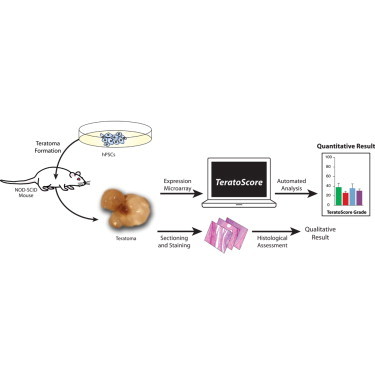
Summary
Teratoma formation is the gold standard assay for testing the capacity of human pluripotent stem cells to differentiate into all embryonic germ layers. Although widely used, little effort has been made to transform this qualitative assay into a quantitative one. Using gene expression data from a wide variety of cells, we created a scorecard representing tissues from all germ layers and extraembryonic tissues. TeratoScore, an online, open-source platform based on this scorecard, distinguishes pluripotent stem cell-derived teratomas from malignant tumors, translating cell potency into a quantitative measure (http://benvenisty.huji.ac.il/teratoscore.php). The teratomas used for the algorithm also allowed us to examine gene expression differences between tumors with a diploid karyotype and those initiated by aneuploid cells. Chromosomally aberrant teratomas show a significantly different gene expression signature from that of teratomas originating from diploid cells, particularly in central nervous system-specific genes, congruent with human chromosomal syndromes.
Graphical Abstract
Highlights
-
•
A gene scorecard representing human tissues from all germ layers was created
-
•
A quantitative pluripotency test named TeratoScore was based on this scorecard
-
•
TeratoScore distinguishes pluripotent stem cell-derived teratomas from other tumors
-
•
Teratomas derived from aneuploid cells show aberrant tissue expression distribution
Teratoma formation is a gold standard assay when defining pluripotency of cells. In this paper, Benvenisty and colleagues utilized global gene expression analysis to transform the qualitative character of this assay into a quantitative one, replacing histological assessment of pluripotent capabilities. Moreover, they show that teratomas initiated from aneuploid cells show an aberrant tissue expression distribution, characteristic of their developmental defects.
Introduction
Pluripotent stem cells (PSCs) are defined by their ability for self-renewal and the capacity to differentiate into cells of the three embryonic germ layers: ectoderm, endoderm, and mesoderm (Thomson et al., 1998). These traits turn human PSCs (hPSCs) into a promising disease-modeling platform and a unique resource in regenerative medicine. PSC differentiation capacity, or cell potency, can be evaluated using several strategies. The gold standard assay to assess the pluripotency of mouse cells is injecting them into blastocysts, generating chimeric mice with germline transmission. As for hPSCs, several alternatives exist. One suggested approach (named PluriTest) is to analyze the expression profile of the undifferentiated cells and deduce their pluripotency based on similarities to known pluripotent cells (Müller et al., 2011). While this approach is promising, it does not validate a cell capacity to differentiate toward all lineages. Furthermore, this approach ignores the possibility that mutations in tissue master regulators (not analyzed in the assay) will affect the potency of the cells to differentiate. Numerous in vitro differentiation protocols have been published, enabling direct differentiation toward cells from all three germ layers. However, this strategy is seldom used, as it is highly expensive, time consuming, and complicated. Furthermore, there is no standard set of direct differentiation protocols sufficient to determine cell potency (Müller et al., 2012). In contrast, in vitro spontaneous differentiation into embryoid bodies (EBs) is a commonly used method to assess hPSC differentiation capability (Itskovitz-Eldor et al., 2000). Although efforts have been made to standardize EB formation and analyze the differentiation capacity into the different germ layers, a commonly accepted method has yet to emerge (Müller et al., 2012).
Teratoma formation is currently the most abundant technique used to evaluate hPSC potency. In this assay, undifferentiated hPSCs are injected into an immune-deficient mouse, forming a heterogeneous tumor composed of terminally differentiated cells from all germ layers. The tumors are then sectioned and stained using H&E and evaluated by an experienced histologist providing qualitative examples for differentiation toward each lineage. Sections can also be stained with antibodies against tissue-specific markers, providing a more direct qualitative approach.
Almost no effort has been made to quantitatively measure teratoma cell composition (Müller et al., 2010), although it might be highly indicative of the initiating cell potency and variation. Gene expression data from teratomas can assess their composition and enable comparison of tumors originating from different PSC lines. However, this kind of analysis and quantitative evaluation had been so far halted by the lack of publicly published expression data.
Using teratoma gene expression data initiated from various hPSC lines, we have developed a method to assess teratoma tissue and lineage composition, hence evaluating the initiating cell’s potency. The TeratoScore algorithm uses in vivo expression profiles to calculate and provide each cell line with a quantitative measure for pluripotency. Further comparison between the expression profiles of teratomas with a normal karyotype and ones with chromosomal aberrations revealed an abnormal tissue-enriched gene expression, suggesting that they can aid in disease modeling.
Results
In order to evaluate cell composition of hPSC-derived teratomas, we have analyzed teratomas from 12 different cell lines (Table S1). Five of the teratomas were generated from diploid hPSCs. Another five were from hPSCs with trisomy of chromosome 12, and four teratomas were initiated from hPSCs with trisomy of chromosome 21. Teratomas were subjected to serial sectioning and H&E staining in order to validate the representation of all three germ layers as previously shown (Ben-David et al., 2014; Biancotti et al., 2010; Chin et al., 2009; Lavon et al., 2008; Narwani et al., 2010). In many histological analyses, structures from all three embryonic germ layers were found in the same section, supporting the notion that different cell types exist in close proximity in the teratomas (Figure S1). RNA from different areas of the teratomas was extracted and analyzed by Affymetrix Human Genome U133 Plus 2.0.
Our aim was to quantitatively analyze cell and lineage differentiation of the teratomas derived from the hPSCs, as an indicator for the cells’ potency. An algorithm assessing cell-specific differentiation of PSCs named CellNet was recently published by Cahan et al. (2014). While CellNet algorithm is extremely useful when dealing with a homogenous population of differentiated cells, when tested by us to examine the differentiation of teratomas, it has proven limited in assessing the complexity of hPSC-derived tumors (Figure S2). Although all three germ layer structures were histologically found in the teratomas (Figure S1), CellNet showed some similarity to undifferentiated ESCs by no clear enrichment of any specific tissues (Figure S2A). This was not the case when measuring homogenous tumors, such as hepatocellular carcinoma, where CellNet correctly identified the tumor composition. A more careful analysis of CellNet’s results revealed that while some tissues, such as liver, were slightly enriched in the teratomas, the heterogeneous tumors still provided similar scores to the undifferentiated cells in most measured tissues (Figure S2B). These results encouraged us to develop a new platform to assess teratoma composition, reflecting the pluripotent capacity of human stem cells that initiated them.
Using previously published expression data sets of 26 different tissues and cell types (Table S2), we generated a list of 100 genes reflecting tissues from all three germ layers as well as placental genes, reflecting an extraembryonic tissue (Table S3). Genes were chosen according to tissue specificity, lack of expression in pluripotent cells, and sufficient expression in an initial database of karyotypically normal teratomas (n = 5). An additional set of ten pluripotent markers was also generated, as a control for undifferentiated PSCs. Tissue expression specificity was validated using the Amazonia! online database, which represents expression profile of over 300 cell and tissue types (Le Carrour et al., 2010). The scorecard includes genes specific for the central and peripheral nervous system as well as skin-specific genes to represent the ectoderm; genes specific to the joint, heart, skeletal muscle, adipose tissue, kidney, and bone marrow to represent the mesoderm; and genes specific to the gut, liver, lung, and pancreas representing the endoderm. We also included a subset of placental genes, representing an extraembryonic tissue. Following the gene list assembly, we looked at their expression in teratomas compared with undifferentiated hPSCs and previously published expression data of tumors from seven different tissues (Figure 1). Teratomas show expression of all tissues from all germ layers as well as placental genes. Importantly, the expression of the different lineages in these tumors was similar. As expected, undifferentiated hPSCs show little to no expression of the tissue-specific genes and a high expression of pluripotency markers. Similarly, tumors originating from different tissues and composed mainly of a single cell type were successfully detected by the scorecard as expressing their specific tissue and lineage. Our scorecard showed specific ectoderm lineage expression for tumors such as medulloblastoma and basal cell carcinoma, specific mesoderm lineage expression for tumors such as liposarcoma and acute lymphoblastic leukemia, specific endoderm lineage expression for tumors such as hepatocellular carcinoma and colon cancer, and the extraembryonic tissue expression for choriocarcinoma (Figure 1). Congruent results were obtained when the equivalent normal tissues were measured for the same scorecard (Figure S3).
Figure 1.
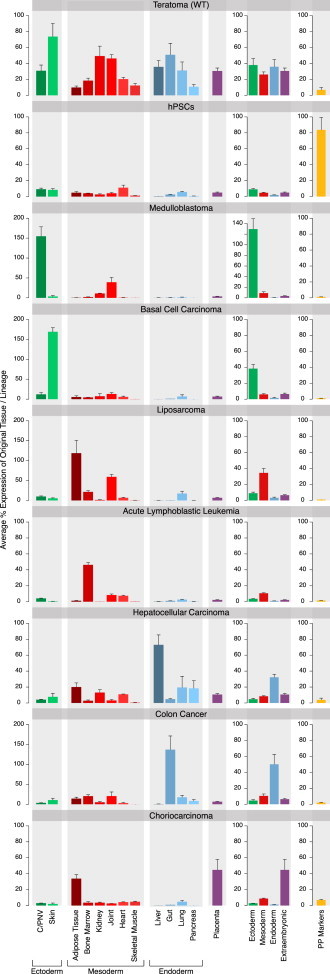
Average Tissue and Lineage Expression in Different Tumors and hPSCs
Average tissue gene expression was calculated in respect of its expression values in the original tissue. Lineage expression was calculated as the mean expression of all genes representing tissues in that lineage (n = 2 for basal cell carcinoma and choriocarcinoma; n = 5 for teratomas, medulloblastoma, liposarcoma, acute lymphoblastic leukemia, hepatocellular carcinoma, and colon cancer; n = 10 for hPSCs). Error bars represent SE. Teratomas show expression of all measured tissues and a similar expression of all lineages, while other tumors show a specific tissue and lineage expression. hPSCs show little to no expression of all the tissues. Pluripotent markers are highly expressed in hPSCs and show low to no expression in all tumors. C/PNV, central or peripheral nervous system.
In order to quantitatively estimate the pluripotency of the tumor initiating cells, we calculated the mean gene expression of each lineage and extraembryonic tissue, multiplied them, and divided by 1,000, producing a single score. This analysis, termed TeratoScore, essentially estimates the differentiation potency of the tumor initiating cells—the very goal of teratoma formation. In other words, TeratoScore validates the potential of cells to differentiate toward all three germ layers, hence being qualified as pluripotent cells. Comparing the TeratoScore outcomes of 14 different teratomas with these of tissue-specific tumors reveals that it can successfully detect the hPSC-derived tumors (Figure 2), with average grades of 1,186 ± 334 (n = 14) compared with 7.3 ± 2.6 (n = 34) of the cell-specific tumors and hPSCs. Of 29 tissue-specific tumors, only one had a grade higher than 50 (colon cancer, TeratoScore value of 77). In contrast, only 1 teratoma in the 14 examined had a grade lower than 100, suggesting that an outcome higher than 100 sufficiently implies a pluripotent cell line-derived tumor. A TeratoScore value lower than 50 thus indicates a tissue-specific tumor, and a value between 50 and 100 was a borderline indicator of a tumor originated from pluripotent cells (Figure 2). Teratoma composition analysis and the unified outcome using TeratoScore are available using our online resource at http://benvenisty.huji.ac.il/teratoscore.php by uploading an Affymetrix U133 Plus 2.0 microarray expression CEL file.
Figure 2.
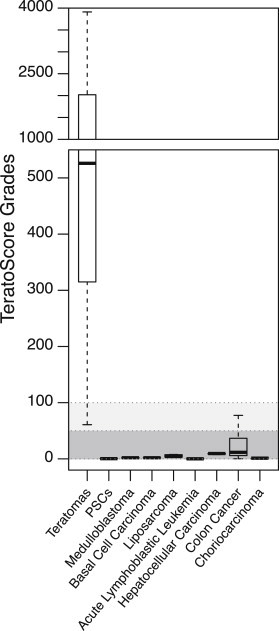
TeratoScore Grades of Different Tumors and PSCs
TeratoScore grades were calculated as the multiplication of the mean lineage gene expression of each tumor divided by 1,000. While teratomas show high and diverse TeratoScore grades (n = 14, mean = 1,186, SEM = 334), all other tumors and PSCs produce significantly lower ones (n = 34, mean = 7.3, SEM = 2.6, p < 10−6).
As this is the first time a substantial amount of expression data regarding teratomas is combined, we were interested in identifying changes between teratomas with a diploid karyotype and teratomas initiated by aneuploid cells. We first examined the differences using the gene scorecard composing the TeratoScore (Figure 3). While some differences were statistically significant, the overall tissue and lineage expression between normal and aberrant cell lines was similar. Teratomas created by hPSCs with a trisomy of chromosome 12 show a significant decrease in adipose tissue and bone marrow-specific gene expression compared with normal cell teratomas (p < 0.02 and p < 0.05, respectively) and lower kidney-specific gene expression compared with normal and trisomy 21 teratomas (p < 0.05 and p < 0.03, respectively). TeratoScore values calculated using these results were 1,490 ± 580 (n = 5) for euploid hPSC-derived teratomas, 1,798 ± 825 (n = 4) for teratomas derived from trisomy 21 hPSC, and 392 ± 117 (n = 5) for trisomy 12-initiated teratomas. Curiously, the mean TeratoScore value of teratomas derived from trisomy 12 hPSC is significantly lower than that of teratomas derived from euploid cells (p = 0.05).
Figure 3.
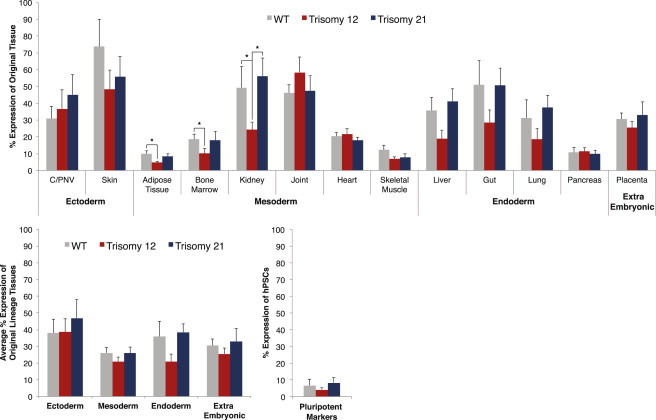
Average Tissue and Lineage Expression in WT and Chromosomally Aberrant Teratomas
Average gene expression was calculated in respect of its expression values in the original tissue. Lineage expression was calculated as the mean expression of all genes representing tissues in that lineage (∗p < 0.05). WT (n = 5) and chromosomally aberrant teratomas (trisomy 12, n = 5; trisomy 21, n = 4) show a similar tissue and lineage expression, with exception for significantly lower expression of genes specific for kidney, adipose tissue, and bone marrow in teratomas with trisomy of chromosome 12 compared with WT. Error bars represent SE.
A live newborn with full trisomy 12 was never reported, suggesting that it is embryonic lethal in humans. However, individuals carrying mosaic tetrasomy of 12p were reported showing a variety of abnormalities (Segel et al., 2006). This condition, known as Pallister-Killian mosaic syndrome, is extremely rare and has a range of developmental and behavioral outcomes (Kostanecka et al., 2012). Individuals carrying chromosome 21 trisomy also show a distinct set of abnormalities alongside intellectual disabilities, a condition known as Down syndrome (Patterson, 2009). With such an atypical in vivo development, we hypothesized that teratomas originating from cells carrying these mutations will show a more substantial differential gene expression. We suspected that the lack of significant changes originated from the small number of examined genes. While our 100-gene platform is sufficient for the TeratoScore analysis and defining a cell as pluripotent, it is not capable of identifying minor expression changes and tissue distribution between euploid and aneuploid cells.
We therefore used another more sensitive bioinformatic strategy, establishing lists of the 200 most expressed genes in each tissue and looking for the ones most altered in chromosomally aberrant teratomas (Figure 4). In this analysis, trisomy 12-derived teratomas show a significant number of upregulated genes enriched for joint, spinal cord, brain, and fetal brain (p < 0.001) and a significant number of downregulated genes enriched for skeletal muscle, lung, small intestine, colon, fetal liver, and liver (p < 0.001) compared with normal karyotype teratomas. This wide set of gene expression alteration might suggest a pleiotropic effect of trisomy 12 on developmental patterns, contributing to its embryonic lethality. In contrast, teratomas initiated from cells with trisomy 21 show a more modest distortion of gene expression with a significant number of upregulated genes enriched for brain, fetal brain, and spinal cord (p < 0.001), as well as for joint, and a significant number of downregulated genes for skeletal muscle only compared with normal karyotype teratomas. The upregulation in brain genes in these teratomas is compatible with the neural phenotypes (such as mental impairment) observed in Down syndrome patients.
Figure 4.
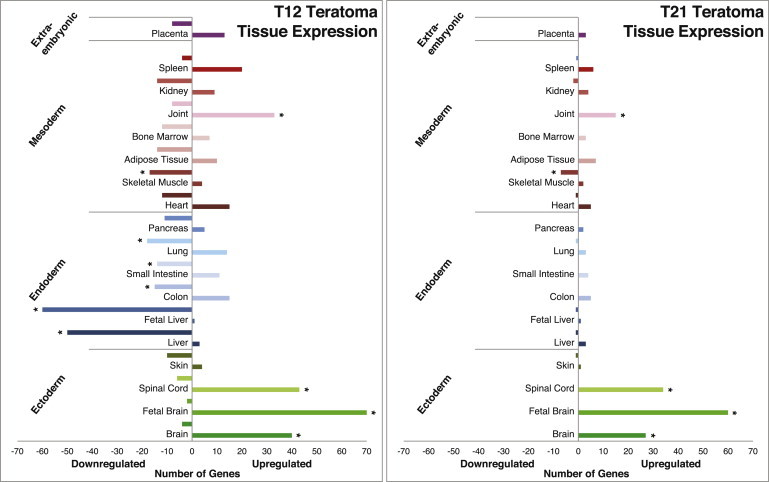
Tissue Classification of Genes Expressed Differently between WT and Aberrant Teratomas
Number of genes differentially expressed in chromosomally aberrant teratomas compared with teratomas initiated from diploid cells. Gene were considered differentially expressed when changed over 2-fold compared with control (∗p < 0.001). Teratomas from cells carrying trisomy of chromosome 12 (T12, n = 5) showed a significant number of upregulated genes enriched for brain, fetal brain, spinal cord, and joint as well as downregulated genes enriched for liver and fetal liver, colon, small intestine, lung, and skeletal muscle compared with WT teratomas (n = 5). In contrast, teratomas from cells carrying trisomy of chromosome 21 (T21, n = 4) showed significant number of downregulated genes specific for skeletal muscle only, with a significant number of upregulated genes enriched for brain, fetal brain, spinal cord, and joint.
Discussion
Teratoma formation is the most abundant method to demonstrate hPSC differentiation capacity and a critical step when defining its potency (Müller et al., 2010). TeratoScore transforms this qualitative obligatory milestone into a quantitative tool. Although alternatives have been suggested to replace teratoma formation, they all suffer from imperfections. The computational method PluriTest has shown promising results identifying PSCs and differentiating them from cancer stem cells and cells that have underwent differentiation (Müller et al., 2011). While this method appears simpler than in vivo or in vitro differentiations, it can only determine a cell resemblance to PSCs in their undifferentiated state. The uniqueness of PSCs, however, is in their ability to transform from the pluripotent state to terminally differentiated cell types. These differentiation processes require master regulators that are not expressed in PSCs. A mutation in one of these genes will not be exposed using PluriTest, claiming that a cell is pluripotent even though it cannot differentiate toward all germ layers. Differentiation per se can be evaluated using directed and spontaneous in vitro protocols. Directed differentiation protocols toward many cell types and tissues have been published, ranging from neurons to pancreatic progenitors (reviewed in Schuldiner and Benvenisty, 2003; Tabar and Studer, 2014). Each protocol requires a set of specific factors and environment, presenting expensive and complex challenges to assess cell potency. Furthermore, there is no agreed-upon assembly of protocols which successful utilization is sufficient in determining cells’ pluripotency. In contrast, spontaneous differentiation is widely used and relatively simple to perform. EB formation has been suggested as a good replacement for teratoma formation. However, EBs do not always show complex differentiated structures and usually encapsulate a core of undifferentiated cells. When compared with teratomas, EBs show a lesser extant of mature tissue differentiation (Ozolek and Castro, 2011)—a difference that might provide a limited estimation of the differentiation capabilities. Teratoma formation enables differentiation to a wide variety of cells and tissues, presenting mature and complex structures. Although it is far from being a perfect assay, teratoma formation is still the best evaluator of pluripotent assessment.
Using expression data from teratomas, we were able to identify a large variety of differentiated tissues within them. This allowed us to analyze a repertoire of differentiated cell types in each embryonic germ layer. Teratomas initiated from PSCs express characteristic genes of all analyzed tissues and a similar expression of all lineages. Other tumors are typically composed of uniform and homogenous tissues, as seen by our 100-gene scorecard. We show that tumors originating from all lineages as well as from extraembryonic tissues express the respective genes in an exclusive manner. Weighing this expression creates a score that successfully separates PSC-derived mature tumors from tumors originating from other cell types. While this separation is evident, it is unclear why some teratomas provide a much higher grade in the TeratoScore analysis. We were not able to find a common ground for high or low scores, looking at karyotype integrity or genetic identity. In our study, we have analyzed two sets of different teratomas generated from the same cell types (CSES22 and CSES45; see Table S1). The TeratoScore of both pairs of teratomas was higher than 100, affirming the cell lines’ pluripotency, but the numbers were somewhat different (160 and 315 for CSES22-initiated tumors and 234 and 429 CSES45-initiated tumors). Examining a much larger teratoma sample from various sources and passages, including partially reprogrammed cells, might aid in identifying a biological origin for TeratoScore performance variation. It is possible that some of the variance originates from TeratoScore’s limited sensitivity. Although we show that a 100-gene scorecard is enough to discover the existence or absence of tissues and lineage, it is insufficient in identifying possible lineage differentiation biases that might affect the TeratoScore outcome. This is also true for the measurement of pluripotency markers’ expression. Pluripotent foci were previously shown to exist in teratomas initiated from trisomy 12 cells (Ben-David et al., 2014). While a small subset of pluripotent markers could not detect residual undifferentiated PSCs, analysis of the top 200 genes expressed in hPSCs revealed that 25 of them were upregulated T12 teratomas compared with WT teratomas (p < 0.06, following Bonferroni correction for multiple testing) compared with only 5 in T21 teratomas (p < 0.8).
Several studies have analyzed the differentiation propensity of hPSCs in vitro, suggesting that different cell lines have differentiation bias, resulting in unequal ability to differentiate toward specific cell types (Bock et al., 2011; Boulting et al., 2011; Osafune et al., 2008). A method to evaluate these biases has been proposed (Bock et al., 2011), and their origins have been suggested to be both genetic and epigenetic (reviewed in Cahan and Daley, 2013). Furthermore, the International Stem Cell Initiative (ISCI) (Andrews et al., 2005) is currently conducting a large-scale study to compare different methods to analyze the pluripotency of human stem cells. Our aim in generating TeratoScore was to give a quantitative measure to differentiation of PSCs in vivo as a way to evaluate the pluripotency of the cells. Teratomas have long been argued to be an intriguing developmental model; however, the extent of their resemblance to typical development is under debate (Ozolek and Castro, 2011). Using karyotypically abnormal cells, we were able to show significant changes in tissue-enriched gene expression. Although these changes were not very significant in the 100-gene scorecard made to identify pluripotent origins, they were highly significant when looking at the top 200 genes expressed in each tissue. Additional copies of chromosomes 12 or 21 caused a substantial increase in expression of genes related to the CNS. This upregulation may indicate these aneuploidies influence in vivo, for it is known that both conditions cause substantial intellectual disabilities (Chapman and Hesketh, 2000; Segel et al., 2006). Furthermore, case studies have reported joint and muscle malfunction in patients with Pallister-Killian mosaic syndrome (Speleman et al., 1991)—another significant trait in our analysis. It is important to note that we could not find a report of patients bearing a complete trisomy of chromosome 12. We can cautiously assume that this aberration is lethal—perhaps due to extant of developmental abnormalities implied in our analysis. In contrast, trisomy 21 seems to cause a significant downregulation in skeletal muscle-specific genes only—perhaps correlating with the successful birth of Down syndrome patients and their typical muscle hypotonia (Cisterna et al., 2013).
It is therefore still unclear how well a teratoma can reflect developmental processes in vivo. More data regarding these and other genetic conditions need to be collected before reaching final conclusions in this matter. However, we find these results encouraging in reflecting the implications chromosomal aberrations have on embryonic development.
Our scorecard and analysis are currently based on a specific microarray platform, but they might be applicable to other methods as well. Our 100-gene scorecard can be measured using a standard quantitative real-time PCR platform, generating a TaqMan low-density array (TLDA). As RNA sequencing is becoming more common and less expensive, it will also allow a reevaluation of our scorecard and TeratoScore analysis. This might enable a more elaborated analysis of teratomas, alongside additional biological material and data.
Our algorithm provides a fast and quantitative evaluation of cell potency. As teratomas are a reliable test for cell differentiation capacity, it is crucial to use this type of quantitative tools to evaluate and compare them. TeratoScore provides an estimation of tissue expression and lineage distribution, as well as a grade determining pluripotency. We show that teratomas originating from karyotypically aberrant cells present an aberrant gene expression, indicating that they might serve disease research and modeling.
Experimental Procedures
Cell Culture
hPSCs were cultured and karyotyped using standard conditions as previously described (Ben-David et al., 2014). See also Supplemental Experimental Procedures.
Tumor Formation and Analysis
Teratomas derived from hPSCs were generated as previously described (Ben-David et al., 2014). Half of the tumor pieces were mixed together and taken for RNA extraction, while the rest were saved for H&E staining, carried out as previously described (Kopper et al., 2010). See also Supplemental Experimental Procedures. All experimental procedures in animals were in accordance with institutional guidelines.
Microarray Analysis
Microarray data of the various body tissues, hPSC, and previously published teratomas and tumors were downloaded from the GEO database (Table S2). An expression ratio was calculated between each tissue and the average of all other body tissues, indicating tissue specificity. Scorecard genes for each tissue were chosen by having a high tissue specificity (over 3-fold) and by being expressed in teratoma but not in hPSCs. Average expression for each lineage was calculated as the mean expression of all genes representing that lineage. TeratoScore grades were calculated as the multiplication of these means and dividing this product by 103. Top-200 tissue-enriched genes were chosen as the top unique genes in the tissue/body ratio mentioned above. Statistical calculations and figures were made in R programming language (http://www.r-project.org). See also Supplemental Experimental Procedures.
Acknowledgments
The authors would like to thank Uri Weissbein, Ido Sagi, and Carmel Braverman-Gross for critically reading the manuscript. N.B. is the Herbert Cohn Chair in Cancer Research. This work was partially supported by the Israel Science Foundation (grant number 269/12), by The Rosetrees Trust, and by The Azrieli Foundation.
Published: June 9, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Accession Numbers
The NCBI GEO accession numbers for the microarray data reported in this paper are GEO: GSM1576685 and GSE51455 (Table S1).
Supplemental Information
Expression profiles of tissues representing the three germ layers, the placenta, and embryonic stem cells used to develop the TeratoScore are listed with their GEO sample and series identifications. Tumors expression profiles used to compare TeratoScores are also listed with their GEO sample and series identifications.
One-hundred genes were selected to represent tissues from all three germ layers and extraembryonic tissue in order to calculate tumors’ TeratoScore. Genes enriched for each tissue were selected computationally and verified as being tissue specific using Amazonia! online database (Le Carrour et al., 2010). Tissues with similar gene expression were merged into a single larger category. An additional set of ten genes representing pluripotent markers was selected. These genes are not taken into account when calculating TeratoScore. Listed are gene symbols, approved names, and chromosomal locations.
Tissue-specific genes differentially expressed in chromosomally aberrant teratomas compared with teratomas initiated from diploid cells. Of the top 200 expressed genes in each tissue, a gene was considered differentially expressed when changed over 2-fold (either upregulated or downregulated) compared with control.
References
- Andrews P.W., Benvenisty N., McKay R., Pera M.F., Rossant J., Semb H., Stacey G.N., Steering Committee of the International Stem Cell Initiative The International Stem Cell Initiative: toward benchmarks for human embryonic stem cell research. Nat. Biotechnol. 2005;23:795–797. doi: 10.1038/nbt0705-795. [DOI] [PubMed] [Google Scholar]
- Ben-David U., Arad G., Weissbein U., Mandefro B., Maimon A., Golan-Lev T., Narwani K., Clark A.T., Andrews P.W., Benvenisty N., Carlos Biancotti J. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat. Commun. 2014;5:4825. doi: 10.1038/ncomms5825. [DOI] [PubMed] [Google Scholar]
- Biancotti J.C., Narwani K., Buehler N., Mandefro B., Golan-Lev T., Yanuka O., Clark A., Hill D., Benvenisty N., Lavon N. Human embryonic stem cells as models for aneuploid chromosomal syndromes. Stem Cells. 2010;28:1530–1540. doi: 10.1002/stem.483. [DOI] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z.D., Ziller M., Croft G.F., Amoroso M.W., Oakley D.H. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulting G.L., Kiskinis E., Croft G.F., Amoroso M.W., Oakley D.H., Wainger B.J., Williams D.J., Kahler D.J., Yamaki M., Davidow L. A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., Daley G.Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., Li H., Morris S.A., Lummertz da Rocha E., Daley G.Q., Collins J.J. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R.S., Hesketh L.J. Behavioral phenotype of individuals with Down syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 2000;6:84–95. doi: 10.1002/1098-2779(2000)6:2<84::AID-MRDD2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Chin M.H., Mason M.J., Xie W., Volinia S., Singer M., Peterson C., Ambartsumyan G., Aimiuwu O., Richter L., Zhang J. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisterna B., Costanzo M., Scherini E., Zancanaro C., Malatesta M. Ultrastructural features of skeletal muscle in adult and aging Ts65Dn mice, a murine model of Down syndrome. Muscles Ligaments Tendons J. 2013;3:287–294. [PMC free article] [PubMed] [Google Scholar]
- Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- Kopper O., Giladi O., Golan-Lev T., Benvenisty N. Characterization of gastrulation-stage progenitor cells and their inhibitory crosstalk in human embryoid bodies. Stem Cells. 2010;28:75–83. doi: 10.1002/stem.260. [DOI] [PubMed] [Google Scholar]
- Kostanecka A., Close L.B., Izumi K., Krantz I.D., Pipan M. Developmental and behavioral characteristics of individuals with Pallister-Killian syndrome. Am. J. Med. Genet. A. 2012;158A:3018–3025. doi: 10.1002/ajmg.a.35670. [DOI] [PubMed] [Google Scholar]
- Lavon N., Narwani K., Golan-Lev T., Buehler N., Hill D., Benvenisty N. Derivation of euploid human embryonic stem cells from aneuploid embryos. Stem Cells. 2008;26:1874–1882. doi: 10.1634/stemcells.2008-0156. [DOI] [PubMed] [Google Scholar]
- Le Carrour T., Assou S., Tondeur S., Lhermitte L., Lamb N., Rème T., Pantesco V., Hamamah S., Klein B., De Vos J. Amazonia!: An Online Resource to Google and Visualize Public Human whole Genome Expression Data. Open Bioinformatics J. 2010;4:5–10. [Google Scholar]
- Müller F.J., Goldmann J., Löser P., Loring J.F. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell. 2010;6:412–414. doi: 10.1016/j.stem.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Müller F.J., Schuldt B.M., Williams R., Mason D., Altun G., Papapetrou E.P., Danner S., Goldmann J.E., Herbst A., Schmidt N.O. A bioinformatic assay for pluripotency in human cells. Nat. Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F.J., Brandl B., Loring J.F. Assessment of human pluripotent stem cells with PluriTest. In: The Stem Cell Research Community, editor. StemBook. Harvard Stem Cell Institute; Cambridge, MA: 2012. [PubMed] [Google Scholar]
- Narwani K., Biancotti J.C., Golan-Lev T., Buehler N., Hill D., Shifman S., Benvenisty N., Lavon N. Human embryonic stem cells from aneuploid blastocysts identified by pre-implantation genetic screening. In Vitro Cell. Dev. Biol. Anim. 2010;46:309–316. doi: 10.1007/s11626-010-9303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K., Caron L., Borowiak M., Martinez R.J., Fitz-Gerald C.S., Sato Y., Cowan C.A., Chien K.R., Melton D.A. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Ozolek J.A., Castro C.A. Teratomas derived from embryonic stem cells as models for embryonic development, disease, and tumorigenesis. In: Kallos M., editor. Embryonic Stem Cells-Basic Biology to Bioengineering. InTech; 2011. [Google Scholar]
- Patterson D. Molecular genetic analysis of Down syndrome. Hum. Genet. 2009;126:195–214. doi: 10.1007/s00439-009-0696-8. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Benvenisty N. Factors controlling human embryonic stem cell differentiation. Methods Enzymol. 2003;365:446–461. doi: 10.1016/s0076-6879(03)65031-7. [DOI] [PubMed] [Google Scholar]
- Segel R., Peter I., Demmer L.A., Cowan J.M., Hoffman J.D., Bianchi D.W. The natural history of trisomy 12p. Am. J. Med. Genet. A. 2006;140:695–703. doi: 10.1002/ajmg.a.31143. [DOI] [PubMed] [Google Scholar]
- Speleman F., Leroy J.G., Van Roy N., De Paepe A., Suijkerbuijk R., Brunner H., Looijenga L., Verschraegen-Spae M.R., Orye E. Pallister-Killian syndrome: characterization of the isochromosome 12p by fluorescent in situ hybridization. Am. J. Med. Genet. 1991;41:381–387. doi: 10.1002/ajmg.1320410321. [DOI] [PubMed] [Google Scholar]
- Tabar V., Studer L. Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nat. Rev. Genet. 2014;15:82–92. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression profiles of tissues representing the three germ layers, the placenta, and embryonic stem cells used to develop the TeratoScore are listed with their GEO sample and series identifications. Tumors expression profiles used to compare TeratoScores are also listed with their GEO sample and series identifications.
One-hundred genes were selected to represent tissues from all three germ layers and extraembryonic tissue in order to calculate tumors’ TeratoScore. Genes enriched for each tissue were selected computationally and verified as being tissue specific using Amazonia! online database (Le Carrour et al., 2010). Tissues with similar gene expression were merged into a single larger category. An additional set of ten genes representing pluripotent markers was selected. These genes are not taken into account when calculating TeratoScore. Listed are gene symbols, approved names, and chromosomal locations.
Tissue-specific genes differentially expressed in chromosomally aberrant teratomas compared with teratomas initiated from diploid cells. Of the top 200 expressed genes in each tissue, a gene was considered differentially expressed when changed over 2-fold (either upregulated or downregulated) compared with control.


