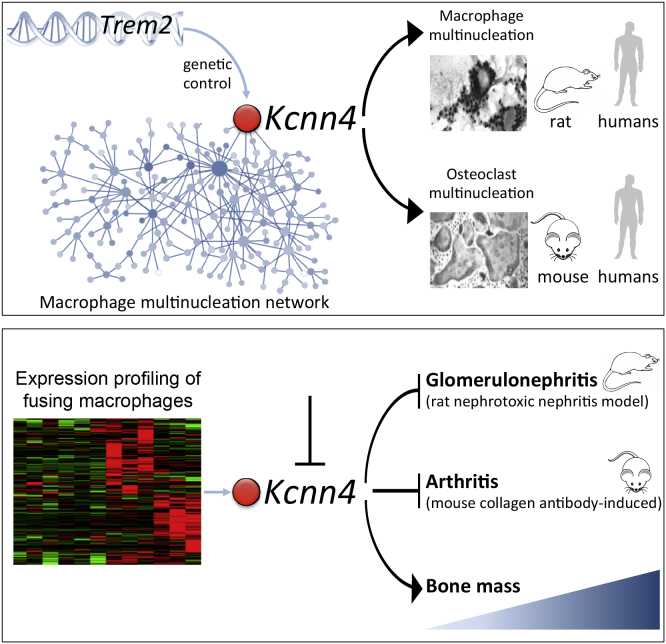
Summary
Macrophages can fuse to form osteoclasts in bone or multinucleate giant cells (MGCs) as part of the immune response. We use a systems genetics approach in rat macrophages to unravel their genetic determinants of multinucleation and investigate their role in both bone homeostasis and inflammatory disease. We identify a trans-regulated gene network associated with macrophage multinucleation and Kcnn4 as being the most significantly trans-regulated gene in the network and induced at the onset of fusion. Kcnn4 is required for osteoclast and MGC formation in rodents and humans. Genetic deletion of Kcnn4 reduces macrophage multinucleation through modulation of Ca2+ signaling, increases bone mass, and improves clinical outcome in arthritis. Pharmacological blockade of Kcnn4 reduces experimental glomerulonephritis. Our data implicate Kcnn4 in macrophage multinucleation, identifying it as a potential therapeutic target for inhibition of bone resorption and chronic inflammation.
Graphical Abstract
Highlights
-
•
We identified a gene network that regulates macrophage multinucleation and includes Kcnn4
-
•
Kcnn4 can be targeted in two inflammatory conditions with macrophage multinucleation
-
•
Kcnn4 regulates bone mass under physiological conditions
-
•
Kcnn4 is a drug target for which inhibitors reached phase III of clinical trials
Kang et al. establish the genetic determinants of macrophage multinucleation, which is involved in the formation of osteoclasts in bone or multinucleated giant cells during inflammatory reaction. They identify genetic variation on rat chromosome 9 linked to the expression of Kcnn4 as part of a macrophage multinucleation gene network. Kcnn4 is a calcium-activated potassium channel, and the authors show that it regulates macrophage multinucleation and bone mass. Blockade of Kcnn4 results in reduced susceptibility to glomerulonephritis and arthritis.
Introduction
Macrophages are versatile and plastic cells that have the ability to fuse and give rise to multinucleate cells (Chen and Olson, 2005). Macrophage fusion leads to the formation of osteoclasts in bone and multinucleate giant cells (MGCs) in chronic inflammatory diseases (Brodbeck and Anderson, 2009; Helming and Gordon, 2009; Vignery, 2005). Thus, both osteoclasts and MGCs are derived from the fusion of macrophages. Multinucleation represents an essential step in osteoclast formation and activation because mononucleate osteoclasts are unable to resorb bone efficiently (Helming and Gordon, 2009; Vignery, 2005, 2008) and the genes that are critical for osteoclast activity are also fundamental to MGC function (Bühling et al., 2001; da Costa et al., 2005; Yagi et al., 2005; Zhu et al., 2007). Despite the well-characterized role of osteoclasts in bone homeostasis, macrophage activation associated with MGC formation in inflammatory diseases remains poorly understood.
Genetic studies in mice and humans suggest that germline sequence variation can regulate macrophage multinucleation and that this can influence disease pathogenesis. In a murine model of chronic tuberculosis infection, genetic loci were associated with production of inflammatory mediators by macrophages (Kramnik et al., 2000; Pichugin et al., 2009), suggesting the presence of a heritable genetic component affecting MGC function, which, in turn, mediates susceptibility to granulomatous diseases. In humans, deletions or loss-of-function mutations in either the TYROBP (DAP12) or TREM2 genes are causally associated with Nasu-Hakola disease (Paloneva et al., 2000, 2002), where defective multinucleation in osteoclasts results in impaired bone resorption (Humphrey et al., 2004). In addition, human genome-wide association studies of MGC-associated diseases such as tuberculosis (Thye et al., 2012) and granulomatosis with polyangiitis (Lyons et al., 2012) identified common sequence variations associated with these disorders. In rheumatoid arthritis (RA), osteoclast differentiation and activation lead to bone erosion associated with prolonged inflammation (McInnes and Schett, 2011). However, to date, genetic studies failed to identify genes and pathways involved in osteoclast activation resulting from multinucleation. Hence, the key determinants and molecular pathways of multinucleation in macrophages and the resulting pathophysiological effects remain largely unexplored.
Results
Identification of a Multinucleation Gene Network in Macrophages
In order to investigate the genetic components of MGC formation, we took advantage of strain-specific phenotypic differences in spontaneous formation of MGCs in the rat, which have not been described previously. We observed that when bone-marrow-derived macrophages (BMDMs) from Wistar Kyoto (WKY) rats and Lewis (LEW) rats are cultured in vitro, WKY macrophages spontaneously form MGCs in contrast with what is observed in LEW (Figures 1A and S1). These strains have been widely studied for their differential susceptibility to experimental glomerulonephritis through macrophage activation (Aitman et al., 2006; Behmoaras et al., 2008, 2010; Page et al., 2012), and WKY rats also show MGCs in their glomeruli following the induction of nephrotoxic nephritis (NTN) (Figure S1). These marked phenotypic differences suggested genetic determinants of the process underlying MGC formation in the rat, which we set out to investigate by profiling genome-wide expression levels in macrophages of 200 backcross rats derived from WKY and LEW.
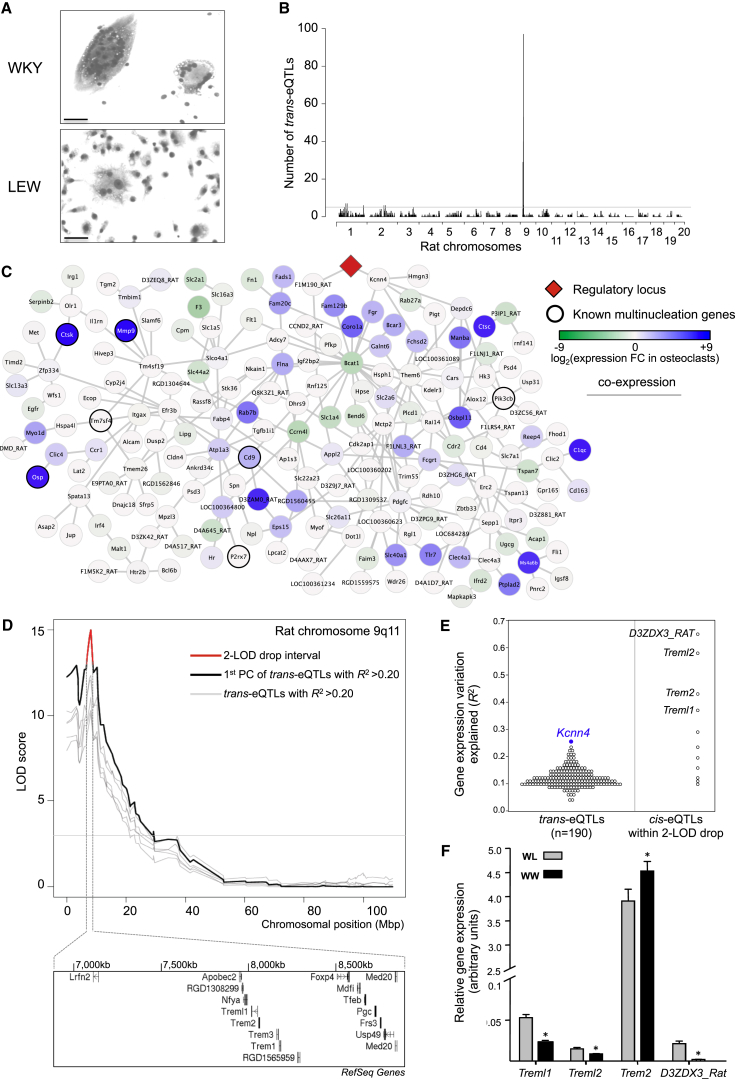
Figure 1.
Identification of Kcnn4 within a Genetically Regulated Macrophage Multinucleation Network
(A) Genetic determinants of macrophage multinucleation were explored in WKY and LEW bone-marrow-derived macrophages (BMDMs). WKY macrophages fuse spontaneously to form multinucleate giant cells (MGCs) in vitro and show a marked phenotypic difference when compared to LEW macrophages, which form very few MGCs at day 6 of cell differentiation (original bars, 50 μm).
(B) eQTL analysis of the backcross (BC) BMDMs identifies a unique master regulatory locus on rat chromosome 9q11. Genome-wide distribution of trans-eQTLs shows a single locus regulating the expression of 190 transcripts in trans, suggesting master regulation of the trans-eQTL cluster. The detailed eQTL hot spots with the corresponding SNP positions are reported in Table 1.
(C) Gene coexpression network of the 190 trans-eQTLs is enriched for osteoclast-expressed genes where the master regulatory locus is highlighted in red. Each gene in the network is represented as a circle (node), and the fold change in expression refers to overexpression (blue) or underexpression (green) in osteoclasts as compared to average expression in other cell types (Supplemental Experimental Procedures). Known multinucleation genes (Mmp9, Ctsk, P2rx7, etc.) are indicated with thick circles. The edges represent coexpression between the two transcripts as identified by ARACNE (Margolin et al., 2006).
(D) Genetic linkage showing the principal component of all the trans-cluster transcripts (LOD > 10) together with trans eQTLs with variation in gene expression explained by the SNP (R2) >0.2. The 2-LOD drop interval (in red) and the underlying positional candidates within 2 Mb are also shown. The annotated gene names are according to RefSeq.
(E) The cis-eQTLs within the 2-LOD drop interval (in red) are shown together with the trans cluster. Treml1, Trem2, D3ZDX3_Rat, Treml2 are positional candidates (R2 > 0.25) and among the trans-eQTLs, Kcnn4 (in blue) is the most significant trans-eQTL.
(F) Quantitative real-time PCR analysis on the positional candidates in BC BMDMs according to their genotype (WKY homozygous, WW; WKY/LEW heterozygous, WL). Note that the expression levels of Trem2 are increased at least 90-fold when compared to other positional candidates.
Error bars indicate SEM, ∗p < 0.01. See also Figures S1 and S2.
We used mRNA expression level as a quantitative trait to carry out genome-wide linkage analysis using a panel of single nucleotide polymorphisms (SNPs) throughout the rat genome. Using multivariate Bayesian regression approaches (Bottolo et al., 2011b), we identified a set of 2,357 transcripts showing significant linkages to discrete genetic loci (posterior probability >80%), which are designated as expression quantitative trait loci (eQTLs, Figure S2). The majority of transcripts were regulated by local genetic variation forming cis-eQTLs (67%); however, we also identified many trans-regulated transcripts (trans-eQTLs) across the rat genome (Figure 1B). As previously reported (Hubner et al., 2005; Langley et al., 2013), trans-eQTLs can form clusters that are commonly referred to as trans-eQTLs hot spots (Breitling et al., 2008) (Table 1). Notably, we identified a large eQTL hot spot that mapped to three SNPs spanning a genomic region of 1.2 Mbp located on rat chromosome 9q11, which regulated in trans the expression of 190 transcripts (Table 1; Figure 1B). These 190 transcripts formed a gene coexpression network, where each gene is regulated in trans by the same genetic locus (Figure 1C). Cell-type enrichment analysis using a mouse gene expression atlas showed that the gene network is enriched for osteoclast genes (enrichment p value = 4 × 10−7, Z test for relative overexpression of the network genes in a tissue/cell type, see Supplemental Experimental Procedures), suggesting a role for the network in macrophage multinucleation (Figure 1C). Closer inspection of the network genes revealed two major determinants of osteoclast activity (Mmp9 and cathepsin K, reviewed in Helming and Gordon, 2009) as well as several reported regulators of macrophage multinucleation such as P2rx7 (Lemaire et al., 2006), Tm7sf4 (also known as DC-STAMP [Yagi et al., 2005]), osteopontin (Osp) (Tsai et al., 2005), Pik3cb (also known as PI3K [Peng et al., 2010]), tetraspanin Cd9 (Takeda et al., 2003), and its binding partner Igsf8. We hypothesize that other genes of the large coexpression network might regulate macrophage multinucleation, therefore defining a “Macrophage Multinucleation Network” or MMnet.
Table 1.
eQTL Hot Spots Identified in Backcross BMDMs Derived from WKY and LEW
| SNP | Chr. | Position (Mb) | Transcripts Mapping to Each SNP | Five Most Strongly Regulated Genes |
|---|---|---|---|---|
| Rn34_1071564105 | 1 | 71564105 | 7 | RGD1562091 / Zfp61 / Vasp / F1LU75_RAT / Slc26a11 |
| Rn34_1071580649 | 1 | 71580649 | 6 | Gemin7l1 / TMED5_RAT / LOC691921 / Kcnn4 / Trove2 |
| Rn34_1084462932 | 1 | 84462932 | 7 | F1M372_RAT / D3ZBY1_RAT / Flna / Plek / Fn1 |
| Rn34_1253288627 | 1 | 253288627 | 6 | Blnk / LOC689756 / Oasl / Apoc1 / Egr3 |
| Rn34_2146742392 | 2 | 146742392 | 6 | RGD1359508 / Dtwd2 / Hrh1 / RGD1561662 / Lce1l |
| Rn34_2164452465 | 2 | 164452465 | 6 | Sucnr1 / Tuft1 / Mgrn1 / Dlgap4 / Ptch1 |
| Rn34_9003266206 | 9 | 3266206 | 29 | Phlda1 / Pak6 / Xylt1 / Slc30a1 / RGD1565705 |
| Rn34_9003796216 | 9 | 3796216 | 9 | RGD1566226 / D4AE99_RAT / Slc35e4 / RGD1311946 / Hprt1 |
| Rn34_9004824426 | 9 | 4824426 | 15 | Plk3 / Capn5 / Bag3 / F1LTZ3_RAT / Ccl22 |
| Rn34_9005917596 | 9 | 5917596 | 7 | Tgfb2 / Leprot / RGD1310371 / Creb3l2 / RTN1_RAT |
| Rn34_9006863573a | 9 | 6863573 | 40 | Tspan7 / Rdh10 / F1LRS4_RAT / Plcd1 / D3ZC56_RAT |
| Rn34_9007659609a | 9 | 7659609 | 53 | D3ZC56_RAT / D3ZC56_RAT / Ms4a6b / Myo1d / Bend6 |
| Rn34_9008096016a | 9 | 8096016 | 97 | Kcnn4 / Slco4a1 / Flna / Bcat1 / Atp1a3 |
| Rn34_9010210028 | 9 | 10210028 | 6 | D4ACH5_RAT / St6gal1 / Myh9 / Plek / E9PU51_RAT |
n = 200. eQTL hot spots were defined as significant overrepresentation of trans-eQTLs (i.e., any SNP is associated with more than five transcripts; see Experimental Procedures).
SNPs delineate a genomic region of 1.2 Mb linked to the expression of 190 transcripts, which were located elsewhere in the genome and regulated in trans (i.e., trans-eQTLs).
To prioritize master regulator genes at the rat chromosome 9 locus, we carried out fine mapping analysis and, among others, identified the Triggering receptor expressed in myeloid cells (Trem) gene family cluster within the 2-LOD drop support interval of the peak of linkage (Figure 1D). At this locus, the Trem family genes showed the strongest cis-acting genetic regulation in macrophages (variation in gene expression explained by the regulatory locus; R2 > 0.25, Figures 1E), and, among all genes in the MMnet, Kcnn4 was the most significant trans-eQTL (Figure 1E). Quantitative real-time PCR analysis of the expression of the Trem family genes in backcross macrophages confirmed their cis-regulation and prioritized Trem2 as the most highly expressed gene in rat macrophages (>90-fold more expression compared to all other genes in the Trem cluster, Figure 1F).
Identification of Trem2 as a Master Genetic Regulator of the MMnet
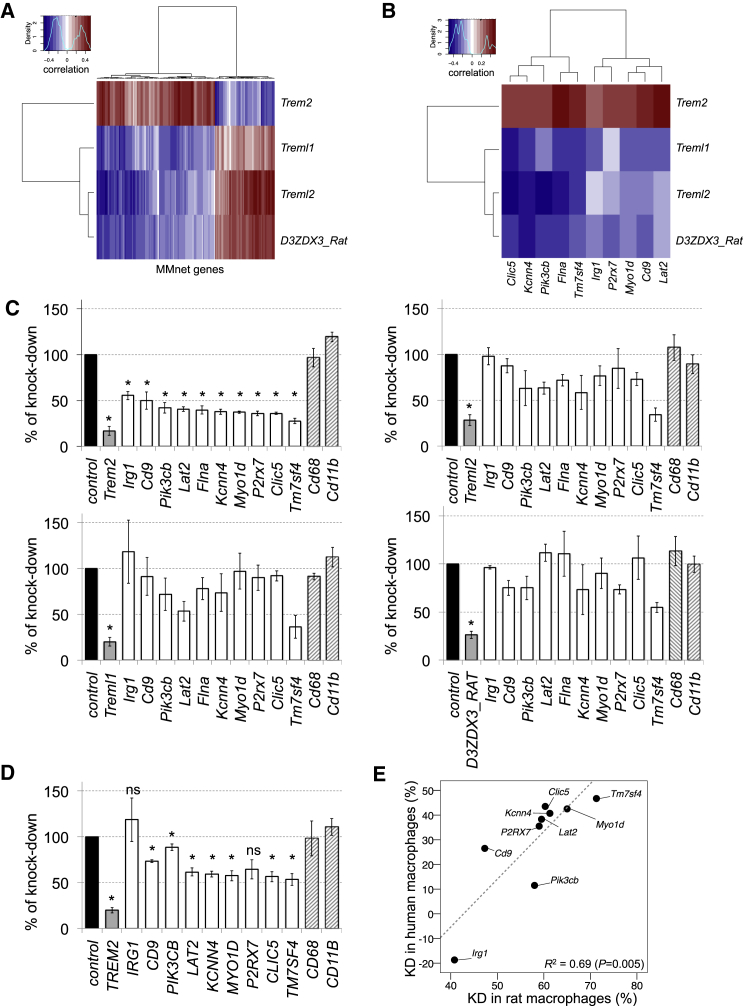
We observed a positive correlation between the expression of Trem2 and 125 (66%) MMnet genes and a negative correlation between Treml1, D3ZDX3_Rat, Treml2, and the network genes (Figure 2A). We selected ten genes from the MMnet, including the most strongly trans-regulated gene (Kcnn4, R2 = 0.25), genes previously reported to play a role in macrophage multinucleation (P2rx7 [Lemaire et al., 2006], Tm7sf4 [Yagi et al., 2005], Pik3cb [Peng et al., 2010], tetraspanin Cd9 [Takeda et al., 2003]) and additional trans-regulated MMnet genes with varying genetic effects (R2 ranging from ∼0.1 to 0.22). These genes recapitulate the correlation pattern observed between the Trem cluster genes and the MMnet (Figure 2B). We carried out RNAi experiments to further prioritize candidates within the Trem gene cluster as potential master regulators of the MMnet, by using small interfering RNA (siRNA) against Treml1, Trem2, D3ZDX3_Rat, and Treml2 genes that were differentially expressed between WKY and LEW macrophages (Figure 1F), followed by quantitative real-time PCR assessment of ten trans-regulated genes from the network. In keeping with the positive correlation between Trem2 and the MMnet genes (Figures 2A and 2B), we found significant downregulation of the trans-regulated genes after Trem2 silencing (Figure 2C). On the contrary, silencing of Treml1, D3ZDX3_Rat, and Treml2 resulted in a weaker and variable downregulation of the network genes (Figure 2C). This downregulation was not in accordance with the expected transcriptional response based on the negative correlation between Treml1, D3ZDX3_Rat, Treml2, and each trans-eQTL gene expression (Figure 2B). These in vitro experiments prioritized Trem2 as a master regulator gene of the MMnet in the rat. We then explored whether silencing of TREM2 in monocyte-derived macrophages had a similar effect on the trans-eQTL network genes in humans (Figure 2D). We showed that there was a significant correlation between the percentage of knockdown in human MDMs and rat BMDMs following Trem2 knockdown (Figure 2E). These data suggest that the Trem2-regulated MMnet genes identified in rat macrophages are similarly regulated in human macrophages.
Figure 2.
Trem2 Is a Master Genetic Regulator of Macrophage Multinucleation Network
(A) Heatmap of the correlations between macrophage mRNA levels of positional candidates (rows) and all trans-regulated genes of MMnet, showing that Trem2 was positively correlated with the majority (66%) of MMnet transcripts. Red indicates positive correlation and blue indicates negative correlations.
(B) Heatmap of the correlations between macrophage mRNA levels of positional candidates (rows) and ten trans-regulated genes representative of the larger MMnet selected for the siRNA knockdown experiments in rat BMDMs. Red indicates positive correlation and blue indicates negative correlations.
(C) siRNA against Trem2, Treml2, Treml1, D3ZDX3_Rat in rat macrophages followed by measurement of 10 transcripts belonging to the chromosome 9q11 trans cluster by quantitative real-time PCR. Cd68 and Cd11b expression levels were assessed as control transcripts that do not belong to MMnet. Given the correlation patterns shown in Figure 2B, a one-sample t test was used to test for directionality of the effect in the transcriptional response. Knockdown of Trem2 led to significant (∼50%, p < 0.001) downregulation of all genes tested, whereas knockdown of Treml2, Treml1, D3ZDX3_Rat did not result in a significant alteration in the expression levels of MMnet genes (p > 0.1 for all MMnet genes tested). At least n = 3 rats were used in each experiments. Error bars indicate SEM, ∗p < 0.001.
(D) siRNA-mediated knockdown of TREM2 in human monocyte derived macrophages (MDMs) and the quantitative real-time PCR analysis of ten transcripts from the MMnet. MDMs are from buffy coats from four healthy donors. Error bars indicate SEM, ∗p < 0.05 compared to control (scrambled).
(E) Knockdown of TREM2 in human MDMs resulted in transcriptional downregulation of MMnet genes, which significantly correlated with downregulation observed in rat BMDMs (R2 = 0.69, p = 0.005).
Taken together, genome-wide eQTL analysis and in vitro experiments uncovered a large gene network regulating macrophage multinucleation, which is under trans-acting genetic control by the Trem2 gene in rats and humans.
Kcnn4 Is Implicated in Macrophage Multinucleation in Rodents and Humans
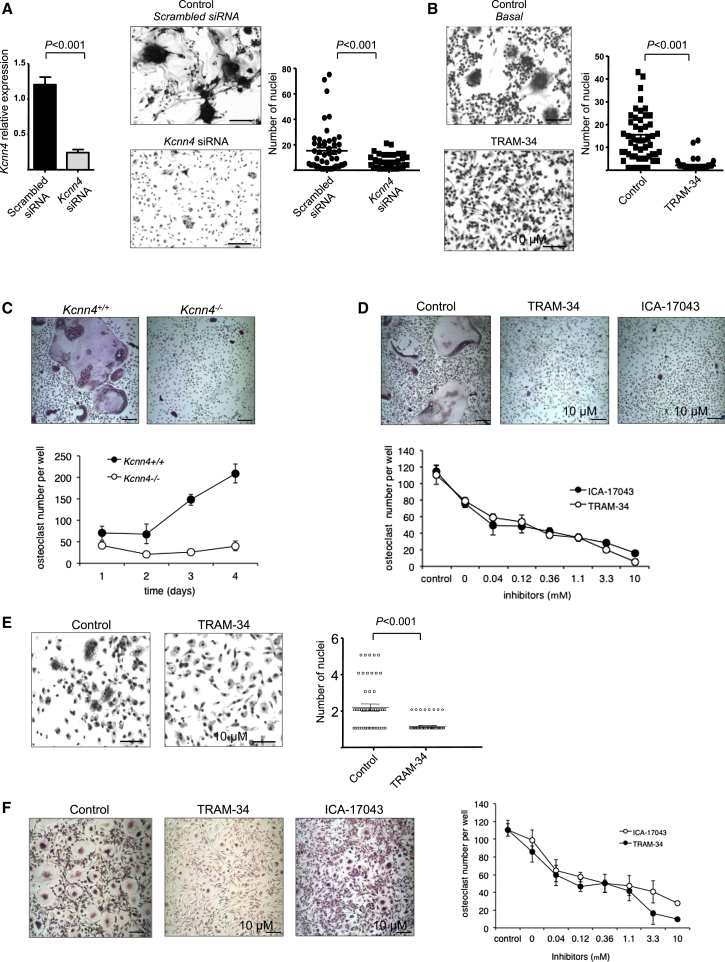
Because multinucleate osteoclasts and MGCs originate from macrophages (Vignery, 2005), our data suggest that MMnet controlled by Trem2 may contain novel genes regulating macrophage multinucleation. We therefore hypothesized that the most significant trans-eQTL in the network (R2 = 0.25, p = 3.8 × 10−16, Figure 1E), Kcnn4, encoding an intermediate-conductance calcium-activated potassium channel, regulates multinucleation in macrophages. Kcnn4 expression peaked during macrophage multinucleation (Figure S3) and a separate time-dependent microarray analysis of fusing rat alveolar macrophages and human monocytes revealed Kcnn4 as a differentially expressed gene, which is strongly induced at the onset of fusion (Figure S3). We therefore investigated whether Kcnn4 determines multinucleation in osteoclasts and MGCs, two cell types resulting from multinucleation in physiological and pathological conditions, respectively. To this aim, we used two selective distinct pharmacological blockers (ICA-17043 and TRAM-34) of Kcnn4 in human and rodent osteoclasts and MGCs and confirmed the effect of these blockers with targeted gene deletion and RNAi for Kcnn4 (Figure 3). Transfection with siRNA designed against Kcnn4 (Figure 3A), selective pharmacological blockade (Figures 3B and 3D), and targeted gene deletion (Figure 3C) of Kcnn4 consistently decreased macrophage multinucleation in rodents. Similar results were obtained in human osteoclasts and MGCs (Figures 3E and 3F), indicating that Kcnn4 regulates macrophage multinucleation in rodents and humans. The inhibitory effect of TRAM-34 on macrophage multinucleation is reversible and TRAM-34 does not affect macrophage differentiation (Figure S3) but changes the macrophage transcriptome assessed by high-throughput sequencing of mRNA (RNA-seq, Figure S3). Furthermore, in order to assess whether the absence of Kcnn4 affects preosteoclast formation, we have quantified TRAP+ mononuclear cells isolated from Kcnn4+/+ and Kcnn4−/− mice at day 3, 4, and 5 of cell differentiation and showed that early steps of osteoclast differentiation do not seem to be affected by Kcnn4 deficiency (Figure S3).
Figure 3.
Kcnn4 Regulates Macrophage Multinucleation in Rodents and Humans
(A) WKY BMDMs were cultured in Lab-Tek chambers and incubated with either scrambled or Kcnn4 siRNA, and cells were fixed for the assessment of the multinucleation. Kcnn4 expression levels were measured by quantitative real-time PCR and normalized to Hprt expression, showing 80% knockdown in macrophages following incubation with Kcnn4 siRNA (left panel). Silencing of Kcnn4 led to a marked reduction in macrophage multinucleation as the number of nuclei in macrophages transfected with Kcnn4 siRNA was significantly reduced (right panel) when compared to controls (mean ± SEM; n = 4 rats). Original bars, 50 μm. p < 0.001 determined by Kruskal-Wallis test.
(B) When macrophages were cultured in the presence of TRAM-34 (10 μM), the number of nuclei per 100 macrophages was significantly reduced when compared to cells incubated with the media only (control, basal cells). The results are representative of five independent experiments. Original bars, 50 μm. p < 0.001 determined by Kruskal-Wallis test.
(C) BMDMs from 6-week-old Kcnn4+/+ and Kcnn4−/− mice were cultured in the presence of macrophage colony stimulating factor (M-CSF) (25 ng/ml) and RANKL (20 ng/ml) for 4 days to induce the differentiation of osteoclasts (upper panel). The formation of osteoclasts measured as the number of TRAP+ multinucleated osteoclasts per well increased with time in Kcnn4+/+ cells but not in Kcnn4−/− cells. Mean ± SD; n = 5; the osteoclast number/well was significantly different (p < 0.001) at all time points; the results are representative of three independent experiments. Original bars, 100 μm.
(D) Mouse BMDMs were cultured in the presence of M-CSF (25 ng/ml) and RANKL (40 ng/ml) and treated with TRAM-34 or ICA-17043 (both 10 μM, upper panel). TRAM-34 and ICA-17043 treatment on mouse osteoclasts reduced the number of osteoclasts per well in a dose-dependent manner (lower panel, mean ± SD; n = 5). Original bars, 100 μm.
(E) Human-monocyte-derived macrophages were cultured in Lab-Tek chambers, and spontaneous MGC formation was observed in some buffy coats, and this was significantly reduced when cells were incubated with TRAM-34 (10 μM). At least three separate buffy coats were used to differentiate human macrophages. Original bars, 50 μm. p < 0.001 determined by Kruskal-Wallis test.
(F) Human peripheral blood monocytes were cultured in the presence of M-CSF (20 ng/ml) and RANKL (10 ng/ml) for 5 days to induce the differentiation of osteoclasts. Cells were treated with TRAM-34 or ICA-17043 (both 10 μM, left panel). Quantification of osteoclast number per well shows that TRAM-34 and ICA-17043 inhibited the formation of monocyte-derived human osteoclasts in a dose-dependent manner (mean ± SD; n = 4). Original bars, 100 μm.
See also Figure S3.
Kcnn4 Modulates Bone Turnover
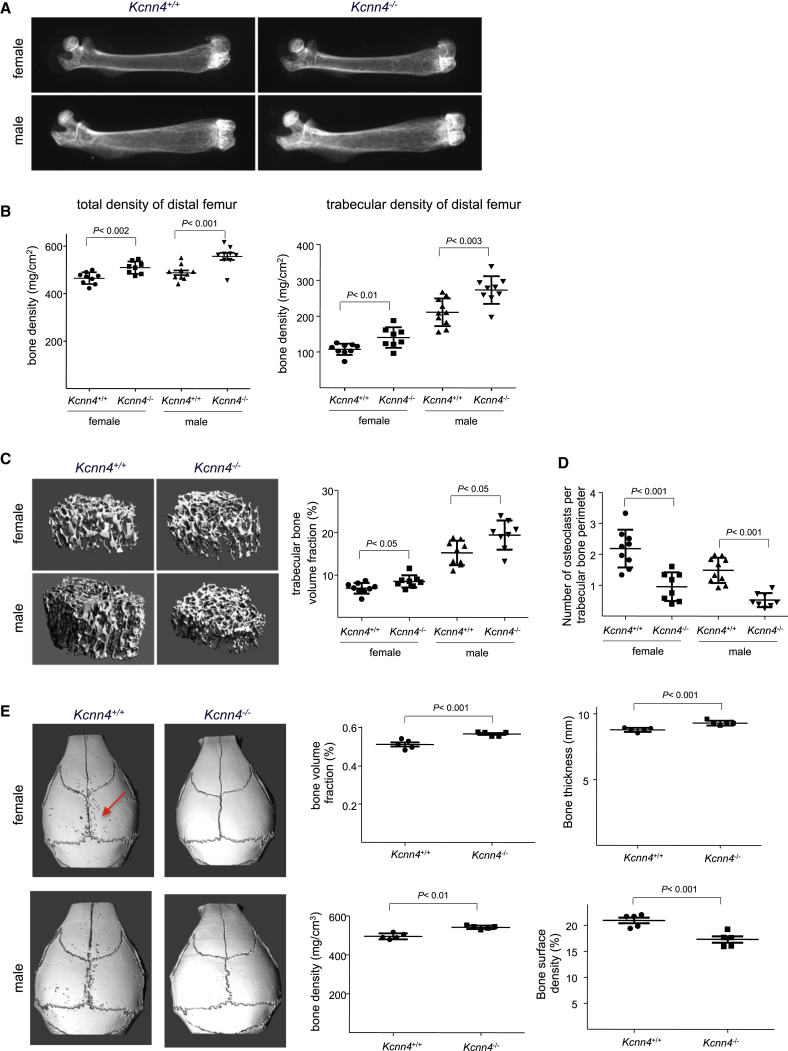
Given our data implicating Kcnn4 in macrophage multinucleation and because defects in osteoclast formation impact bone resorption (Teitelbaum, 2011), we tested whether Kcnn4 deficiency affects bone mass. Femurs from Kcnn4+/+ and Kcnn4−/− female and male mice were analyzed by X-ray, peripheral quantitative computed tomography (pQCT), and microcomputed tomography (microCT). These analyses consistently revealed increased bone mass and density in both female and male Kcnn4−/− mice compared with Kcnn4+/+ mice (Figure 4). In particular, femurs from Kcnn4−/− mice showed increased (1) X-ray absorption (Figure 4A), (2) total bone mineral density (9% and 14% increases in females and males, respectively), (3) trabecular bone density (30% and 29% in females and males), and (4) trabecular bone volume fraction (27% and 26% in females and males) (Figures 4B and 4C). Accordingly, Kcnn4−/− mice had reduced numbers of TRAP-positive osteoclasts (Figure S4) and a reduced number of osteoclasts per unit of bone surface (Figures 4D and S4; Table S1), thus supporting a role for Kcnn4 in osteoclast formation in vivo. Furthermore, Kcnn4−/− mice also displayed increased bone formation and mineral apposition rates compared with Kcnn4+/+ mice, indicating the high bone mass phenotype in Kcnn4−/− mice results from effects on both bone formation and bone resorption (Table S1). We next investigated whether osteoclast activity was decreased in Kcnn4−/− mice in an inflammation-induced calvaria bone resorption provocation model. This study revealed that Kcnn4−/− mice lacked the normal osteoclast-mediated bone resorption response to inflammation (Figure 4E). We also showed that deletion of Kcnn4 results in reduced osteoclastic resorption in vitro (Figure S4). Taken together, these data indicate that Kcnn4 participates in the control of bone turnover by regulating functional osteoclast formation and suggest that its effects on bone formation may be mediated via a primary role in osteoclasts.
Figure 4.
Kcnn4 Modulates Bone Homeostasis via Osteoclasts
(A) Genetic deletion of Kcnn4 increases bone mass as shown by X-ray analysis performed on femurs from 8-week-old Kcnn4+/+ and Kcnn4−/− female and male mice. Note the stronger radio-opacity in femurs from Kcnn4−/− mice when compared with Kcnn4+/+ mice.
(B) Peripheral quantitative computed tomography (pQCT) analysis of femoral bones showing total bone density and trabecular bone density of distal femurs from 8-week-old Kcnn4−/− mice compared with Kcnn4+/+ mice (mean ± SD; n = 7–9).
(C) Representative images of microcomputed tomography (microCT) analysis of distal femurs (left panel) from 8-week-old female and male Kcnn4+/+ and Kcnn4−/− mice. Trabecular bone volume fraction from Kcnn4+/+ and Kcnn4−/− female and male mice is shown in the right panel (mean ± SD; n = 7–9).
(D) Histomorphometry analysis of tibiae from 8-week-old Kcnn4+/+ and Kcnn4−/− female and male mice showing the number of osteoclasts per trabecular bone perimeter (mean ± SD; n = 10).
(E) Microcomputed tomography analysis of calvaria from 8-week-old Kcnn4−/− and Kcnn4+/+ male and female mice that had received a 2 μl of injection of 25 μg LPS on the right calvaria. Mice were sacrificed 5 days later. Note the low abundance of resorption lacunae in Kcnn4−/− mice when compared with wild-type. Calvarial bone volume fraction, bone thickness and bone density were higher, whereas bone surface density was lower in Kcnn4−/− mice when compared with wild-type (right panel; mean ± SD; n = 5).
Inactivation of Kcnn4 Reduces Severity of Glomerulonephritis and Bone Erosion in Arthritis
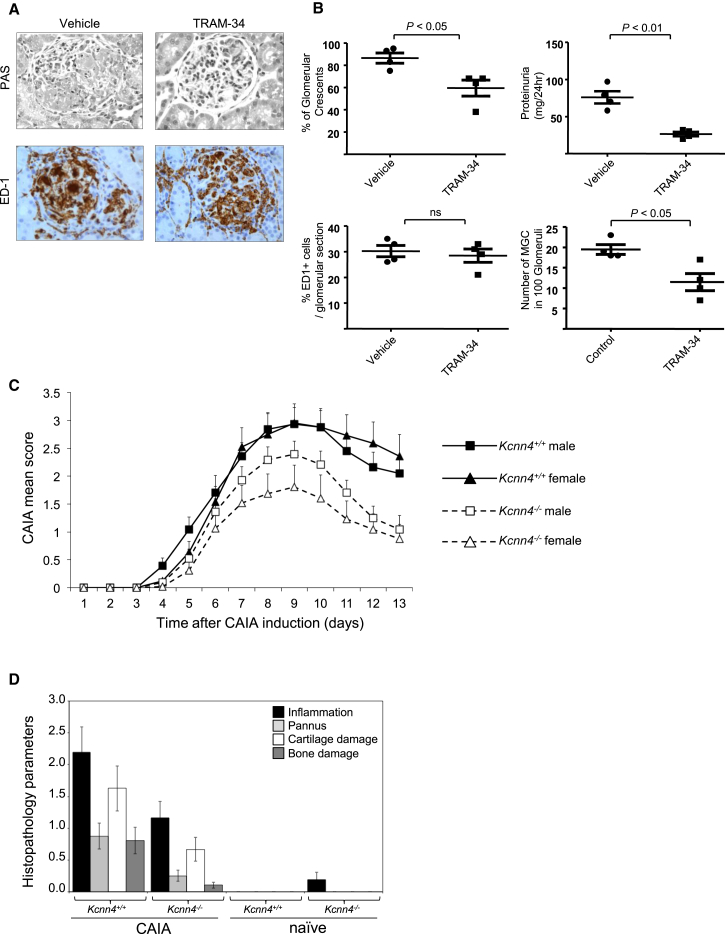
To study the role of Kcnn4 in macrophage multinucleation in pathological conditions, we used two inflammatory models in rodents, characterized by MGC and osteoclast activation: glomerulonephritis and arthritis, respectively. In crescentic glomerulonephritis (Crgn), we blocked Kcnn4 with its selective inhibitor TRAM-34 (Wulff et al., 2000) in the WKY NTN model characterized by glomerular MGCs (Figure S1; Lan et al., 1998). In the WKY NTN model, the severe and reproducible glomerular macrophage infiltration following nephrotoxic serum injection (Tam et al., 1999) is also characterized by approximately 20% of glomeruli presenting MGCs (Figures 5 and S1). It is therefore possible to study the effect of TRAM-34 on glomerular MGC formation and renal injury assessed by proteinuria. TRAM-34 treatment of WKY rats with NTN led to significant reduction of glomerular crescents, glomerular MGC formation, and proteinuria without affecting the number of macrophages infiltrating the glomeruli (Figures 5A and 5B). To evaluate the role of Kcnn4 in bone erosion in inflammatory arthritis, we assessed the effect of Kcnn4 deficiency in anticollagen antibody-induced arthritis (CAIA) and showed that Kcnn4−/− mice have reduced joint inflammation and tissue damage (Figures 5C, 5D, and S5). Following CAIA serum bone resorption (CTX) and bone formation (P1NP) markers were measured in Kcnn4+/+ and Kcnn4−/− mice. Kcnn4 deficiency resulted in impaired osteoclastic bone resorption that was not affected by CAIA but did not affect osteoblastic bone formation (Figure S5). This is consistent with the results obtained in the lipopolysaccharide (LPS)-induced calvaria model (Figure 4E) showing reduced osteoclast activity in vivo. Taken together, the results obtained from the LPS and CAIA experiments suggest that reduced bone loss in Kcnn4−/− mice can be attributed to decreased osteoclast activity resulting from impaired multinucleation.
Figure 5.
Reduced Severity of Glomerulonephritis and Arthritis by Kcnn4 Inactivation
(A) Glomerular immunohistochemistry showing the effect of specific Kcnn4 blocker TRAM-34 in renal injury in the WKY nephrotoxic nephritis model. Following nephrotoxic serum injection, glomerular crescents (PAS) and percentage of ED1+ macrophages were assessed in control (vehicle) and TRAM-34-treated rats (n = 4 rats were used per group of treatment). Original bars, 20 μm.
(B) Quantification of glomerular crescents, proteinuria, percentage of ED1+ cells and number of glomerular MGCs.
(C) Collagen antibody-induced arthritis (CAIA) was induced in 2-month-old male and female Kcnn4−/− and Kcnn4+/+ mice (mean ± SD; n = 12–15). Arthritic severity was monitored daily using a visual scoring system as detailed in Supplemental Experimental Procedures. Based on inflammation scoring, Kcnn4 deletion reduced inflammation in males and females, though the prevention was more efficient in female mice. From day 8 onward, the CAIA mean score is significantly different between Kcnn4+/+ and Kcnn4−/− animals for both genders (p < 0.001).
(D) Histopathomorphometry analysis of the paws and the ankles of Kcnn4+/+ and Kcnn4−/− naive and CAIA-induced female mice. All the histomorphometric parameters (inflammation, pannus, cartilage damage, and bone damage) were compared to CAIA Kcnn4+/+ mice and are significant (mean ± SEM; n = 10–12 mice in each group; p < 0.05). Similar results were obtained in male mice (data not shown).
See also Figure S5.
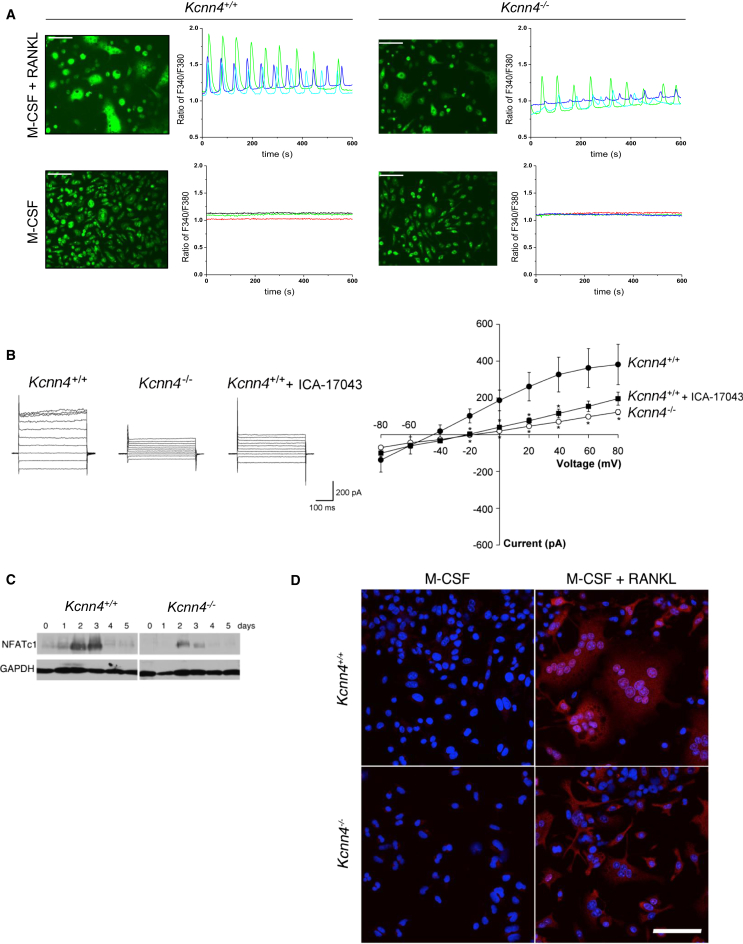
Kcnn4 Regulates Ca2+ Signaling in Macrophage Multinucleation
In order to investigate the mechanism by which Kcnn4 regulates multinucleation, we focused on intracellular Ca2+ levels because of the established link between K+ channels and Ca2+ influx during lymphocyte activation (Feske et al., 2012). We assessed whether pharmacological blockade or gene deletion of Kcnn4 results in disturbed Ca2+ signaling and oscillations and revealed that there are decreased Ca2+ oscillations in Kcnn4−/− osteoclasts compared to Kcnn4+/+ (Figure 6A). We also detected differences between Kcnn4+/+ and Kcnn4−/− macrophages in patch clamp experiments, including a shift in observed reversal potential following gene deletion or inhibition of Kcnn4 (Figure 6B). One of the major downstream effects of receptor activator of nuclear factor kappa-B ligand (RANKL)-induced activation of Ca2+ signaling is nuclear translocation of dephosphorylated nuclear factor of activated T cells, cytoplasmic 1 (NFATc1) (Takayanagi, 2007), and, consistent with this, there was reduced expression of NFATc1 in Kcnn4−/− osteoclasts (Figure 6C). The reduced NFATc1 expression was associated with decreased nuclear translocation in Kcnn4−/− bone-marrow-derived macrophages in response to RANKL (Figure 6D). Similarly, WKY BMDMs treated with TRAM-34 exhibit reduced Nfatc1 expression associated with decreased multinucleation (data not shown). To elucidate the mechanisms by which Kcnn4 affects osteoclast differentiation, we stimulated macrophages with RANKL or LPS for increasing time periods and found that RANKL-induced p38 and ERK activation were not modified by Kcnn4 deficiency (data not shown). In contrast, RANKL-induced Akt activation was reduced (Figure S6). This result was confirmed in human macrophages treated with TRAM-34 (Figure S6). Because Akt has been shown to be a critical mediator of cell proliferation and survival in a variety of cell types (Scheid and Woodgett, 2001), our results showing impaired Akt phosphorylation in association with Kcnn4 deficiency suggested that Kcnn4 may regulate apoptosis. Supporting this hypothesis, deletion of Kcnn4 was associated with increased apoptosis in osteoclasts (Figure S6).
Figure 6.
Kcnn4 Regulates Ca2+-NFATc1 Signaling in Multinucleate Macrophages
(A) Ca2+ oscillations were recorded by intracellular Ca2+ imaging using fura-2 in multinucleate osteoclasts from Kcnn4+/+ and Kcnn4−/− mice. Fluorescence images of cells is shown on the left panel, whereas traces of change in fura-2 fluorescence ratio in single cells treated with M-CSF only or M-CSF and RANKL for 72 hr are shown on the right panel. Note the decrease in Ca2+ oscillations associated with Kcnn4 deficiency, and the absence of Ca2+ oscillations in the presence of M-CSF alone, independent of Kcnn4. These experiments were repeated multiple times with similar results, and one representative result is shown. Original bars, 50 μm.
(B) Representative current responses to increasing voltage steps (−100 mV to +80 mV; 400 ms) observed in mouse BMDMs from Kcnn4+/+ and Kcnn4−/− mice treated with M-CSF (25 ng/ml) and RANKL (40 ng/ml) for 5 days (left panel). I–V plot demonstrates the average current/voltage relationship observed (right panel). Data points for Kcnn4−/− and Kcnn4+/+ macrophages treated with the Kcnn4 inhibitor ICA-17043 (1 μM) were statistically different from Kcnn4+/+ in the range of −20 mV to +80 mV (mean ± SD, ∗p < 0.05; n = 10), with the exception of Kcnn4+/+ + ICA-17043 at + 60 mV (mean ± SD, p = 0.06; n = 10) and +80 (mean ± SD, p = 0.11; n = 10). There was no significant difference between Kcnn4−/− macrophages and Kcnn4+/+ treated with ICA-17043.
(C) NFATc1 protein levels in BMDMs from Kcnn4+/+ and Kcnn4−/− mice. BMDMs isolated from Kcnn4+/+ and Kcnn4−/− mice were cultured in the presence of M-CSF (25 ng/ml) and RANKL (40 ng/ml) and subjected to western blot analysis at the indicated times using antibodies directed against NFATc1. GAPDH served as an internal control for equal loading of proteins on a SDS-PAGE protein gel. Note the lower abundance of NFATc1 in Kcnn4−/− macrophages compared with Kcnn4+/+ cells. This figure is representative of several experiments performed with similar results.
(D) BMDMs isolated from Kcnn4−/− and Kcnn4+/+ mice were treated with M-CSF (25 ng/ml) alone or supplemented with RANKL (40 ng/ml) for 5 days and subjected to immunocytochemistry using anti-NFATc1 antibody. Note the decrease in immunoreactive NFATc1 in the nuclei of Kcnn4−/− macrophages treated with RANKL compared with Kcnn4+/+ cells.
Original bars, 100 μm. See also Figure S6.
Discussion
Using systems genetics approaches, our study demonstrates how genomic sequence variation affects a unique regulatory network that is associated with macrophage multinucleation in the rat. By conducting an eQTL study in the primary macrophages of a backcross rat population derived from two inbred rat strains with contrasting phenotypes (“markedly fusogenic WKY macrophages” versus “lowly fusogenic LEW macrophages”), we have identified a unique gene regulatory network (MMnet) associated with macrophage multinucleation in the rat. We show that Trem2 regulate MMnet, which was enriched for genes regulating osteoclast formation in the rat, a result that was confirmed in human macrophages. Consistent with the previously established role of Trem2 in macrophage fusion (Helming et al., 2008), activation (Turnbull et al., 2006), and osteoclast formation (Cella et al., 2003; Humphrey et al., 2006; Otero et al., 2012), here, we demonstrate that the strongest trans-eQTL in the Trem2-controlled gene network, Kcnn4, is a key determinant of macrophage multinucleation in rodent and human osteoclasts and MGCs. We independently confirmed the role of Kcnn4 as a potential modulator of multinucleation by microarray analysis of fusing rat macrophages and human monocytes that form osteoclasts. We show that Kcnn4 inhibitors prevent the formation of monocyte-derived human osteoclasts and MGCs, in addition to mouse bone-marrow-derived osteoclasts and rat MGCs.
The intermediate-conductance Ca2+-activated K+ channel Kcnn4 (also known as Kca3.1 or IK1) is activated in response to increases in intracellular Ca2+ through calmodulin bound to its intracellular C terminus (Lam and Wulff, 2011). The blockade of this channel by a selective small molecule inhibitor (TRAM-34) was first shown to have immunosuppressive effects on T lymphocytes (Wulff et al., 2000) and further tested on T cell-mediated diseases such as inflammatory bowel disease, experimental autoimmune encephalomyelitis, rheumatoid arthritis, and asthma (reviewed in Lam and Wulff [2011]). Although Kcnn4 is expressed in macrophages, its role in macrophage multinucleation has not previously been reported. Treatment with TRAM-34 improved atherosclerosis in ApoE−/− mice aortas and carotid arteries by decreasing macrophage infiltration and by possibly inhibiting their migration (Toyama et al., 2008). Similarly, TRAM-34 treatment reduced ED1+ macrophages and microglia in the rat brain following ischemia/reperfusion stroke (Chen et al., 2011b), and, more recently, it has been shown that blockade of Kcnn4 reduced F4/80+ cells in a streptozotocin-induced diabetic mouse model (Huang et al., 2013).
Although it had been reported that Ca2+-activated K+ currents are involved in the regulation of osteoclast movement and spreading on bone substrates, and that increase in intracellular Ca2+ blocks osteoclast bone resorption in vitro (Valverde et al., 2005), the specific contribution of Kcnn4 in inflammation-induced osteoclast development in vivo was not previously reported. The effect of Kcnn4 on bone mass is modest compared to its critical regulatory role in osteoclast and macrophage multinucleation. This is consistent with previous findings demonstrating a relatively mild effect of macrophage fusion marker (DC-STAMP) on bone mass (Yagi et al., 2005). At this stage, the bone formation abnormality observed in Kcnn4−/− mice remains incompletely understood and additional studies to investigate further will require detailed analysis of Kcnn4+/+ and Kcnn4−/− mice at various ages during development and adulthood, as well as cell-specific gene targeting approaches because it was previously used for studying the effect of cathepsin-k deletion on bone formation (Lotinun et al., 2013).
We also show that Kcnn4 affects macrophage multinucleation without having an effect on preosteoclast formation, a result previously documented for Atp6v0d2, a regulator of osteoclast fusion (Lee et al., 2006). Here, we show that Kcnn4-deficient mice fail to mount an osteoclastic response to local injection of LPS into calvaria. Local osteoclast formation and bone loss are a major component of the pathogenesis of metabolic and inflammatory bone diseases (Boyle et al., 2003). Thus, Kcnn4, which was previously reported as a potential target for immunosuppression (Cahalan and Chandy, 1997; Jensen et al., 2001), autoimmune diseases (Wulff et al., 2003), and periodontal disease (Valverde et al., 2005), also represents a potential new target for the prevention of inflammation-related bone loss.
The WKY NTN model of Crgn is characterized by glomerular MGC formation (Kaneko et al., 2003) and the renal injury is entirely dependent on macrophage infiltration and activation (Aitman et al., 2006; Behmoaras et al., 2008, 2010; D’Souza et al., 2013; Deplano et al., 2013; Hull et al., 2013; Page et al., 2012). We show that blocking Kcnn4 in vivo by administration of TRAM-34 significantly reduces glomerular injury in the WKY NTN model. Although we did not observe different numbers of glomerular macrophages between TRAM-34 and vehicle-treated animals, there was a reduction in glomerular MGCs, which suggests that Kcnn4 blockade reduced multinucleation in vivo by preventing disease. In keeping with this, we show that Kcnn4 deletion leads to a reduced severity in chronic inflammatory arthritis characterized by osteoclast activation. Collagen-induced arthritis is characterized by focal collections of osteoclasts at sites of bone destruction, and previous reports showed that osteoclast depletion ameliorated bone erosion in this model (Romas et al., 2002; Sims et al., 2004). Our results showing reduced bone loss in anticollagen antibody-induced arthritis in Kcnn4−/− mice support these findings. However, the effect of Kcnn4 deletion in the CAIA model could be driven by either inflammation or osteoclast-mediated bone erosion/resorption. Further studies using osteoclast-specific Kcnn4 knockout mice will be crucial in dissecting the relative contribution of inflammation and osteoclast-driven responses in this model.
The measurements of intracellular Ca2+ reveal that Kcnn4 deficiency reduces the amplitude of Ca2+ oscillations, but not their frequency, suggesting that K+ currents through the Kcnn4 channel contribute to the sustained Ca2+ oscillations mainly via the regulation of membrane potential, rather than Ca2+ mobilization from intracellular Ca2+ stores. Absence of Kcnn4 appears to affect several downstream signaling pathways, including NFATc1 via calcium signaling, and Akt activation, which together lead to decrease survival of multinucleate macrophages.
In summary, using a systems genetics approach, we identified a trans-regulated multinucleation network in macrophages and provided compelling evidence that at least one gene within the network (Kcnn4) regulates macrophage multinucleation in bone homeostasis. We also show that Kcnn4 can be effectively targeted to decrease severity of inflammatory conditions characterized by MGC or osteoclast activation.
Experimental Procedures
Animals, Genetic Crosses, and Genotyping
Wistar-Kyoto (WKY/NCrl) rats and Lewis (LEW/Crl) were purchased from Charles River UK. Two hundred Backcross (BC) rats were produced by breeding WKY with LEW rats (congenic WKY rats were used where the previously identified crescentic glomerulonephritis QTLs on chromosome 13 and 16 were introgressed from the LEW rats [Behmoaras et al., 2010]). The (WKY x LEW) F1 animals were backcrossed to the congenic WKY rats. For genotyping, DNA was isolated from the BC rat spleens using a standard phenol-chloroform extraction. DNA (250 ng) was used for NspI and StyI fragmentation according to Affymetrix SNP6.0 GeneChip instructions. Custom designed Whole Genome Rat Genotyping arrays (RATDIV arrays, MDC) that contain 803,484 SNPs covering the rat genome (based on Rn3.4 version) were used. SNP calling was performed using the BRLMM algorithm and led to the identification of 785,247 SNPs with >95% call rate (see Supplemental Experimental Procedures for additional details). Kcnn4−/− mice were generated on a mixed 129J/C57BL6 background, bred, and genotyped as previously described (Begenisich et al., 2004). The animals were housed in standard caging on a 12-hr-light cycle and were offered free access to rodent chow (Harlan Teklad #2018 Rodent Diet) and water.
Bone X-Ray Radiography, pQCT, microCT, and Histomorphometry in Mice
Excised femurs were subjected to X-ray using a MX-20 (Faxitron X-ray) at 30 kV for 3 s. X-ray films were scanned using an Epson Perfection 4870. Bone density was determined as described previously (Ballica et al., 1999) by Peripheral Quantitative Computed Tomography (pQCT, XCT Research M; Norland Medical Systems) of a virtual 1 mm cross-section of the distal femur 0.25 mm proximal to the growth plate. In addition, distal femurs were scanned with a Microcomputed Tomography (microCT) scanner (MicroCT 40; Scanco) with a 2,048 × 2,048 matrix and isotropic resolution of 12 μm3 voxel size. 3D trabecular measurements in the secondary spongiosa were made directly, as previously described (Li et al., 2005). Femurs and tibiae from Kcnn4+/+ and Kcnn4−/− mice were dehydrated in a graded ethanol series and embedded without decalcification in methylmethacrylate, as we described previously (Baron et al., 1984). Four-micrometer-thick cross-sections of the distal femur were stained with Villanueva Mineralized Bone Stain for static histomorphometric analysis (see Supplemental Experimental Procedures), whereas 8-μm-thick sections were left unstained for dynamic bone histomorphometric parameters.
All procedures were approved by the Yale University Institutional Animal Care and Use Committee. The care and treatment of experimental mice complied with all applicable federal guidelines and was approved by the Institutional Animal Care and Use Committee at Yale University.
Microarray Expression Profiling, eQTL Mapping, and Network Analysis
Total RNA was extracted from BMDMs from 200 backcross rats using TRIzol (Invitrogen), and gene expression was measured using Affymetrix 1.0 ST Rat Gene array. WKY-LEW next-generation sequencing data (Illumina Hiseq 2000) was used to remove SNP-containing probes prior to normalization. Arrays were normalized using RMA normalization from the Affymetrix package (Irizarry et al., 2003). Normalization step included Norm-exp background correction and quantile normalization. Probes signal were summarized at a transcript level using median polish. The resulting data were log transformed (Figure S2). Remaining batch effects were removed using ComBat (Chen et al., 2011a; Johnson et al., 2007) with nonparametric prior. All other parameters were left to default. The first two principal components of the normalized data before and after correcting for batch effects are shown in Figure S2. The eQTL mapping was performed using ESS++ (Bottolo et al., 2011a, 2011b). Fixed effects on each individual were added as covariates in the variable selection process to account for potential outliers or genotyping errors. eQTL analysis details, network inference, and cell-type enrichment analysis are described in Supplemental Experimental Procedures.
Nephrotoxic Nephritis in Rats and Collagen Antibody-Induced Arthritis in Mice
The effect of TRAM-34 on nephrotoxic nephritis (NTN) in the WKY rat was assessed as follows: eight male WKY rats were injected with the nephrotoxic serum (NTS) as previously described (D’Souza et al., 2013). A group of four rats received intraperitoneal injections of TRAM-34 (40 mg/kg) in Miglyol 812 neutral oil (Kemcare) at 1 μl/g, twice a day with 10 hr intervals for 7 days. The control group (n = 4) had IP injections of Miglyol (vehicle). Rats were killed 7 days following injection of NTS, and glomerular crescents and proteinuria were measured as previously described (D’Souza et al., 2013). Chronic inflammatory arthritis was induced in 8-week-old male and female Kcnn4+/+ and Kcnn4−/− mice by intraperitoneal (i.p.) injection of 7 mg (700 μl) of Arthrogen monoclonal antibodies (ArthoMAB) blend (Millipore) on day 0. This cocktail of monoclonal antibodies is directed against epitopes recognized in the region CB11 of collagen type II. On day 3, 50 μg (<100 μl volume) of LPS was administered i.p. Beginning on day 4, the animals were monitored daily for the onset and development of collagen-antibody-induced arthritis (CAIA), and the injection site on each animal was evaluated for signs of infection, such as heat, redness, and/or exudation. Mice that felt as “cold,” which likely underwent cachexia in response to LPS, received subcutaneous injection of 500 μl of warm Ringer solution. Arthritic severity was monitored daily using a visual scoring system (Supplemental Experimental Procedures). The histopathological evaluation of CAIA is described in Supplemental Experimental Procedures.
Author Contributions
A.V., J.L., G.N., E.P., and J.B. designed the study. H.K. completed the cell biology experiments on human and mouse osteoclasts. A.K. performed the sample preparation for microarrays, RNAi experiments, RNA-seq, and cell biology experiments on rat and human macrophages with contributions from J.B. and J.H.K. M.R. has performed the eQTL mapping and network analysis with contributions from E.P. X.X. analyzed the role of Kcnn4 in the formation of human and mouse osteoclasts and contributed to histomorphometry analysis and the calvaria assay. Q.Z. completed the phenotyping of the mouse bones and contributed to the histomorphometry analysis of the mouse femurs and to the CAIA study. Z.D. performed the in vivo studies in rats and analyzed the results with J.B. and H.T.C. M.K. completed the pilot experiments on human and mouse osteoclasts. J.C.S. contributed to the in vivo experiments in mice. J.H.K., J.H.D.B., and G.R.W. performed ELISA for bone turnover markers. P.K.S. analyzed RNA-seq studies. J.R.G. completed the Ca++ flux studies. L.G. performed the microarrays on rat macrophages. J.E.M. provided the Kcnn4−/− mice. A.H. performed microarray experiments of rat alveolar macrophages and human osteoclasts. J.D. performed compound inhibition studies in human multinucleate giant cells. J.Z. performed microCT analysis. D.S. performed the CAIA study. A.K.B. performed expression analysis of mouse osteoclasts. J.B., E.P., and A.V. coordinated the project and wrote the manuscript with contributions from W.C., H.K., T.J.A., H.T.C., J.E.M., J.H.D.B., G.R.W., and J.L.
Acknowledgments
The authors are grateful to Veterinary Clinical Services at the Yale School of Medicine and to the Yale Core Center for Musculoskeletal Diseases, in particular, to Nancy Troiano for her assistance in processing the mouse bones for histomorphometry analysis. We also thank Bolder Biopath (Boulder, CO) for processing the arthritic mouse bones for histology, and grading the pathology. We would like to thank Jennifer Smith for her excellent technical assistance and Kathrin Saar (MDC, Berlin) for genotyping protocols and data analysis. We gratefully acknowledge funding from Kidney Research UK (RP9/2013) (J.B.), Wellcome Trust (WT092523MA) (J.B.). We also acknowledge funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement no. HEALTH-F4- 2010-241504 (EURATRANS) (E.P. and M.R.) and the Medical Research Council (E.P.). A.H., J.D., J.Z., D.S., A.K.B., G.N., and J.L. are employees of Boehringer Ingelheim. This work was supported in part by a grant from Boehringer Ingelheim.
Published: August 14, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Agnès Vignery, Email: agnes.vignery@yale.edu.
Enrico Petretto, Email: enrico.petretto@imperial.ac.uk.
Jacques Behmoaras, Email: jacquesb@imperial.ac.uk.
Accession Numbers
The ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) accession number for the rat-bone-marrow-derived macrophage microarray data reported in this paper is E-MTAB-2719.
Supplemental Information
References
- Aitman T.J., Dong R., Vyse T.J., Norsworthy P.J., Johnson M.D., Smith J., Mangion J., Roberton-Lowe C., Marshall A.J., Petretto E. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- Ballica R., Valentijn K., Khachatryan A., Guerder S., Kapadia S., Gundberg C., Gilligan J., Flavell R.A., Vignery A. Targeted expression of calcitonin gene-related peptide to osteoblasts increases bone density in mice. J. Bone Miner. Res. 1999;14:1067–1074. doi: 10.1359/jbmr.1999.14.7.1067. [DOI] [PubMed] [Google Scholar]
- Baron R., Tross R., Vignery A. Evidence of sequential remodeling in rat trabecular bone: morphology, dynamic histomorphometry, and changes during skeletal maturation. Anat. Rec. 1984;208:137–145. doi: 10.1002/ar.1092080114. [DOI] [PubMed] [Google Scholar]
- Begenisich T., Nakamoto T., Ovitt C.E., Nehrke K., Brugnara C., Alper S.L., Melvin J.E. Physiological roles of the intermediate conductance, Ca2+-activated potassium channel Kcnn4. J. Biol. Chem. 2004;279:47681–47687. doi: 10.1074/jbc.M409627200. [DOI] [PubMed] [Google Scholar]
- Behmoaras J., Bhangal G., Smith J., McDonald K., Mutch B., Lai P.C., Domin J., Game L., Salama A., Foxwell B.M. Jund is a determinant of macrophage activation and is associated with glomerulonephritis susceptibility. Nat. Genet. 2008;40:553–559. doi: 10.1038/ng.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behmoaras J., Smith J., D’Souza Z., Bhangal G., Chawanasuntoropoj R., Tam F.W., Pusey C.D., Aitman T.J., Cook H.T. Genetic loci modulate macrophage activity and glomerular damage in experimental glomerulonephritis. J. Am. Soc. Nephrol. 2010;21:1136–1144. doi: 10.1681/ASN.2009090968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottolo L., Chadeau-Hyam M., Hastie D.I., Langley S.R., Petretto E., Tiret L., Tregouet D., Richardson S. ESS++: a C++ objected-oriented algorithm for Bayesian stochastic search model exploration. Bioinformatics. 2011;27:587–588. doi: 10.1093/bioinformatics/btq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottolo L., Petretto E., Blankenberg S., Cambien F., Cook S.A., Tiret L., Richardson S. Bayesian detection of expression quantitative trait loci hot spots. Genetics. 2011;189:1449–1459. doi: 10.1534/genetics.111.131425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle W.J., Simonet W.S., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Breitling R., Li Y., Tesson B.M., Fu J., Wu C., Wiltshire T., Gerrits A., Bystrykh L.V., de Haan G., Su A.I., Jansen R.C. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 2008;4:e1000232. doi: 10.1371/journal.pgen.1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck W.G., Anderson J.M. Giant cell formation and function. Curr. Opin. Hematol. 2009;16:53–57. doi: 10.1097/MOH.0b013e32831ac52e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühling F., Reisenauer A., Gerber A., Krüger S., Weber E., Brömme D., Roessner A., Ansorge S., Welte T., Röcken C. Cathepsin K—a marker of macrophage differentiation? J. Pathol. 2001;195:375–382. doi: 10.1002/path.959. [DOI] [PubMed] [Google Scholar]
- Cahalan M.D., Chandy K.G. Ion channels in the immune system as targets for immunosuppression. Curr. Opin. Biotechnol. 1997;8:749–756. doi: 10.1016/s0958-1669(97)80130-9. [DOI] [PubMed] [Google Scholar]
- Cella M., Buonsanti C., Strader C., Kondo T., Salmaggi A., Colonna M. Impaired differentiation of osteoclasts in TREM-2-deficient individuals. J. Exp. Med. 2003;198:645–651. doi: 10.1084/jem.20022220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.H., Olson E.N. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- Chen C., Grennan K., Badner J., Zhang D., Gershon E., Jin L., Liu C. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS ONE. 2011;6:e17238. doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.J., Raman G., Bodendiek S., O’Donnell M.E., Wulff H. The KCa3.1 blocker TRAM-34 reduces infarction and neurological deficit in a rat model of ischemia/reperfusion stroke. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2011;31:2363–2374. doi: 10.1038/jcbfm.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza Z., McAdoo S.P., Smith J., Pusey C.D., Cook H.T., Behmoaras J., Aitman T.J. Experimental crescentic glomerulonephritis: a new bicongenic rat model. Dis. Model. Mech. 2013;6:1477–1486. doi: 10.1242/dmm.012328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa C.E., Annels N.E., Faaij C.M., Forsyth R.G., Hogendoorn P.C., Egeler R.M. Presence of osteoclast-like multinucleated giant cells in the bone and nonostotic lesions of Langerhans cell histiocytosis. J. Exp. Med. 2005;201:687–693. doi: 10.1084/jem.20041785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplano S., Cook H.T., Russell R., Franchi L., Schneiter S., Bhangal G., Unwin R.J., Pusey C.D., Tam F.W., Behmoaras J. P2X7 receptor-mediated Nlrp3-inflammasome activation is a genetic determinant of macrophage-dependent crescentic glomerulonephritis. J. Leukoc. Biol. 2013;93:127–134. doi: 10.1189/jlb.0612284. [DOI] [PubMed] [Google Scholar]
- Feske S., Skolnik E.Y., Prakriya M. Ion channels and transporters in lymphocyte function and immunity. Nat. Rev. Immunol. 2012;12:532–547. doi: 10.1038/nri3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helming L., Gordon S. Molecular mediators of macrophage fusion. Trends Cell Biol. 2009;19:514–522. doi: 10.1016/j.tcb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Helming L., Tomasello E., Kyriakides T.R., Martinez F.O., Takai T., Gordon S., Vivier E. Essential role of DAP12 signaling in macrophage programming into a fusion-competent state. Sci. Signal. 2008;1:ra11. doi: 10.1126/scisignal.1159665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Shen S., Ma Q., Chen J., Gill A., Pollock C.A., Chen X.M. Blockade of KCa3.1 Ameliorates Renal Fibrosis Through the TGF-beta1/Smad Pathway in Diabetic Mice. Diabetes. 2013 doi: 10.2337/db13-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner N., Wallace C.A., Zimdahl H., Petretto E., Schulz H., Maciver F., Mueller M., Hummel O., Monti J., Zidek V. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- Hull R.P., Srivastava P.K., D’Souza Z., Atanur S.S., Mechta-Grigoriou F., Game L., Petretto E., Cook H.T., Aitman T.J., Behmoaras J. Combined ChIP-Seq and transcriptome analysis identifies AP-1/JunD as a primary regulator of oxidative stress and IL-1β synthesis in macrophages. BMC Genomics. 2013;14:92. doi: 10.1186/1471-2164-14-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey M.B., Ogasawara K., Yao W., Spusta S.C., Daws M.R., Lane N.E., Lanier L.L., Nakamura M.C. The signaling adapter protein DAP12 regulates multinucleation during osteoclast development. J. Bone Miner. Res. 2004;19:224–234. doi: 10.1359/JBMR.0301234. [DOI] [PubMed] [Google Scholar]
- Humphrey M.B., Daws M.R., Spusta S.C., Niemi E.C., Torchia J.A., Lanier L.L., Seaman W.E., Nakamura M.C. TREM2, a DAP12-associated receptor, regulates osteoclast differentiation and function. J. Bone Miner. Res. 2006;21:237–245. doi: 10.1359/JBMR.051016. [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jensen B.S., Strøbaek D., Olesen S.P., Christophersen P. The Ca2+-activated K+ channel of intermediate conductance: a molecular target for novel treatments? Curr. Drug Targets. 2001;2:401–422. doi: 10.2174/1389450013348173. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kaneko Y., Sakatsume M., Xie Y., Kuroda T., Igashima M., Narita I., Gejyo F. Macrophage metalloelastase as a major factor for glomerular injury in anti-glomerular basement membrane nephritis. J. Immunol. 2003;170:3377–3385. doi: 10.4049/jimmunol.170.6.3377. [DOI] [PubMed] [Google Scholar]
- Kramnik I., Dietrich W.F., Demant P., Bloom B.R. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2000;97:8560–8565. doi: 10.1073/pnas.150227197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J., Wulff H. The Lymphocyte Potassium Channels Kv1.3 and KCa3.1 as Targets for Immunosuppression. Drug Dev. Res. 2011;72:573–584. doi: 10.1002/ddr.20467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H.Y., Yu X.Q., Yang N., Nikolic-Paterson D.J., Mu W., Pichler R., Johnson R.J., Atkins R.C. De novo glomerular osteopontin expression in rat crescentic glomerulonephritis. Kidney Int. 1998;53:136–145. doi: 10.1046/j.1523-1755.1998.00748.x. [DOI] [PubMed] [Google Scholar]
- Langley S.R., Bottolo L., Kunes J., Zicha J., Zidek V., Hubner N., Cook S.A., Pravenec M., Aitman T.J., Petretto E. Systems-level approaches reveal conservation of trans-regulated genes in the rat and genetic determinants of blood pressure in humans. Cardiovasc. Res. 2013;97:653–665. doi: 10.1093/cvr/cvs329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Rho J., Jeong D., Sul J.Y., Kim T., Kim N., Kang J.S., Miyamoto T., Suda T., Lee S.K. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 2006;12:1403–1409. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- Lemaire I., Falzoni S., Leduc N., Zhang B., Pellegatti P., Adinolfi E., Chiozzi P., Di Virgilio F. Involvement of the purinergic P2X7 receptor in the formation of multinucleated giant cells. J. Immunol. 2006;177:7257–7265. doi: 10.4049/jimmunol.177.10.7257. [DOI] [PubMed] [Google Scholar]
- Li H., Cuartas E., Cui W., Choi Y., Crawford T.D., Ke H.Z., Kobayashi K.S., Flavell R.A., Vignery A. IL-1 receptor-associated kinase M is a central regulator of osteoclast differentiation and activation. J. Exp. Med. 2005;201:1169–1177. doi: 10.1084/jem.20041444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotinun S., Kiviranta R., Matsubara T., Alzate J.A., Neff L., Lüth A., Koskivirta I., Kleuser B., Vacher J., Vuorio E. Osteoclast-specific cathepsin K deletion stimulates S1P-dependent bone formation. J. Clin. Invest. 2013;123:666–681. doi: 10.1172/JCI64840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons P.A., Rayner T.F., Trivedi S., Holle J.U., Watts R.A., Jayne D.R., Baslund B., Brenchley P., Bruchfeld A., Chaudhry A.N. Genetically distinct subsets within ANCA-associated vasculitis. N. Engl. J. Med. 2012;367:214–223. doi: 10.1056/NEJMoa1108735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin A.A., Nemenman I., Basso K., Wiggins C., Stolovitzky G., Dalla Favera R., Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes I.B., Schett G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- Otero K., Shinohara M., Zhao H., Cella M., Gilfillan S., Colucci A., Faccio R., Ross F.P., Teitelbaum S.L., Takayanagi H., Colonna M. TREM2 and β-catenin regulate bone homeostasis by controlling the rate of osteoclastogenesis. J. Immunol. 2012;188:2612–2621. doi: 10.4049/jimmunol.1102836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page T.H., D’Souza Z., Nakanishi S., Serikawa T., Pusey C.D., Aitman T.J., Cook H.T., Behmoaras J. Role of novel rat-specific Fc receptor in macrophage activation associated with crescentic glomerulonephritis. J. Biol. Chem. 2012;287:5710–5719. doi: 10.1074/jbc.M111.260695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloneva J., Kestilä M., Wu J., Salminen A., Böhling T., Ruotsalainen V., Hakola P., Bakker A.B., Phillips J.H., Pekkarinen P. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat. Genet. 2000;25:357–361. doi: 10.1038/77153. [DOI] [PubMed] [Google Scholar]
- Paloneva J., Manninen T., Christman G., Hovanes K., Mandelin J., Adolfsson R., Bianchin M., Bird T., Miranda R., Salmaggi A. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Malhotra S., Torchia J.A., Kerr W.G., Coggeshall K.M., Humphrey M.B. TREM2- and DAP12-dependent activation of PI3K requires DAP10 and is inhibited by SHIP1. Sci. Signal. 2010;3:ra38. doi: 10.1126/scisignal.2000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichugin A.V., Yan B.S., Sloutsky A., Kobzik L., Kramnik I. Dominant role of the sst1 locus in pathogenesis of necrotizing lung granulomas during chronic tuberculosis infection and reactivation in genetically resistant hosts. Am. J. Pathol. 2009;174:2190–2201. doi: 10.2353/ajpath.2009.081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romas E., Sims N.A., Hards D.K., Lindsay M., Quinn J.W., Ryan P.F., Dunstan C.R., Martin T.J., Gillespie M.T. Osteoprotegerin reduces osteoclast numbers and prevents bone erosion in collagen-induced arthritis. Am. J. Pathol. 2002;161:1419–1427. doi: 10.1016/S0002-9440(10)64417-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid M.P., Woodgett J.R. PKB/AKT: functional insights from genetic models. Nat. Rev. Mol. Cell Biol. 2001;2:760–768. doi: 10.1038/35096067. [DOI] [PubMed] [Google Scholar]
- Sims N.A., Green J.R., Glatt M., Schlict S., Martin T.J., Gillespie M.T., Romas E. Targeting osteoclasts with zoledronic acid prevents bone destruction in collagen-induced arthritis. Arthritis Rheum. 2004;50:2338–2346. doi: 10.1002/art.20382. [DOI] [PubMed] [Google Scholar]
- Takayanagi H. The role of NFAT in osteoclast formation. Ann. N Y Acad. Sci. 2007;1116:227–237. doi: 10.1196/annals.1402.071. [DOI] [PubMed] [Google Scholar]
- Takeda Y., Tachibana I., Miyado K., Kobayashi M., Miyazaki T., Funakoshi T., Kimura H., Yamane H., Saito Y., Goto H. Tetraspanins CD9 and CD81 function to prevent the fusion of mononuclear phagocytes. J. Cell Biol. 2003;161:945–956. doi: 10.1083/jcb.200212031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam F.W., Smith J., Morel D., Karkar A.M., Thompson E.M., Cook H.T., Pusey C.D. Development of scarring and renal failure in a rat model of crescentic glomerulonephritis. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 1999;14:1658–1666. doi: 10.1093/ndt/14.7.1658. [DOI] [PubMed] [Google Scholar]
- Teitelbaum S.L. The osteoclast and its unique cytoskeleton. Ann. N Y Acad. Sci. 2011;1240:14–17. doi: 10.1111/j.1749-6632.2011.06283.x. [DOI] [PubMed] [Google Scholar]
- Thye T., Owusu-Dabo E., Vannberg F.O., van Crevel R., Curtis J., Sahiratmadja E., Balabanova Y., Ehmen C., Muntau B., Ruge G. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat. Genet. 2012;44:257–259. doi: 10.1038/ng.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama K., Wulff H., Chandy K.G., Azam P., Raman G., Saito T., Fujiwara Y., Mattson D.L., Das S., Melvin J.E. The intermediate-conductance calcium-activated potassium channel KCa3.1 contributes to atherogenesis in mice and humans. J. Clin. Invest. 2008;118:3025–3037. doi: 10.1172/JCI30836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A.T., Rice J., Scatena M., Liaw L., Ratner B.D., Giachelli C.M. The role of osteopontin in foreign body giant cell formation. Biomaterials. 2005;26:5835–5843. doi: 10.1016/j.biomaterials.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Turnbull I.R., Gilfillan S., Cella M., Aoshi T., Miller M., Piccio L., Hernandez M., Colonna M. Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol. 2006;177:3520–3524. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- Valverde P., Kawai T., Taubman M.A. Potassium channel-blockers as therapeutic agents to interfere with bone resorption of periodontal disease. J. Dent. Res. 2005;84:488–499. doi: 10.1177/154405910508400603. [DOI] [PubMed] [Google Scholar]
- Vignery A. Macrophage fusion: the making of osteoclasts and giant cells. J. Exp. Med. 2005;202:337–340. doi: 10.1084/jem.20051123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignery A. Macrophage fusion: molecular mechanisms. Methods Mol. Biol. 2008;475:149–161. doi: 10.1007/978-1-59745-250-2_9. [DOI] [PubMed] [Google Scholar]
- Wulff H., Miller M.J., Hansel W., Grissmer S., Cahalan M.D., Chandy K.G. Design of a potent and selective inhibitor of the intermediate-conductance Ca2+-activated K+ channel, IKCa1: a potential immunosuppressant. Proc. Natl. Acad. Sci. USA. 2000;97:8151–8156. doi: 10.1073/pnas.97.14.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff H., Calabresi P.A., Allie R., Yun S., Pennington M., Beeton C., Chandy K.G. The voltage-gated Kv1.3 K(+) channel in effector memory T cells as new target for MS. J. Clin. Invest. 2003;111:1703–1713. doi: 10.1172/JCI16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M., Miyamoto T., Sawatani Y., Iwamoto K., Hosogane N., Fujita N., Morita K., Ninomiya K., Suzuki T., Miyamoto K. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 2005;202:345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.W., Price N.M., Gilman R.H., Recarvarren S., Friedland J.S. Multinucleate giant cells release functionally unopposed matrix metalloproteinase-9 in vitro and in vivo. J. Infect. Dis. 2007;196:1076–1079. doi: 10.1086/521030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.