PCR. We found that most of these genes were indeed upregulated, including the cell cycle-associated gene PTTG1 and the apoptosis-related gene BAX (Table S4). Interestingly, PD-1, which is expressed on the activated T cell to attenuate the T cell receptor signaling pathway, was also included (Table5). Thus, CD14+ monocytes and CD4+T cells were the meaningfully affected subpopulations of peripheral blood cells in PDAC patients.
Table 2.
Biological process networks for 261 genes whose expression in CD14+ peripheral blood cells was significantly altered between patients with pancreatic ductal adenocarcinoma and healthy volunteers
| Networks | Total | P-value | False discovery rate | In data | Network objects from active data |
|---|---|---|---|---|---|
| Blood coagulation | 94 | 3.09E-06 | 4.33E-04 | 11 | α-IIb/β-3 integrin, PAR1, thrombospondin 1, TFPI, Galpha(q)-specific nucleotide-like GPCRs, P2Y1, ITGB3, sCD40L, GP-IB beta, protein C, CD40L(TNFSF5) |
| Inflammation_NK cell cytotoxicity | 164 | 1.32E-04 | 9.25E-03 | 12 | KIR2DL4, KLRK1 (NKG2D), SAP, PPP2R2B, NKG2C, KIR3DL1, IP3 receptor, NKG2A, histone H1, IgG1, CD94, KLRC4 (NKG2F) |
| Inflammation_Interferon signaling | 110 | 4.12E-04 | 1.92E-02 | 9 | CCL5, PPAR-γ, IFITM2, PKR, IFI17, IFI27, IFI6, IL-18R1, IFI44 |
| Cell adhesion_Platelet-endothelium-leucocyte interactions | 174 | 3.05E-03 | 8.79E-02 | 10 | CCL5, α-IIb/β-3 integrin, DNAM1, thrombospondin 1, PDGF-B, 08p22/MSR1(CD204), GP-IB β, protein C, CD40L(TNFSF5), JAM3 |
| Cell cycle_Mitosis | 179 | 3.74E-03 | 0.087907 | 10 | ASPM, MCAK, PKR, cyclin B, cyclin B2, survivin, securin, CAP-G/G2, histone H1, AF15q14 |
| Inflammation_Complement system | 73 | 0.00376744 | 0.087907 | 6 | C2, Factor H, C2b, C2a, Factor I, clusterin |
| Cell cycle_Core | 115 | 0.00935692 | 0.172396 | 7 | CAP-G, MCM6, cyclin B, cyclin B2, survivin, securin, CDK6 |
| Cell cycle_G2–M | 206 | 0.00985118 | 0.172396 | 10 | Histone H1.5, p38 MAPK, CAP-G, cyclin B, PDGF-B, cyclin B2, securin, p38delta (MAPK13), CAP-G/G2, histone H1 |
| Development_Regulation of angiogenesis | 223 | 0.01649253 | 0.256551 | 10 | Ephrin-B receptors, ephrin-A receptors, Galpha(q)-specific peptide GPCRs, EDNRB, thrombospondin 1, RhoB, IP3 receptor, PKC, IL-18R1, clusterin |
| Cell cycle_S phase | 149 | 0.03378318 | 0.438589 | 7 | Histone H1.5, MCM6, cyclin B, cyclin B2, securin, histone H1, ChAF1 subunit B |
Table 3.
Significant genes with upregulated expression in CD14+ peripheral blood cells from patients with pancreatic ductal adenocarcinoma
| P-value | Fold-change (PK/healthy) | Symbol | Description | Accession† | Defined gene list |
|---|---|---|---|---|---|
| 5.10E-06 | 1.666666667 | DLC1 | Deleted in liver cancer 1 | NM_001164271 | |
| 2.21E-05 | 2.083333333 | HPGD | Hydroxyprostaglandin dehydrogenase 15-(NAD) | NM_000860 | |
| 2.99E-05 | 1.851851852 | EPS8 | Epidermal growth factor receptor pathway substrate 8 | NM_004447 | |
| 4.11E-05 | 1.612903226 | DENND1B | DENN/MADD domain containing 1B | NM_001142795 | |
| 4.23E-05 | 1.851851852 | AMIGO2 | Adhesion molecule with Ig-like domain 2 | NM_001143668 | |
| 0.0000435 | 2.631578947 | MSR1 | Macrophage scavenger receptor 1 | NM_002445 | Phagosome |
| 8.58E-05 | 1.724137931 | EPSTI1 | Epithelial stromal interaction 1 (breast) | NM_001002264 | |
| 0.0000927 | 2.083333333 | ARID5B | AT rich interactive domain 5B (MRF1-like) | NM_001244638 | |
| 0.0001720 | 1.587301587 | EIF2AK2 | Eukaryotic translation initiation factor 2-alpha kinase 2 | NM_001135651 | Bone remodelling, double-stranded RNA-induced gene expression, inactivation of GSK3 by AKT causes accumulation of β-catenin in alveolar macrophages, regulation of EIF2, Toll-like receptor pathway, hepatitis c, protein processing in endoplasmic reticulum |
| 0.0001827 | 1.666666667 | FBXO38 | F-box protein 38 | NM_001271723 | |
| 0.0002219 | 3.333333333 | UTY | Ubiquitously transcribed tetratricopeptide repeat containing, Y-linked | NM_001258249 | |
| 0.0002584 | 1.694915254 | FKBP5 | FK506 binding protein 5 | NM_001145775 | |
| 3.47E-04 | 1.639344262 | LRP12 | Low density lipoprotein receptor-related protein 12 | NM_001135703 | |
| 0.0003545 | 16.12903226 | DDX3Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | NM_001122665 | RIG-I-like receptor signaling pathway |
| 0.0004157 | 17.85714286 | RPS4Y2 | Ribosomal protein S4, Y-linked 2 | NM_001039567 | |
| 4.29E-04 | 2.777777778 | FAM20A | Family with sequence similarity 20, member A | NM_001243746 | |
| 0.0004546 | 2.040816327 | CLU | Clusterin | NM_001831 | |
| 4.59E-04 | 17.54385965 | RPS4Y1 | Ribosomal protein S4, Y-linked 1 | NM_001008 | Ribosome |
| 5.09E-04 | 1.5625 | VWCE | Von Willebrand factor C and EGF domains | NM_152718 | |
| 5.50E-04 | 1.639344262 | CDK6 | Cyclin-dependent kinase 6 | NM_001145306 | Cell cycle: G1/s checkpoint, cyclins and cell cycle regulation, estrogen-responsive protein EFP controls cell cycle and breast tumors growth, influence of Ras and Rho proteins on G1 to S transition, cell cycle, chronic myeloid leukemia, glioma, melanoma, non-small-cell lung cancer, p53 signaling pathway, pancreatic cancer, pathways in cancer, small-cell lung cancer |
| 5.57E-04 | 1.612903226 | P2RY1 | Purinergic receptor P2Y, G-protein coupled, 1 | NM_002563 | Neuroactive ligand–receptor interaction |
| 0.0005792 | 2.083333333 | PRDM1 | PR domain containing 1, with ZNF domain | NM_001198 | |
| 0.0005939 | 1.785714286 | IFI44 | Interferon-induced protein 44 | NM_006417 | |
| 6.98E-04 | 1.515151515 | MT2A | Metallothionein 2A | NM_005953 | |
| 0.0007509 | 1.694915254 | LY6E | Lymphocyte antigen 6 complex, locus E | NM_001127213 | |
| 0.0008281 | 1.612903226 | BAMBI | BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) | NM_012342 | |
| 0.0008473 | 1.754385965 | C2 | Complement component 2 | NM_000063 | Classical complement pathway, complement pathway, lectin-induced complement pathway, complement and coagulation cascades, Staphylococcus aureus infection, systemic lupus erythematosus |
| 1.09E-03 | 2 | TTTY15 | Testis-specific transcript, Y-linked 15 (non-protein coding) | NR_001545 | |
| 1.27E-03 | 1.851851852 | NGFRAP1 | Nerve growth factor receptor (TNFRSF16) associated protein 1 | NM_014380 | Neurotrophin signaling pathway |
| 0.0013756 | 2.222222222 | PDK4 | Pyruvate dehydrogenase kinase, isozyme 4 | NM_002612 | |
| 1.52E-03 | 2.564102564 | ZFY | Zinc finger protein, Y-linked | NM_001145275 | |
| 1.64E-03 | 1.515151515 | CABLES1 | Cdk5 and Abl enzyme substrate 1 | NM_001100619 | |
| 1.68E-03 | 1.694915254 | TNIK | TRAF2 and NCK interacting kinase | NM_001161560 | |
| 0.0018725 | 1.538461538 | CHAF1B | Chromatin assembly factor 1, subunit B (p60) | NM_005441 | BTG family proteins and cell cycle regulation |
| 0.0018950 | 1.724137931 | BTG3 | BTG family, member 3 | NM_001130914 | RNA degradation |
| 0.0019480 | 1.612903226 | HDGFRP3 | Hepatoma-derived growth factor, related protein 3 | NM_016073 | |
| 2.08E-03 | 2.631578947 | PPARG | Peroxisome proliferator-activated receptor gamma | NM_005037 | Basic mechanism of action of PPARa, PPARb(d) and PPARg and effects on gene expression, nuclear receptors in lipid metabolism and toxicity, role of PPAR-γ coactivators in obesity and thermogenesis, visceral fat deposits and the metabolic syndrome, Huntington's disease, osteoclast differentiation, pathways in cancer, PPAR signaling pathway, thyroid cancer |
| 2.21E-03 | 1.587301587 | MAF | V-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | NM_001031804 | |
| 2.31E-03 | 1.639344262 | TFDP2 | Transcription factor Dp-2 (E2F dimerization partner 2) | NM_001178138 | Cell cycle |
| 0.0023519 | 1.5625 | FOXC1 | Forkhead box C1 | NM_001453 | |
| 2.36E-03 | 1.587301587 | PLAC8 | Placenta-specific 8 | NM_001130715 | |
| 2.41E-03 | 1.666666667 | BEX1 | Brain expressed, X-linked 1 | NM_018476 | |
| 0.0024547 | 1.515151515 | PIM1 | Pim-1 oncogene | NM_001243186 | Acute myeloid leukemia, Jak-Stat signaling pathway |
| 0.0024567 | 2.173913043 | CST7 | Cystatin F (leukocystatin) | NM_003650 | |
| 0.0025775 | 1.960784314 | PCSK6 | Proprotein convertase subtilisin/kexin type 6 | NM_002570 | |
| 0.0029609 | 1.639344262 | REST | RE1-silencing transcription factor | NM_001193508 | Huntington's disease |
| 0.0029864 | 1.666666667 | FKBP11 | FK506 binding protein 11, 19 kda | NM_001143781 | |
| 0.0029868 | 1.639344262 | OASL | 2′-5′-oligoadenylate synthetase-like | NM_001261825 | |
| 0.0033994 | 1.754385965 | CD226 | CD226 molecule | NM_006566 | Cell adhesion molecules |
| 3.85E-03 | 1.538461538 | PNMA1 | Paraneoplastic Ma antigen 1 | NM_006029 |
PK, pancreatic cancer patients.
Table 4.
Biological process networks for 496 genes whose expression in CD4+ peripheral blood cells was significantly altered. between pancreas cancer patients and healthy volunteers
| Networks | Total | P-value | False discovery rate | In data | Network objects from active data |
|---|---|---|---|---|---|
| Cell cycle_G2–M | 206 | 6.19E-08 | 9.48E-06 | 26 | Histone H1.5, INCENP, BUB1, lamin B, UBE2C, cyclin A2, CAP-G, ETS2, GADD45 α, CAP-C, Ceb1, cyclin A, CAP-G/G2, Chk1, PLK1, KNSL1, HDAC4, MAPKAPK2, cyclin B, cyclin B2, securin, lamin B1, histone H1, GADD45 β, 14-3-3, 14-3-3 eta |
| Cell cycle_S phase | 149 | 1.87E-07 | 1.43E-05 | 21 | Histone H1.5, BUB1, Cdt1, AHR, PCNA, cyclin A2, CDC18L (CDC6), CDH1, geminin, GADD45 α, cyclin A, PLK1, E2F1, cyclin B, cyclin B2, TEP1, separase, securin, DOC-1, histone H1, GADD45 β |
| Cell cycle_Mitosis | 179 | 1.07E-06 | 5.45E-05 | 22 | INCENP, BUB1, MCAK, PKR, CAP-C, cyclin A, CAP-G/G2, PLK1, KNSL1, ASPM, PBK, HZwint-1, tubulin α, cyclin B, cyclin B2, separase, survivin, securin, α-centractin, histone H1, AF15q14, 14-3-3 eta |
| Cell cycle_Core | 115 | 1.44E-06 | 5.52E-05 | 17 | INCENP, BUB1, Cdt1, CDC18L (CDC6), CAP-G, CDH1, CAP-C, cyclin A, PLK1, E2F1, p19, cyclin B, E2F2, cyclin B2, separase, survivin, securin |
| Apoptosis_Apoptotic nucleus | 159 | 3.27E-05 | 0.000999 | 18 | histone H1.5, AHR, lamin B, PKR, Bcl-6, HMG2, GADD45 α, Chk1, E2F1, tBid, ELMO2, tubulin α, separase, lamin B1, histone H1, Bid, GADD45 β, clusterin |
| Cell cycle_G1–S | 163 | 0.000152 | 0.003865 | 17 | BCAT1, BTG3, PCNA, cyclin A2, CDH1, ETS2, GADD45 α, TYSY, Ceb1, cyclin A, Chk1, PLK1, E2F1, p19, GADD45 β, 14-3-3, 14-3-3 eta |
| Cytoskeleton_Spindle microtubules | 109 | 0.000245 | 0.004556 | 13 | INCENP, BUB1, MCAK, UBE2C, sororin, PLK1, KNSL1, HZwint-1, tubulin α, cyclin B, cyclin B2, separase, securin |
| DNA damage_Checkpoint | 124 | 0.000254 | 0.004556 | 14 | PCNA, cyclin A2, heme oxygenase 1, GADD45 α, cyclin A, Chk1, E2F1, cyclin B, cyclin B2, separase, securin, GADD45 β, 14-3-3, 14-3-3 eta |
| Inflammation_Interferon signaling | 110 | 0.000268 | 0.004556 | 13 | PKR, IFNGR1, IFI6, MxA, IFN-α, IL-18R1, IFI27, TIMP1, FasR(CD95), PML, IFI44, ISG15, SERPINB9 |
| Apoptosis_Apoptotic mitochondria | 77 | 0.002527 | 0.038666 | 9 | NIP2, RIPK2, PUMA, Bax, tBid, Bid, endophilin B1, 14-3-3, 14-3-3 eta |
Table 5.
Significant genes with upregulated expression in CD4+ peripheral blood cells of patients with pancreatic ductal adenocarcinoma
| P-value | Fold-change (PK/healthy) | Symbol | Description | Accession no. | Defined gene list |
|---|---|---|---|---|---|
| 3.00E-07 | 1.612903226 | LPP | LIM domain containing preferred translocation partner in lipoma | NM_001167671 | |
| 3.00E-07 | 1.754385965 | PTTG1 | Pituitary tumor-transforming 1 | NM_004219 | Cell cycle, oocyte meiosis |
| 4.00E-07 | 2.040816327 | PRDM1 | PR domain containing 1, with ZNF domain | NM_001198 | |
| 4.00E-07 | 1.666666667 | GMNN | Geminin, DNA replication inhibitor | NM_001251989 | |
| 5.00E-07 | 1.754385965 | EIF2AK2 | Eukaryotic translation initiation factor 2-alpha kinase 2 | NM_001135651 | Bone remodelling, double-stranded RNA-induced gene expression, inactivation of Gsk3 by AKT causes accumulation of β-catenin in alveolar macrophages, regulation of eIF2, Toll-Like receptor pathway, hepatitis C, protein processing in endoplasmic reticulum |
| 2.20E-06 | 1.5625 | PAK2 | p21 protein (Cdc42/Rac)-activated kinase 2 | NM_002577 | Agrin in postsynaptic differentiation, FAS signaling pathway (CD95), Fc epsilon receptor I signaling in mast cells, HIV-I Nef: negative effector of Fas and TNF, MAPKinase signaling pathway, TNFR1 signaling pathway, axon guidance, ErbB signaling pathway, focal adhesion, MAPK signaling pathway, regulation of actin cytoskeleton, renal cell carcinoma, T cell receptor signaling pathway |
| 2.50E-06 | 1.851851852 | EPSTI1 | Epithelial stromal interaction 1 (breast) | NM_001002264 | |
| 2.70E-06 | 2 | CASC5 | Cancer susceptibility candidate 5 | NM_144508 | |
| 2.90E-06 | 1.694915254 | SLA | Src-like-adaptor | NM_001045556 | |
| 3.00E-06 | 1.694915254 | SAR1A | SAR1 homolog A (S. cerevisiae) | NM_001142648 | Protein processing in endoplasmic reticulum |
| 3.00E-06 | 1.538461538 | PML | Promyelocytic leukemia | NM_002675 | Regulation of transcriptional activity by PML, acute myeloid leukemia, endocytosis, pathways in cancer, ubiquitin-mediated proteolysis |
| 3.20E-06 | 1.639344262 | MT1E | Metallothionein 1E | NM_175617 | |
| 3.90E-06 | 1.492537313 | LOC442157 | Heterogeneous nuclear ribonucleoprotein L pseudogene | ||
| 4.30E-06 | 1.666666667 | BAX | BCL2-associated X protein | NM_004324 | Apoptotic signaling in response to DNA damage, ceramide signaling pathway, hypoxia and p53 in the cardiovascular system, p53 signaling pathway, regulation of BAD phosphorylation, role of mitochondria in apoptotic signaling, amyotrophic lateral sclerosis, apoptosis, colorectal cancer, Huntington's disease, neurotrophin signaling pathway, p53 signaling pathway, pathways in cancer, Prion diseases, protein processing in endoplasmic reticulum |
| 4.70E-06 | 1.5625 | PTPRC | Protein tyrosine phosphatase, receptor type, C | NM_001267798 | Activation of Csk by cAMP-dependent protein kinase inhibits signaling through the T cell receptor, B lymphocyte cell surface molecules, Lck and Fyn tyrosine kinases in initiation of TCR activation, T cytotoxic cell surface molecules, T helper cell surface molecules, cell adhesion molecules, Fc γR-mediated phagocytosis, primary immunodeficiency, T cell receptor signaling pathway |
| 4.80E-06 | 1.5625 | MUC1 | Mucin 1, cell surface associated | NM_001018016 | |
| 5.40E-06 | 1.5625 | MT1X | Metallothionein 1X | NM_005952 | |
| 5.70E-06 | 1.666666667 | HPGD | Hydroxyprostaglandin dehydrogenase 15-(NAD) | NM_000860 | |
| 6.00E-06 | 2 | CENPN | Centromere protein N | NM_001100624 | |
| 6.00E-06 | 1.538461538 | POMP | Proteasome maturation protein | NM_015932 | Proteasome |
| 6.10E-06 | 1.612903226 | FZR1 | Fizzy/cell division cycle 20 related 1 (Drosophila) | NM_001136197 | Cell cycle, progesterone-mediated oocyte maturation, ubiquitin-mediated proteolysis |
| 6.20E-06 | 1.612903226 | HMGB2 | High mobility group box 2 | NM_001130688 | Apoptotic DNA fragmentation and tissue homeostasis, granzyme A-mediated apoptosis pathway |
| 6.70E-06 | 1.666666667 | BATF | Basic leucine zipper transcription factor, ATF-like | NM_006399 | |
| 7.50E-06 | 2.127659574 | CPT1A | Carnitine palmitoyl-transferase 1A (liver) | NM_001031847 | Mitochondrial carnitine palmitoyltransferase system, reversal of insulin resistance by leptin, adipocytokine signaling pathway, fatty acid metabolism, PPAR signaling pathway |
| 7.60E-06 | 2.083333333 | UBE2C | Ubiquitin-conjugating enzyme E2C | NM_007019 | Ubiquitin-mediated proteolysis |
| 8.40E-06 | 2.272727273 | TK1 | Thymidine kinase 1, soluble | NM_003258 | Drug metabolism – other enzymes, metabolic pathways, pyrimidine metabolism |
| 8.70E-06 | 1.818181818 | HDGFRP3 | Hepatoma-derived growth factor, related protein 3 | NM_016073 | |
| 8.70E-06 | 1.538461538 | MT1H | Metallothionein 1H | NM_005951 | |
| 9.20E-06 | 3.846153846 | TTTY15 | Testis-specific transcript, Y-linked 15 (non-protein coding) | NR_001545 | |
| 1.02E-05 | 1.538461538 | DNAJC3 | Dnaj (Hsp40) homolog, subfamily C, member 3 | NM_006260 | Double-stranded RNA-induced gene expression, protein processing in endoplasmic reticulum |
| 1.05E-05 | 1.587301587 | MT1L | Metallothionein 1L (gene/pseudogene) | NR_001447 | |
| 1.08E-05 | 1.5625 | ACTR2 | ARP2 actin-related protein 2 homolog (yeast) | NM_001005386 | |
| 1.09E-05 | 1.851851852 | HERC5 | HECT and RLD domain containing E3 ubiquitin protein ligase 5 | NM_016323 | |
| 1.11E-05 | 2.325581395 | KIAA0101 | KIAA0101 | NM_001029989 | |
| 1.19E-05 | 26.31578947 | DDX3Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | NM_001122665 | RIG-I-like receptor signaling pathway |
| 1.20E-05 | 1.785714286 | CDCA8 | Cell division cycle associated 8 | NM_001256875 | |
| 1.25E-05 | 1.612903226 | DDB2 | Damage-specific DNA binding protein 2, 48kda | NM_000107 | Nucleotide excision repair, p53 signaling pathway, ubiquitin-mediated proteolysis |
| 1.29E-05 | 1.754385965 | ALCAM | Activated leukocyte cell adhesion molecule | NM_001243280 | Cell adhesion molecules |
| 1.31E-05 | 1.515151515 | RNF11 | Ring finger protein 11 | NM_014372 | |
| 1.33E-05 | 1.515151515 | CCNK | Cyclin K | NM_001099402 | |
| 1.33E-05 | 1.470588235 | REEP3 | Receptor accessory protein 3 | NM_001001330 | |
| 1.35E-05 | 2.127659574 | MT1M | Metallothionein 1M | NM_176870 | |
| 1.54E-05 | 1.666666667 | FAS | Fas (TNF receptor superfamily, member 6) | NM_000043 | Antigen-dependent B cell activation, bystander B cell activation, CTL-mediated immune response against target cells, FAS signaling pathway (CD95), HIV-induced T cell apoptosis, HIV-I Nef: negative effector of Fas and TNF, IL-2 receptor β chain in T cell activation, keratinocyte differentiation, regulation of transcriptional activity by PML, stress induction of HSP regulation, African trypanosomiasis, allograft rejection, Alzheimer's disease, apoptosis, autoimmune thyroid disease, Chagas disease (American trypanosomiasis), cytokine–cytokine receptor interaction, graft-versus-host disease, MAPK signaling pathway, natural killer cell-mediated cytotoxicity, p53 signaling pathway, pathways in cancer, type I diabetes mellitus |
| 1.55E-05 | 1.724137931 | HERPUD1 | Homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | NM_001010989 | Protein processing in endoplasmic reticulum |
| 1.58E-05 | 1.666666667 | LPGAT1 | Lysophosphatidylglycerol acyltransferase 1 | NM_014873 | Glycerophospholipid metabolism |
| 1.67E-05 | 1.612903226 | FKBP5 | FK506 binding protein 5 | NM_001145775 | |
| 1.71E-05 | 1.886792453 | PDCD1 | Programmed cell death 1 | NM_005018 | Cell adhesion molecules, T cell receptor signaling pathway |
| 1.76E-05 | 37.03703704 | RPS4Y2 | Ribosomal protein S4, Y-linked 2 | NM_001039567 | |
| 1.79E-05 | 2.325581395 | BIRC5 | Baculoviral IAP repeat containing 5 | NM_001012270 | B cell survival pathway, colorectal cancer, pathways in cancer |
| 1.79E-05 | 2.040816327 | CDCA2 | Cell division cycle associated 2 | NM_152562 |
PK, pancreatic cancer patients.
Increased frequency of CD4+PD-1+ subpopulation in PBMCs of PDAC patients
CD4+PD-1+ cells infiltrated local PDAC tissues, and PD-1 gene expression was significantly up-regulated in CD4+ T cells of peripheral blood of PDAC patients, we further examined the frequency of PD-1-expressing cells in peripheral blood. Flow cytometry analysis showed that the frequency of CD4+PD-1+ cells, but not CD8+PD-1+ cells, was increased in the PBMCs of PDAC patients (Fig.5a,b); this is consistent with the elevated PD-1 gene expression of CD4+ cells in PDAC patients shown using RTD-PCR (Fig. S2a, Data S2). The frequency of regulatory T cells, phenotypically defined as a CD4+CD25+CD127low/− population,12 was greater in the peripheral blood of PDAC patients (Fig.5c); however, FoxP3 gene expression was not significantly elevated in CD4+ T cells of PDAC patients (Fig. S2b, Doc. S2). The frequencies of CD4+PD-1+ T cells and CD4+CD25+CD127low/− cells were not correlated (Fig.5d). Neither the frequency of CD4+PD-1+ T cells nor CD4+CD25+CD127low/− T cells was associated with cancer progression stages (Fig.5e,f). However, patients whose responsiveness to chemotherapy were progressive disease tended to show a relatively high frequency of CD4+PD-1+ cells in the peripheral blood compared to patients with a diagnosed therapeutic effect of stable disease or partial responsiveness with chemotherapy, whereas this was not observed for CD4+CD25+CD127low/− T cells (Fig.5g,h). We divided PDAC patients into two groups: one with ≥10% CD4+PD-1+ T cells, and the other with <10% of such cells in peripheral blood. The overall survival of the former group was relatively shorter than that of the latter group. However, the P-value (P = 0.111) indicated that statistical significance was not attained. These data suggest that the subpopulation of peripheral CD4+ T cells from PDAC patients contained the important subfraction of activated and exhausted CD4+PD-1+ T cells, which may influence the therapeutic effect of chemotherapy.
Figure 5.
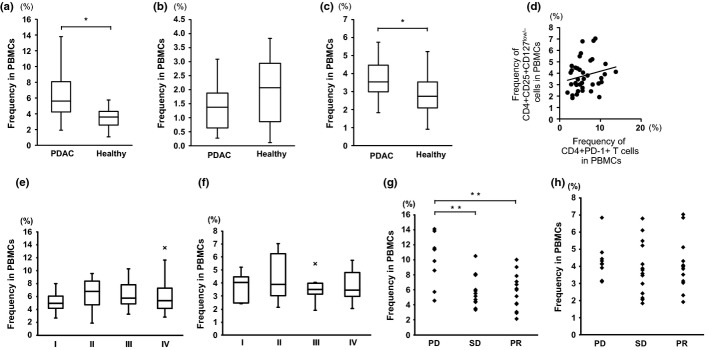
Frequency of CD4+PD-1+ cells and CD4+CD25+CD127low/− cells in PBMCs of pancreatic ductal adenocarcinoma (PDAC) patients (n = 50) and healthy volunteers (n = 27). The frequencies of CD4+PD-1+ cells (a), CD8+PD-1+ cells (b), and CD4+CD25+CD127low/− cells (c) were assessed by flow cytometry. (d) Scattergram of the frequencies of CD4+PD-1+ cells and CD4+CD25+CD127low/− cells in PDAC patients. (e, f) The frequency of CD4+PD-1+ cells (e) and CD4+CD25+CD127low/− cells (f) in the PBMCs of PDAC patients in the context of each clinical stage. (g, h) The chemotherapy responsiveness and frequency of CD4+PD-1+ cells (g) and CD4+CD25+CD127low/− cells (h). PD, progressive disease; PR, partial responsiveness; SD, stable disease. *P < 0.05; **P < 0.01.
Discussion
In the current study, we examined systemic inflammatory conditions of PDAC by analyzing the PDAC tissues, sera, and peripheral blood cells. We observed that the PDAC tissues were remarkably infiltrated by monocytes/macrophages and CD4+ T cells, especially M2-phenotype macrophages and PD-1+ cells. Serum concentrations of IL-6, IL-7, IL-15, MCP-1, and IP-10 were elevated in PDAC patients, suggesting humoral inflammatory mediators related to the macrophages and CD4+ T cells were present in the blood of PDAC patients. In addition, we observed distinctively different gene expression profiles of CD14+ monocytes and CD4+ T cells among subfractions of peripheral blood cells between PDAC patients and healthy volunteers. Cell cycle processes as well as inflammation-associated biological processes were commonly related to upregulated genes in the CD14+ monocytes and CD4+ T cells of PDAC patients. More intriguingly, PD-1, an important molecule that is upregulated in activated T cells and attenuates T cell receptor signaling, was upregulated in the CD4+ T cells of PDAC patients. The frequency of CD4+PD-1+ T cells was increased and correlated with a resistance to chemotherapy.
Pathologically, PDAC tissues were substantially infiltrated by monocytes/macrophages and CD4+T cells. Monocytes/macrophages are generally considered to be involved in non-specific innate immunity; they digest antigens in the presence of pro-inflammatory cytokines.13 In the context of cancer immunity, two important subsets of monocytes/macrophages have been recognized, M1 and M2 macrophages.14 M1 macrophages play a principal role in anticancer immunity, whereas M2 macrophages sustain or promote cancer growth by inhibiting anticancer immunity. We observed a substantial number of CD163+ cells among CD33+ and neutrophil elastase-negative macrophages, suggesting an M2 macrophage phenotype15 in PDAC tissues, which presumably contributes to sustained cancer growth.
Among infiltrated CD4+ T cells in PDAC tissues, the expression of FoxP3 and PD-1 was frequently observed. FoxP3 is a transcriptional factor that is suggestive of activated T cells as well as regulatory T cells.16 PD-1 is a receptor, whose expression is induced in activated T cells, attenuating the signal transduced from a T cell receptor encountered by a cognate antigen.17–19 These inflammatory features of PDAC tissues highlight monocytes/macrophages and CD4+ T cells, especially M2 macrophages and exhausted CD4+PD-1+ T cells, which may contribute to cancer progression.
Previously, we observed that the gene expression profile of total blood cells from patients with digestive cancers was distinct from that of healthy volunteers.11 This is consistent with the current study, in which the gene expression profile of CD14+ monocytes and CD4+ T cells in PBMCs as well as entire blood populations was distinct between PDAC patients and healthy volunteers. Affected genes, most of which were upregulated, were related to cell cycle and inflammation processes in CD14+ monocytes and CD4+ T cells; this suggests that these inflammatory cells in the peripheral blood were in an activated state. Significantly affected genes in CD14+ monocytes were also related to blood coagulation, cell adhesion, and the developmental regulation of angiogenesis, all of which are important biological processes of activated macrophages. However, it remains to be elucidated whether the immunological consequence of activation of monocytes/macrophages in peripheral blood cells anti-cancer or cancer promoting effect inhibits or promotes cancer development.
Affected genes in CD4+ T cells were also related to DNA damage and apoptosis. CD4+ T cells undergo activation-induced cell death through Fas-mediated signaling,20 and Fas (CD95) was among the most upregulated genes. Taken together, gene expression analysis disclosed that myeloid-lineage CD14+ monocytes/macrophages and CD4+ T cells are important affected fractions of immune-mediating cells in the peripheral blood cells of PDAC patients, with the implication of a cancer-associated activated inflammatory condition.
Corresponding to the upregulated expression of the PD-1 gene in the peripheral CD4+T cells of PDAC patients, the frequency of CD4+PD-1+ cells in the peripheral blood of PDAC patients was also increased. Intriguingly, the relatively poor success of chemotherapy correlated with an increased level of CD4+PD-1+ T cells. The overall survival of PDAC patients with ≥10% CD4+PD-1+ T cells was somewhat shorter than that of those with <10% such cells, although statistical significance was not attained. Any underlying role for CD4+PD-1+ T cells in terms of responsiveness to chemotherapy remains to be explored; we observed neither a supporting effect on cancer cell proliferation nor a suppressive effect on IFN-γ-secreting activated cytotoxic T cells in vitro (data not shown). PD-1 attenuates T cell receptor signaling, therefore, CD4+ T cells expressing PD-1 are considered to be exhausted if anticancer inflammation is not induced. An increased level of CD4+PD-1+T cells may reflect the fact that the anticancer inflammation induced during chemotherapy is inadequate. Clinical trials featuring blocking of PD-1-expressing cells (using an anti-PD-1 antibody) to enhance anticancer immune reactions are currently underway; this may be a valuable therapy for lung cancer and melanoma, overcoming immune resistance.21 Although further clinical studies are needed to explore the role played by CD4+PD-1+ T cells in chemotherapy and overall survival, our current finding that CD4+ PD-1+ T cells infiltrate PDAC tissues and increase in proportion among peripheral blood cells suggests that immunotherapy targeting the exhausted PD-1+ population may be a useful novel immunotherapeutic approach toward PDAC.
Concentrations of the macrophage- and T cell-related cytokines and chemokines IL-6, IL-7, IL-15, MCP-1, and IP-10 were significantly elevated in the sera of PDAC patients. Interleukin-15, MCP-1, and IP-10 are produced by monocytes/macrophages, considerably inducing or activating an innate immune reaction.22–24 The biological function of the cytokine IL-7 is maintenance of the naïve T cells as well as T cell proliferation,25 whereas IL-6 is involved in macrophage polarization and in the initiation and proliferation of PDAC.9 Although elevation of these cytokines/chemokines is considered to reflect the inflammatory condition of PDAC, the clinical impact of the elevated serum cytokines and chemokines in PDAC should be further studied in the context of anticancer or cancer-promoting humoral immune reactions.
Although the current study highlighted the importance of focusing on monocytes/macrophages and CD4+ T cells in local PDAC tissues as well as peripheral blood, these immune-mediating populations are heterogeneous and extremely complex. The increased frequency of CD4+PD-1+ T cells, which are presumably an important population affecting host immunity against cancer, suggest that significant subfractions in monocytes/macrophages and the CD4+ T cell population may exist. Additional detailed studies are needed to identify which subfractions are significantly associated with the clinical prognosis of PDAC patients and to verify the clinical impact of the subfractions in well-designed clinical trials.
In conclusion, the current study showed that PDAC is associated with a systemic inflammatory condition, highlighting the presence of activated monocytes/macrophages and CD4+ T cells, which are presumed to be exhausted both in local cancer tissues and peripheral blood. Substantial infiltration of cancer-promoting immune cells, including M2 macrophages, in local cancer tissues was concomitant with the increased expression of PD-1 in T cells as well as the increased frequency of CD4+PD-1+ T cells in the peripheral blood of PDAC patients. This may contribute, in part, to persistent chronic inflammation as a consequence of failure to eliminate cancer despite the host immune response. However, the immune system includes both cancer-promoting immune-mediating cells and anticancer inflammatory cells. Further studies focusing on monocyte/macrophage and CD4+ T cell subfractions in PDAC patients may reveal further details regarding the PDAC immune condition, and such information may be helpful in the development of novel diagnostic markers for detecting PDAC. It may also provide meaningful insights into the development of novel immunological therapeutic approaches for modulating the inflammatory condition in PDAC toward anticancer inflammation.
Acknowledgments
We sincerely thank Ms. Mami Iwasaki for her excellent technical assistance. This study was supported, in part, by a subsidy from the Japanese Ministry of Health, Labor, and Welfare.
Disclosure Statement
The authors have no conflict of interest.
Supporting Information
Data S1. Supporting materials and methods.
Fig. S1. Cytokine and chemokine gene expression in peripheral CD14+ monocytes/macrophages in CD4+ T cells in patients with pancreatic ductal adenocarcinoma.
Fig. S2. Analysis of PD-1 and FoxP3 gene expression in CD4+ T cells in patients with pancreatic ductal adenocarcinoma.
Table S1. Characteristics of study subjects for serum concentration of cytokines/chemokines and flow cytometry analysis of peripheral blood cells.
Table S2. Characteristics of study subjects for gene expression profiles of peripheral blood cell subfractions.
Table S3. Characteristics of study subjects for gene expression profile analysis of CD14+ monocytes and CD4+ T cells in peripheral blood cells.
Table S4. real time detection-PCR (RTD-PCR) analysis of genes whose expression was upregulated by microarray analysis in peripheral blood cells of patients with pancreatic ductal adenocarcinoma.
References
- Matsuda T, Ajiki W, Marugame T, et al. Population-based survival of cancer patients diagnosed between 1993 and 1999 in Japan: a chronological and international comparative study. Jpn J Clin Oncol. 2011;41:40–51. doi: 10.1093/jjco/hyq167. [DOI] [PubMed] [Google Scholar]
- Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–20. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson AS, Tran Cao HS, Tempero MA, Lowy AM. Therapeutic advances in pancreatic cancer. Gastroenterology. 2013;144:1316–26. doi: 10.1053/j.gastro.2013.01.078. [DOI] [PubMed] [Google Scholar]
- Reni M, Cordio S, Milandri C, et al. Gemcitabine versus cisplatin, epirubicin, fluorouracil, and gemcitabine in advanced pancreatic cancer: a randomised controlled multicentre phase III trial. Lancet Oncol. 2005;6:369–76. doi: 10.1016/S1470-2045(05)70175-3. [DOI] [PubMed] [Google Scholar]
- Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–8. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solito S, Marigo I, Pinton L, Damuzzo V, Mandruzzato S, Bronte V. Myeloid-derived suppressor cell heterogeneity in human cancers. Ann N Y Acad Sci. 2014;1319:47–65. doi: 10.1111/nyas.12469. [DOI] [PubMed] [Google Scholar]
- Facciabene A, Motz GT, Coukos G. T-regulatory cells: key players in tumor immune escape and angiogenesis. Cancer Res. 2012;72:2162–71. doi: 10.1158/0008-5472.CAN-11-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesina M, Wörmann SM, Neuhöfer P, Song L, Algül H. Interleukin-6 in inflammatory and malignant diseases of the pancreas. Semin Immunol. 2014;26:80–7. doi: 10.1016/j.smim.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Honda M, Fujinaga H, et al. Common transcriptional signature of tumor-infiltrating mononuclear inflammatory cells and peripheral blood mononuclear cells in hepatocellular carcinoma patients. Cancer Res. 2008;68:10267–79. doi: 10.1158/0008-5472.CAN-08-0911. [DOI] [PubMed] [Google Scholar]
- Honda M, Sakai Y, Yamashita T, et al. Differential gene expression profiling in blood from patients with digestive system cancers. Biochem Biophys Res Commun. 2010;400:7–15. doi: 10.1016/j.bbrc.2010.07.123. [DOI] [PubMed] [Google Scholar]
- Liu W, Putnam AL, Xu-Yu Z, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203:1701–11. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–96. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–6. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Fabriek BO, Dijkstra CD, van den Berg TK. The macrophage scavenger receptor CD163. Immunobiology. 2005;210:153–60. doi: 10.1016/j.imbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37:129–38. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11:3887–95. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T, Chikuma S, Iwai Y, Fagarasan S, Honjo T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat Immunol. 2013;14:1212–8. doi: 10.1038/ni.2762. [DOI] [PubMed] [Google Scholar]
- Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–9. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- Green DR, Droin N, Pinkoski M. Activation-induced cell death in T cells. Immunol Rev. 2003;193:70–81. doi: 10.1034/j.1600-065x.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagaya Y, Bamford RN, DeFilippis AP, Waldmann TA. IL-15: a pleiotropic cytokine with diverse receptor/signaling pathways whose expression is controlled at multiple levels. Immunity. 1996;4:329–36. doi: 10.1016/s1074-7613(00)80246-0. [DOI] [PubMed] [Google Scholar]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 2002;168:3195–204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- Akashi K, Kondo M, Weissman IL. Role of interleukin-7 in T-cell development from hematopoietic stem cells. Immunol Rev. 1998;165:13–28. doi: 10.1111/j.1600-065x.1998.tb01226.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting materials and methods.
Fig. S1. Cytokine and chemokine gene expression in peripheral CD14+ monocytes/macrophages in CD4+ T cells in patients with pancreatic ductal adenocarcinoma.
Fig. S2. Analysis of PD-1 and FoxP3 gene expression in CD4+ T cells in patients with pancreatic ductal adenocarcinoma.
Table S1. Characteristics of study subjects for serum concentration of cytokines/chemokines and flow cytometry analysis of peripheral blood cells.
Table S2. Characteristics of study subjects for gene expression profiles of peripheral blood cell subfractions.
Table S3. Characteristics of study subjects for gene expression profile analysis of CD14+ monocytes and CD4+ T cells in peripheral blood cells.
Table S4. real time detection-PCR (RTD-PCR) analysis of genes whose expression was upregulated by microarray analysis in peripheral blood cells of patients with pancreatic ductal adenocarcinoma.