Summary
Dysfunction of the specialized cardiac conduction system (CCS) is associated with life-threatening arrhythmias. Strategies to derive CCS cells, including rare Purkinje cells (PCs), would facilitate models for mechanistic studies and drug discovery and also provide new cellular materials for regenerative therapies. A high-throughput chemical screen using CCS:lacz and Contactin2:egfp (Cntn2:egfp) reporter embryonic stem cell (ESC) lines was used to discover a small molecule, sodium nitroprusside (SN), that efficiently promotes the generation of cardiac cells that express gene profiles and generate action potentials of PC-like cells. Imaging and mechanistic studies suggest that SN promotes the generation of PCs from cardiac progenitors initially expressing cardiac myosin heavy chain and that it does so by activating cyclic AMP signaling. These findings provide a strategy to derive scalable PCs, along with insight into the ontogeny of CCS development.
Highlights
-
•
A chemical screen was carried out for compounds that induce cardiac conduction cells
-
•
Two ESC reporter lines were used to identify lead hits
-
•
Sodium nitroprusside efficiently generated scalable amounts of PC-like cells
-
•
By activating cAMP signaling, PCs are derived from cardiac progenitors
Evans, Chen, Fishman, and colleagues devised a strategy to enhance the generation of otherwise rare cardiac conduction system cells. They carried out a chemical screen, using murine ESCs, and found a small molecule, sodium nitroprusside, that could efficiently generate cells with a cardiac Purkinje cell (PC) phenotype. The PCs are generated from MHC-expressing cardiac cells, through activation of cAMP signaling.
Introduction
The cardiac conduction system (CCS) is composed of the impulse-generating sino-atrial (SA) and atrio-ventricular (AV) nodes and the impulse-propagating His-Purkinje system, which play critical roles initiating and regulating excitation and contraction of the cardiac chambers. Dysfunction of the CCS caused by disease, congenital malformations, aging, or heritable gene defects results in cardiac arrhythmias, a major cause of morbidity and mortality worldwide (Spooner et al., 2001). Current treatments of cardiac arrhythmias include pharmacological therapy and implantable rhythm-management devices, such as pacemakers and defibrillators. However, these treatments can have low efficacy and also cause additional problems, such as pro-arrhythmia side effects, infection, and device malfunction (Holzmeister and Leclercq, 2011; Woods and Olgin, 2014). Devices placed into pediatric patients must be replaced as heart development proceeds throughout life. Thus, there is a clear and compelling need to develop strategies to treat patients with CCS defects.
Genetic and cell-based therapeutic approaches have focused on generation of biological pacemakers (Cho and Marbán, 2010; Rosen et al., 2004). Embryonic stem cells (ESCs) are a feasible starting material, as they are pluripotent and can be differentiated to any cell lineage. Several ESC reporter lines have been engineered and used to isolate CCS cells, including (1) HCN4p-EGFP+ cells that express transcripts for CCS markers Hcn4, Cav3.2, and Cx40 and display a hyperpolarization-activated depolarizing current (termed the funny current; Morikawa et al., 2010); (2) an ESC reporter line containing a Mink:lacz transgene and a reporter regulated by a chicken Gata6 (cGata6) enhancer; the double-positive cells display pacemaker-like morphology and show pacemaking action potential waveform as well as a funny current (White and Claycomb, 2005); (3) a Shox2 promoter with a Cx30.2 enhancer to generate ESC-derived SHOX2+ and Cx30.2+ cardiomyocytes that express additional CCS markers (Hcn4, Cx45, Cx30.2, Tbx2, and Tbx3) and exhibit pacemaker-nodal phenotype (Hashem and Claycomb, 2013); (4) ESC-derived Nkx2.5:GFP+ cells expressing Hcn1 and Hcn4 that display the funny current (Yano et al., 2008); and (5) an ESC reporter line derived from Contactin2:EGFP BAC transgenic mice (Cntn2:egfp) was recently used to isolate rare cells that display transcriptional signatures and functional properties comparable to endogenous cardiac Purkinje cells (PCs) (Maass et al., 2015). Finally, CD166 has been identified as a CCS cell-surface marker (Scavone et al., 2013), although it is not specific to CCS constituents.
ESCs can also be directed into CCS-like cells using chemicals and small-molecule compounds. For example, vitamin B12 promoted the expression of CCS markers Cx40 and Hcn4 at an early stage of cardiomyocyte differentiation (Saito et al., 2009). Treating ESCs with Ca2+-activated potassium channel (SKCa) activator, 1-ethyl-2-benzimidazolinone (EBIO), or suramin promoted a nodal-like cell phenotype (Kleger et al., 2010; Wiese et al., 2011). Hence, cell-permeable small molecules that modulate functions of specific pathways provide a convenient and efficient approach to control stem/progenitor cell fate. Importantly, these small molecules provide new tools to dissect molecular mechanisms that control embryonic development, therefore facilitating a better understanding for functions of relevant signaling pathways. However, overall efficiency of generating CCS cells using any of the current protocols is poor (typically below 1% of the culture). Thus, developing an efficient strategy to derive CCS cells will not only facilitate developing disease models for mechanistic studies and drug discovery but also provide new cellular materials for regenerative therapy.
Here, we describe a high-throughput screen of ∼5,000 compounds using an ESC line derived from the CCS:lacz reporter mouse, containing a transgene that fortuitously marks cells of the CCS lineage (Rentschler et al., 2001). We discovered that the small molecule sodium nitroprusside (SN) efficiently enhances the generation of CCS cells from ESCs. The screen was validated using an additional reporter line, with GFP expression driven by a Contactin2 (Cntn2) transgene (Maass et al., 2015). The derived GFP+ cells induced by SN display an expression profile, molecular markers, and functional properties that are comparable to endogenous PC of the ventricular CCS. Importantly, we found that SN promotes the generation of PC-like cells through activating cyclic AMP (cAMP) signaling in embryonic cardiac cells, providing new insight into the ontogeny of CCS lineages.
Results
A High-Throughput Screen to Identify Compounds that Promote CCS Fate
The ESC reporter line CCS:Lacz was used to screen for small molecules that promote the generation of CCS cells, in the context of a directed differentiation assay. This reporter line was derived from the CCS:Lacz transgenic mouse strain carrying a β-galactosidase (Lacz) reporter gene that had randomly inserted near the Slco3A1 locus and that shows robust β-galactosidase expression in all cells of the developing and mature CCS, including the SA node, AV node, His bundle, bundle branches, and Purkinje fibers (Rentschler et al., 2001; Stroud et al., 2007). Since CCS components are derived from cardiac progenitors (Laflamme and Murry, 2011), the first step of the assay was to direct the ESCs toward commitment of cardiac mesoderm. Cardiac progenitors are marked by co-expression of FLK1 (KDR, VEGFR2) and PDGFR-α, which is enhanced in the presence of optimized levels of the growth factors BMP4 and ACTIVIN-A (Kattman et al., 2011). Therefore, cytokine induction conditions were first established for the CCS:Lacz ESC line, in which the double-positive (FLK1+ and PDGFR-α+) cell population was efficiently induced (Figure S1). The line was then used to screen under these conditions for subsequent enhanced generation of LacZ expression (see Figure 1A and the Experimental Procedures for details of the assay).
Figure 1.
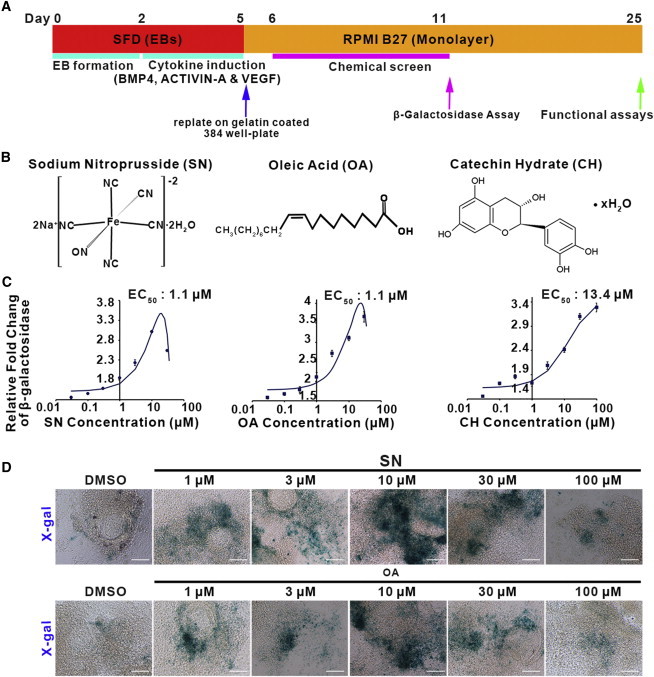
High-Throughput Screening and Characterization of Hit Compounds
(A) Scheme of high-throughput screen of CCS cell differentiation. CCS:Lacz ESCs were suspended in serum-free differentiation (SFD) medium without cytokines for 2 days and allowed to form embryoid bodies (EBs). EBs were then dissociated and reaggregated in SFD medium for 3 days with the defined cardiac mesoderm cytokine induction cocktail. At day 5, EBs were harvested and dissociated and cells re-plated on gelatin-coated 384-well plates at a density of 5,000 cells/well in cardiomyocyte medium (RPMI with B27).
(B) Chemical structures of top hit compounds: sodium nitroprusside (SN), oleic acid (OA), and catechin hydrate (CH).
(C) Efficacy curves of SN, OA and CH. Error bars show SD.
(D) X-gal staining shows β-galactosidase expression under different doses of either SN or OA treatment, as indicated. DMSO was used as a control. β-galactosidase expression is shown in blue.
Scale bar, 200 μm. See also Figure S1.
To perform high throughput screening, we added a single compound from a library containing 4,880 chemicals to each well in a 384-well format. The library is composed of annotated compounds including signaling pathway regulators, kinase inhibitors, and Food and Drug Administration (FDA)-approved drugs. Cells were screened at two concentrations for each compound (10 μM or 1 μM). After 5 days of chemical treatment, cells were lysed to quantify β-galactosidase activity relative to cells treated with DMSO alone, which served as negative controls. 96 compounds caused at least a 2.5-fold increase in β-galactosidase activity compared to DMSO controls and were chosen as primary hits for further analysis (Figure S1). We focused on 15 primary-hit compounds that had effects under 10 μM (Table S1), and these were re-examined using the primary screening platform. Of these, three compounds (SN, oleic acid [OA], and catechin hydrate [CH]) reproducibly enhanced β-galactosidase activity significantly at both concentrations and were therefore chosen for further study.
For validation, these three compounds (Figure 1B) were re-ordered and tested by serial dilution to generate efficacy curves and to determine their half maximal effective concentrations (EC50). Consistently, these three hits enhanced β-galactosidase activity in a dose-dependent manner. Two hit compounds, SN and OA, showed effects at relatively low effective concentration (SN, EC50 = 1.1 μM; OA, EC50 = 1.1 μM), while the EC50 of CH was ∼10-fold higher than the other two compounds (Figure 1C). When cells were stained directly in situ, both SN and OA strongly increased X-Gal staining (Figure 1D). However, CH failed to enhance the X-Gal signal and was therefore not investigated further.
SN Significantly Enhances Generation of CCS Cells
To rule out that the compounds were simply activating expression of the CCS:lacz reporter gene, we used a second CCS reporter line to examine the effect of SN and OA on cardiac progenitors. Contactin2 (Cntn2) is specifically expressed in cells comprising the CCS of the mouse heart including the SA node, AV node, His bundle, bundle branches, and Purkinje fibers (Pallante et al., 2010). The Cntn2:egfp line is derived from Cntn2:egfp BAC transgenic mice that were shown to faithfully reproduce the endogenous Cntn2 expression pattern (Pallante et al., 2010). A full description and validation of this reporter line have been reported recently (Maass et al., 2015). Again, growth factor concentrations were defined using this ESC line to promote a significant double-positive (FLK1+ and PDGFR-α+) cell population, which was found to be similar as for the CCS:Lacz line (Figure S1). The same differentiation protocol was used as for the primary screening (Figure 1A), except that the cells were replated on gelatin-coated plates at differentiation day 4 instead of day 5 to enhance the survival of Cntn2:egfp-derived cardiac progenitors. After 5 days of treatment, SN or OA were removed from the media. With this protocol, beating cell clusters were routinely observed at differentiation day 7–8, and this was not obviously affected by the addition of compounds. At differentiation day 25, cells were co-stained using antibodies to GFP in addition to the cardiac conduction marker HCN4. By immunofluorescence, both SN (Figure 2A) and OA (Figure S2A) highly increased the percentage of cells expressing GFP and HCN4 compared to DMSO controls.
Figure 2.
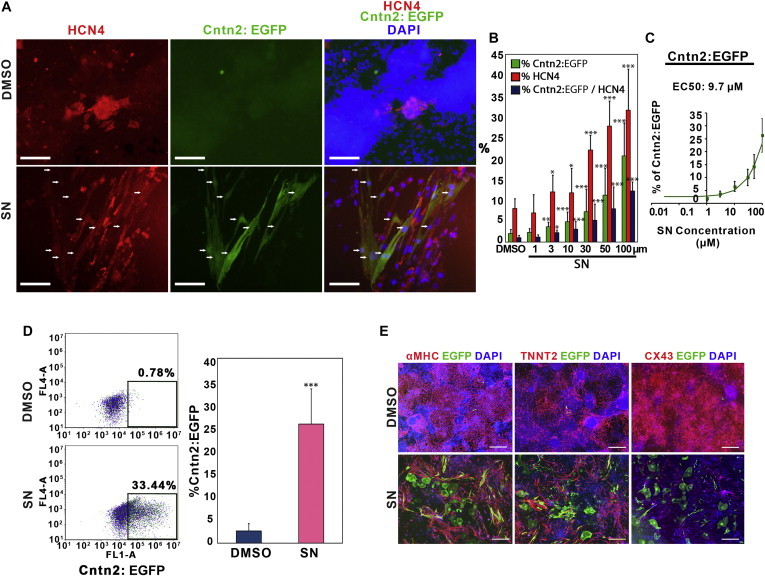
Sodium Nitroprusside Enhances Generation of Cardiac Conduction System Cells
(A) Expression of cardiac conduction system markers, HCN4 and Cntn2:EGFP, was examined by immunofluorescence staining using antibodies against either HCN4 or GFP. Scale bar, 200 μm. DMSO was used as a control. Arrows indicate cells that are double positive deriving from the SN-treated cultures.
(B) Quantification of immunofluorescence staining of HCN4+, Cntn2:EGFP+, and HCN4+/Cntn2:EGFP+ double-positive cells by MetaExpress image analysis. Results represent the combined data from three independent differentiation experiments. Error bars show SD.
(C) Efficacy curve of SN in the Cntn2:egfp cell line. Results represent the combined data from three independent differentiation experiments. Error bars show SD.
(D) Flow Cytometry analysis of Cntn2:EGFP expression. Following 5 days treatment of either SN or DMSO, cells were harvested at day 25 of differentiation. Quantification of Cntn2:EGFP expression is shown in the right panel. Results are from five independent experiments. Error bars show SD.
(E) SN- or DMSO-treated cells co-express cardiomyocyte markers αMHC or TNNT2 with Cntn2:EGFP. Cntn2:EGFP+ cells express relatively weak levels of CX43 (right panel). Scale bar, 200 μm.
Asterisks indicate significant differences compared with DMSO. Statistical significance is indicated: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S2.
The percentages of HCN4+, Cntn2:EGFP+, and HCN4+/Cntn2:EGFP+ double-positive cells were quantified by MetaExpress image analysis. SN significantly increased the percentage of all three classes of cells in a dose-dependent manner (Figure 2B) and with an EC50 of 9.7 μM for generation of Cntn2:EGFP+ cells (Figure 2C), which we note is somewhat higher than found for LacZ expression using the initial reporter line. While OA significantly enhanced HCN4-expressing cells at 100 μM, it failed to increase either Cntn2:EGFP+ or HCN4+/Cntn2:EGFP+ double-positive cells (Figure S2A). Assessed by flow cytometry, SN increased nearly 10-fold the percentage of Cntn2:EGFP+ cells at day 25 of differentiation (25.7% ± 7.6%) compared to DMSO controls (2.6% ± 1.6%) (see representative plots in Figure 2D). Cell cultures were also co-stained for the expression of cardiomyocyte markers: α myosin heavy chain (αMHC), troponin T2 (TNNT2), and connexin 43 (CX43). DMSO-treated cells expressed high levels of all three cardiac markers but generated few Cntn2:EGFP+ cells (Figure 2E, upper panel), showing that the protocol efficiently induced cardiomyocyte differentiation, as expected. In the SN-treated cells, most of the GFP+ cells co-stained for cardiomyocyte markers, although many Cntn2:EGFP+ cells appeared relatively low for levels of αMHC and TNNT2 (Figure 2E). In addition, the Cntn2:EGFP+ cells expressed Cx43 at relatively weak levels compared to DMSO controls (Figure 2E). These results suggest that SN can significantly enhance the differentiation from cardiac progenitors to cardiac conduction cell fate.
There are limitations to using these markers for reading out CCS differentiation. Although the differentiation protocol should not promote neurogenesis, Cntn2 and Hcn4 are expressed in neurons as well as cardiac cells (DiFrancesco, 1985; Fain et al., 1978; Furley et al., 1990). Therefore, it was important to rule out the possibility that SN was promoting neural fate differentiation. After allowing cells to undergo either spontaneous differentiation (which generates some differentiated neurons) or following the cardiac differentiation including SN, cells were stained for expression of TUBB3, a neural marker (Burgoyne et al., 1988). Many TUBB3+ cells were found in samples from ESCs undergoing spontaneous differentiation (Figure S2B, left panel), while in contrast, no Cntn2:EGFP+ or HCN4+ cells co-stained as TUBB3+ cells (Figure S2B, right panel). To further confirm this observation, we quantified the expression levels of additional neuronal genes, including Mash1, Pax3, Sox3, and Tubb3, in RNA derived either from cells undergoing spontaneous differentiation or from SN-induced Cntn2:EGFP+ cells, using real-time qPCR. The results confirmed that SN-induced Cntn2:EGFP+ cells do not express neural genes (Figure S2C). Thus, SN significantly enhances the generation of cells expressing CCS markers HCN4 and CNTN2, and these HCN4+ and Cntn2:EGFP+ cells are not neural cells.
SN-Induced Cntn2:EGFP+ Cells Display a PC-like Action Potential Phenotype
Cntn2:EGFP+ cells derived from the Cntn2:egfp reporter ESC line were shown to display functional properties comparable to endogenous PC, including action potentials, intracellular calcium cycling, and chronotropic behavior (Maass et al., 2015). To confirm that these SN-induced Cntn2:EGFP+ cells display the same functional PC phenotype, we used the whole-cell patch-clamp technique to investigate their electrophysiologic properties. It has been reported that PCs show a unique action potential morphology and prolonged duration compared to ventricular myocytes (Pallante et al., 2010). Hence, the action potential durations were compared between Cntn2:EGFP− and Cntn2:EGFP+ cells from differentiation cultures that had been induced with SN (Figure 3A). Although we did not confirm with an independent reporter, all patched cells from which action potentials were measured are cardiomyocytes and presumably MHC positive. Compared with GFP− cells, the APD50 and APD90 values were found to be increased in Cntn2:EGFP+ cells 2.5-fold and 2.0-fold, respectively (Figure 3B), comparable to previously reported differences between GFP− and GFP+ cells differentiated in the absence of SN (Maass et al., 2015). Also, the action potential amplitude was slightly higher and the maximal rate of rise (dV/dtmax) of the action potential upstroke was greater in Cntn2:EGFP+ cells (Table S2). Since there was no significant difference in the resting potentials between GFP− and GFP+ cells (Table S2), the faster action potential upstrokes suggest higher sodium current expression levels in GFP+ cells. These findings are consistent with the biophysical properties of Purkinje cells (Vaidyanathan et al., 2013), suggesting that the differentiated Cntn2:EGFP cells induced by SN treatment are PC-like cells.
Figure 3.
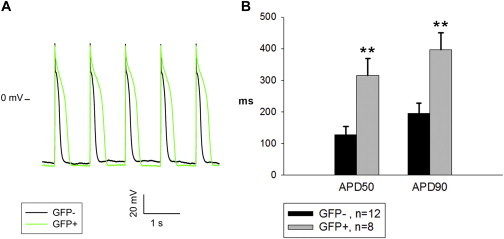
SN-Induced Cntn2:EGFP+ Cells Display a Purkinje-Fiber-like Action Potential Phenotype
Cells from differentiated cultures that had been induced with SN were evaluated in patch-clamp studies. Cells were patched from three independent differentiation experiments.
(A) Representative stimulated AP traces recorded in Cntn2:EGFP− or Cntn2:EGFP+ cells.
(B) Comparison of action potential duration (APD) 50 and 90 between GFP− (n = 12) and GFP+ cells (n = 8) (mean ± SE). Statistical significance is indicated: ∗∗p < 0.01.
SN-Induced PC-like Cells Display Cardiac Conduction System Gene Expression Profiles
To further characterize the phenotype of the SN-induced EGFP+ cells, we measured the transcript profiles of known cardiac-expressed genes in sorted cells. To distinguish cardiac cells from non-cardiac cells in the GFP− cell fraction, we used a dual ESC reporter line harboring the Cntn2:egfp transgene and transduced it with a lentiviral vector carrying the αMhc:mcherry reporter gene (Kita-Matsuo et al., 2009). As shown previously (Maass et al., 2015), we could use fluorescence-activated cell sorting (FACS) to separate differentiating cultures into three cell populations: negative, αMhc:mCherry+ (GFP negative), and Cntn2:EGFP+. We noted that this GFP+ cell population is also Cherry+, although dim. Using stringent gating criteria, when compared to DMSO-treated cells, SN consistently enhanced the generation of the Cntn2:EGFP+ (mCherry-dim) cells, from 0.17% ± 0.1% to 38.3% ± 3.4% from three independent experiments; a representative sort is shown in Figure 4A.
Figure 4.
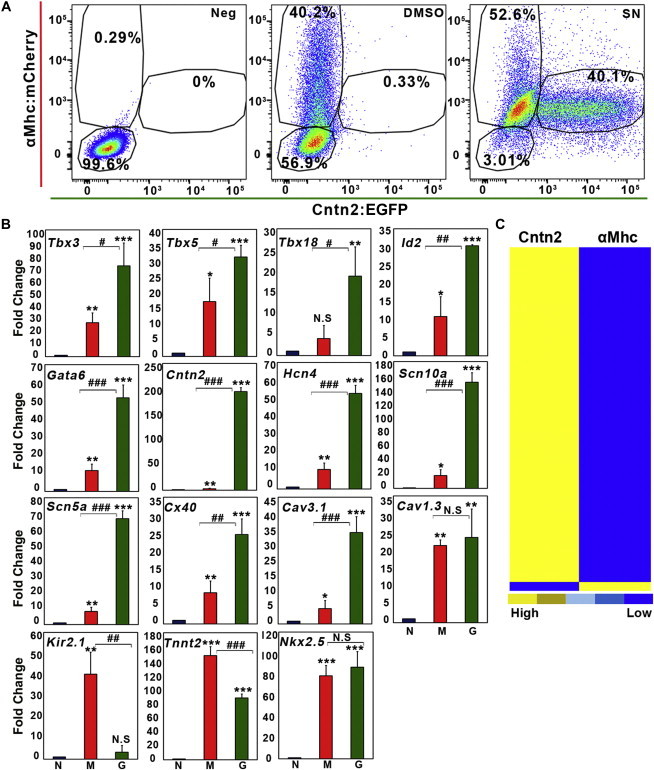
SN Directs Cells to Express Cardiac Conduction System Markers
(A) FACS plots of Cntn2:EGFP+ and αMhc:mCherry+ populations using the αMhc: mcherry/Cntn2:egfp double reporter line. CCS:lacz cells were used as negative control. Cells were harvested at differentiation day 25. FACS plots of negative, DMSO-, and SN-treated cells are shown in the left, middle, and right panels, respectively.
(B) Cardiac conduction system gene expression profiles of three populations (negative, blue bars, normalized to 1; αMhc:mCherry+ [GFP negative], red bars; Cntn2:EGFP+ [mCherry-dim], green bars) analyzed by qPCR. Comparison for significance of either mCherry+ or EGFP+ cell populations with the negative population is indicated by asterisks (∗). Comparison of Cntn2:EGFP+ cells with αMhc:mCherry+ cells are shown by hashtags (#). Error bars show SD. Results are from three independent experiments.
(C) Hierarchical clustering analysis of significantly altered gene expression based on RNA sequencing from αMhc:mCherry+ and Cntn2:EGFP+ cells that were derived from differentiation cultures following induction with SN. αMhc, αMhc:mCherry+; Cntn2, Cntn2:EGFP+ cells.
Statistical significance is indicated: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and #p < 0.05, ##p < 0.01, ###p < 0.001. N.S. indicates not statistically significant.
We used qPCR to analyze the expression of cardiac conduction-associated genes in these three populations, including genes that are reported to be associated with cardiac conduction progenitors (Tbx3, Tbx5, Tbx18, Id2, and Gata6), CCS channels (Hcn4, Cx30.2, Scn5a, Scn10a, Cntn2, Cx40, Cav1.3, and Cav3.1), and cardiomyocyte markers (Nkx2.5, Tnnt2, and Kir2.1). As expected, all of these genes are expressed at significantly higher levels in the αMhc:mCherry+ cells compared to the negative cells (Figure 4B). Notably, all of the transcription factor genes associated with cardiac conduction cells (Tbx3, Tbx5, Tbx18, Id2, and Gata6) were expressed markedly higher in the Cntn2:EGFP+ cells compared to expression levels in the cardiac αMhc:mCherry+ cells. The same was true for PC differentiation channels including Hcn4, Scn5a, Scn10a, and Cntn2. Previous studies showed that Scn5a transcripts are significantly enriched in PC (Maass et al., 2015; Pallante et al., 2010). However, there was no significant difference between these two populations for expression levels of the cardiac marker Nkx2.5 (Figure 4B), consistent with the PC-like cells retaining cardiac lineage fate. We note that the GFP+ cells do express lower levels of some cardiomyocyte transcripts, including Tnnt2, consistent with the relatively low (dim) αMhc:mCherry profile. Kir2.1, normally expressed at high levels in working cardiomyocytes, but not in CCS (Wolf and Berul, 2006; Zaritsky et al., 2001), and the CCS channel Cav3.1 clearly distinguishes the two cardiac cell phenotypes according to the qPCR results (Figure 4B). The transcript profiles of these three cell populations isolated from DMSO-treated control cells are very comparable to the profiles obtained using SN-treated cells (Figure S3). Notably, both DMSO-treated and SN-treated GFP+ cells display 5- to 10-fold higher levels of Hcn4, Scn10a, Scn5a, and Cx40 compared to the αMHC-mCherry+ (GFP−) cardiomyocytes. Thus, SN treatment appears to expand specifically the PC population that is present as a rare cell type in the absence of SN.
To compare the gene expression profiles globally in these cell populations, we performed RNA sequencing. The expression levels of genes in SN-induced αMhc:mCherry+ and Cntn2:EGFP+ (αMhc:mCherrydim) cells were normalized with expression levels in negative cells. Next, the gene list was filtered based on expression levels that are 10-fold different in Cntn2:EGFP+ cells. Hierarchical clustering analysis revealed that the two cell populations are much closer to each other compared to negative cells (not shown) and that most of the differences (Figure 4C) are upregulated in Cntn2:EGFP+ cells (90% are relatively decreased in αMhc:mCherry+ cells). Subsequently, the gene list was filtered based on expression levels that are at least 5-fold higher in either αMhc:mCherry+ (898 genes) or Cntn2:EGFP+ (918 genes). Venn diagram analysis showed that among these filtered gene sets, Cntn2:EGFP+ cells shared relatively few genes with αMhc:mCherry+ cells (127 genes; Figure 5A). Gene Ontology (GO) biological analyses revealed that the genes expressed higher in αMhc:mCherry+ single-positive cells are involved among various processes in muscle tissue development and cardiac cell development (Figure 5B), while those enhanced in the EGFP+ cells are associated with cell responsive processes. To assess which gene sets are enriched in the PC-like cells, we performed gene set enrichment analysis (GSEA) comparing gene sets with published microarray data. We used two gene sets (GSE60987) in which mouse PCs or ventricular myocytes were isolated from adult compound transgenic mice expressing Cntn2:EGFP and αMHC-Cre/floxed tdTomato reporter genes (Kim et al., 2014). As expected, the gene set from αMhc:mCherry+ cells is enriched in cardiomyocyte control samples (Figure 5C, left panel). However, the gene set from Cntn2:EGFP+ cells is highly enriched in adult mouse PCs (Figure 5C, right panel), further confirming that the SN-induced Cntn2:EGFP+ cells express PC gene expression profiles.
Figure 5.
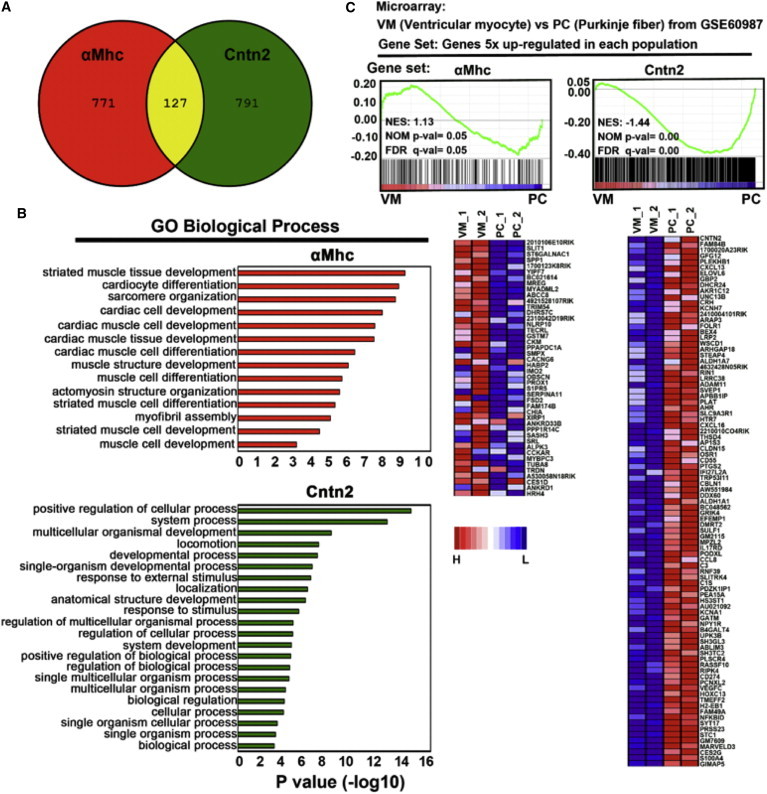
ESC-Derived CCS Cells Display PC Gene Expression Profiling
(A) Venn diagram of gene sets at least 5-fold upregulated in αMhc:mCherry+ (GFP−) or Cntn2:EGFP+ (mCherry-dim) populations.
(B) GO biological process analyses of the same gene sets.
(C) GSEA indicates that the restricted Cntn2:EGFP+ gene set is significantly enriched in adult mouse PC cells. A published dataset (GSE60987) and the two gene sets that are >5-fold upregulated from αMhc:mCherry+ and Cntn2:EGFP+ populations were used. Enriched gene sets are selected based on statistical significance (FDR q value < 0.25 and/or NOM p value < 0.05). Below are heat maps representing enriched gene sets. High and low expression of the indicated genes in αMhc:mCherry+ and Cntn2:EGFP+ populations are represented in red and blue, respectively. PC_1 and 2, two dataset of adult mouse PC cells; VM_1 and 2, two dataset of ventricular myocytes; αMhc, αMhc:mCherry+; Cntn2, Cntn2:EGFP+ cells.
See also Figure S3.
SN Enhances ESC-Derived Cardiac Conduction Cell Differentiation through Modulating the cAMP Signaling Pathway
SN is known to be a short-acting parenteral arterial and venous vasodilator. It has been shown that its effect is through release of nitric oxide (NO) and activation of guanylate cyclase (Friederich and Butterworth, 1995). Moreover, SN has been shown to enhance cAMP levels (Kim et al., 2006; Polte and Schröder, 1998). To test whether SN acts to enhance CCS generation through a cAMP signal, we directed Cntn2:egfp cells to cardiac fate and challenged on day 5 with cAMP activators including zardaverine (ZA), phosphodiesterase (PDE) inhibitor, dibutyryl-cAMP, and forskolin (Figure 6A). Interestingly, all cAMP activators significantly increased the generation of Cntn2:EGFP+ cells (14.7 ± 0.6 for ZA, 9.3 ± 1.1 for dibutyryl-cAMP, and 9.1 ± 1.2 for forskolin; fold change and SD) compared with DMSO controls (1.3 ± 0.6 fold change and SD; Figure 6B). We tested whether the cAMP activators can synergize with SN to enhance cardiac conduction cell fate and found that combining an individual activator with SN significantly enhanced the percentage of cells expressing Cntn2:EGFP compared with each activator alone (20.0 ± 1.9 for ZA, 11.3 ± 1.3 for dibutyryl-cAMP, and 12.8 ± 1.9 for forskolin; fold change and SD). A cAMP inhibitor (H89; also protein kinase A inhibitor) was also tested and found to abrogate the ability of SN to enhance generation of Cntn2:EGFP+ cells (H89 + SN, 2.3 ± 1.2 versus SN, 10.4 ± 1.6; fold change and SD; Figure 6B). These data suggest that cAMP activators enhance PC-like cell differentiation and that cAMP signaling is required for the effect of SN.
Figure 6.
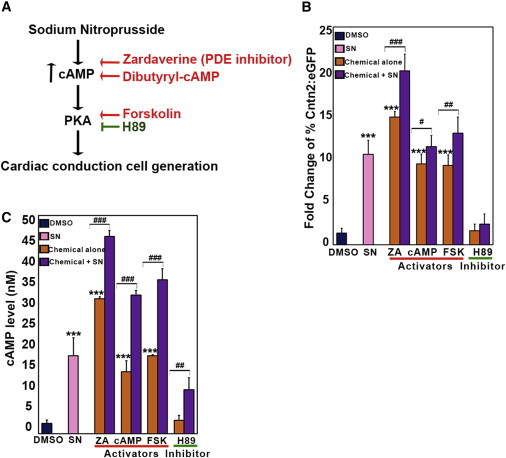
Sodium Nitroprusside Promotes Generation of Cardiac Conduction System Cells through cAMP Signaling
(A) A proposed mechanism of SN action. cAMP activators and inhibitors are shown in red and green, respectively.
(B) Quantification of the immunofluorescence staining of Cntn2:EGFP expression by MetaExpress image analysis. Cells were treated with cAMP activators (ZA, cAMP, and FSK) or inhibitor (H89), respectively. A blue column (DMSO control), pink column (SN treatment), orange columns (cAMP activators or inhibitors treatment), and purple columns (treatment of cAMP activators or inhibitors combined with SN) are shown.
(C) Quantification of cAMP levels. Error bars show SD. Comparison of either the activator or the inhibitor treatments with DMSO control is indicated by an asterisk (∗). Comparison of either activators or inhibitors combined with SN to the activators or inhibitors itself is shown by a hashtag (#). Results are derived from three independent experiments.
Statistical significance is indicated: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and #p < 0.05, ##p < 0.01, ###p < 0.001.
The results suggest that SN may function entirely through this pathway, since cAMP activators enhanced Cntn2:EGFP+ cell generation comparably to SN. Therefore, it was important to confirm whether cAMP levels are in fact increased by these treatments. During the cardiac differentiation protocol, cells were treated at day 5 with SN, cAMP activators, or cAMP inhibitors for 24 hr. Cells were harvested and the cAMP levels measured using a cAMP-Glo assay (Figure 6C). SN significantly enhanced cAMP levels compared to DMSO controls (17.9 ± 4.1 versus 2.4 ± 0.8 nM). As expected, all the cAMP activators also increased cAMP levels (ZA: 30.9 ± 0.5 nM, cAMP:14.2 ± 2.5 nM, and FSK: 17.9 ± 0.2 nM). Combining each activator with SN enhanced approximately 2-fold levels of cAMP (ZA: 45.2 ± 1.3 nM, cAMP: 31.8 ± 1.1 nM, and FSK: 35.2 ± 2.5 nM) compared to treatments with SN or each activator alone. On the contrary, H89 significantly blocked the ability of SN to enhance cAMP levels (10.1 ± 2.7 versus 30.9 ± 0.5; p < 0.005). The cAMP levels consistently correlated with Cntn2:EGFP expression. Taken together, these data support a model in which SN significantly enhances ESC-derived cardiac conduction cell generation through activation of the cAMP signaling pathway.
SN-Induced Cntn2:EGFP+ Cells Are Derived from Cardiomyocytes
The origin of CCS cell lineages remains an area of exploration. Cell lineage studies in the embryonic chick heart showed that Purkinje fibers can be differentiated from cardiomyocytes (Cheng et al., 1999; Gourdie et al., 1995). However, whether or not PC-like cells differentiate from cardiomyocytes in ESC differentiation systems remains unknown, although it is consistent with the co-expression of the αMhc:mCherry reporter. To investigate this issue, we used the αMhc:mcherry/Cntn2:egfp double reporter line to monitor the emergence of differentiated cells from day 12 to day 17 via live imaging. The αMhc:mCherry+ (single-positive) cells could first be imaged at day 8, while the first Cntn2:EGFP+ cells did not emerge until around day 12. Intriguingly, very faint GFP+ cells were first observed coming from weakly fluorescent αMhc:mCherry+ cells (Cherry-low), and these cells gradually over time generated a much stronger GFP signal (Figure 7A; Movie S1). Moreover, to further confirm the live imaging data, a pure population of αMhc:mCherry+ cells was sorted at early differentiation stage day 11 (Figure S4, left panel) and placed back into culture. After 14 days, Cntn2:EGFP+ (mCherry-dim) cells were generated from these sorted αMhc:mCherry+ cells (Figure S4, right panel). These data confirm that ESC-derived PC-like cells are generated from cardiomyocytes.
Figure 7.
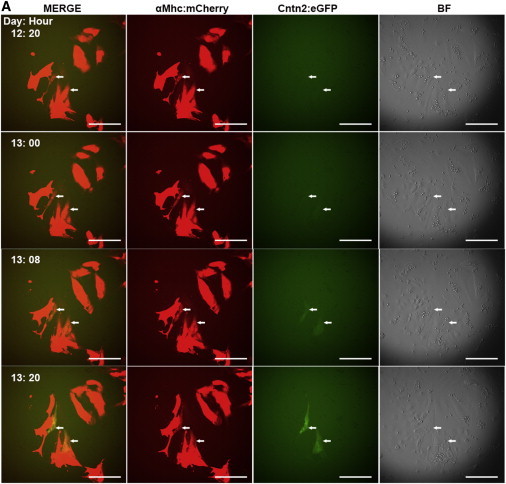
SN-Induced Cntn2:EGFP+ Cells Are Derived from Cardiomyocytes
Using the αMhc:mcherry/Cntn2:egfp double reporter line, differentiated cells were monitored from day 12 to day 17 of differentiation. Images were taken every 4 hr. Faint GFP+ cells are initially seen to emerge from αMhc:mCherry+ cells at day 12 and were followed as GFP expression was increased during 20 hr of culture. Shown are representative panels from an experiment that was reproducible in more than three independent experiments. See also Figure S4.
Discussion
Directed differentiation protocols have been employed to efficiently generate cardiomyocytes from both mouse and human ESCs (Kattman et al., 2011). However, it has remained a challenge to efficiently generate CCS cells using currently established protocols. In this study, we performed a high-throughput screen and identified a small molecule, sodium nitroprusside, which significantly enhances PC-like differentiation efficiency up to 40% of the culture in the context of a standard cardiac differentiation protocol. SN was shown using two different CCS ESC reporter lines to efficiently and reproducibly promote CCS cell generation. Using a αMhc:mcherry/Cntn2:egfp double reporter line, it was possible to perform several complementary comparisons to confirm that SN-treated cells represent a PC-like lineage, including comparison of transcript and protein profiles and action potential properties. Importantly, we found that SN promotes the generation of PC-like cells by activating cAMP signaling, suggesting a key pathway involved in embryonic derivation of the cardiac Purkinje network. By monitoring the differentiation process with live imaging and pre-sorting, we also demonstrated that SN-induced Cntn2:EGFP+ cells are derived from cardiac cells already expressing αMHC during ESC differentiation.
There are no known transcription factors that are unique to CCS or specifically PC fate. However, Tbx3, Tbx5, Tbx18, Id2, and Gata6 are genes previously associated with expression in CCS progenitors and derivatives. For instance, Tbx3 can direct SA node specification and formation. Ectopic expression of Tbx3 in atria represses the atrial phenotype and generates a pacemaker phenotype (Hoogaars et al., 2007). Tbx5 is expressed in the AV bundle and bundle branch, and in mice, Tbx5 haploinsufficiency causes AV node maturation failure and abnormalities of the bundle branch (Moskowitz et al., 2004). Tbx18 has been shown to contribute to SA node formation (Wiese et al., 2009). Id2 is expressed in AV bundle, and Id2-null mice show left bundle branch block (Moskowitz et al., 2007). While Gata6 is expressed throughout the developing heart, it becomes much more restricted in SA and AV nodes as well as the AV bundle in the adult mouse (Edwards et al., 2003). We found that SN-induced Cntn2:EGFP+ cells expressed relatively higher levels of these genes compared to cells that only express αMhc:mCherry. Perhaps the best characterized marker for PC is the Cntn2 gene, and Cntn2:EGFP+ cells that derive from ESC differentiation have been described and functionally characterized as displaying a PC phenotype (Maass et al., 2015). Induction with SN provides a markedly simple strategy to generate scalable numbers of these otherwise rare cells. During live imaging, the EGFP+ cells were seen developing from cells that were already αMhc:mCherry+, and they were also generated by further culturing αMhc:mCherry+-sorted cells, indicating that they derive from cardiac progenitors. Live imaging, flow cytometry, and gene expression profiling data suggest that the Cntn2:EGFP+ cells develop by downregulation of the working cardiomyocyte program. We did not find Cntn2:EGFP+ cells that were also αMhc:mCherry+high; rather, they were all αMhc:mCherry+dim.
Notably, we found that SN enhances PC-like cell generation through the cAMP signaling pathway. Interestingly, consistent with this mechanism, cAMP levels in the developing mouse heart increase through the β2-adrenergic receptor and adenylate cyclase type VI, which then leads to the activation of the HCN4 channel (Biel et al., 2009). Moreover, evidence has been presented to suggest that cAMP can transform ventricular cardiomyocytes into pacemaker-like cells (Ruhparwar et al., 2010; Sastry et al., 2006). Thus, the findings presented here provide useful practical information for developing new regenerative therapeutics and also shed light on the normal process of CCS development.
Experimental Procedures
ESC Culture and Differentiation
CCS:Lacz (Rentschler et al., 2001), Cntn2:egfp (Pallante et al., 2010), and lentivirus-transduced αMhc:mcherry/Cntn2:egfp (Maass et al., 2015) lines were maintained without feeder cells on 0.1% gelatin-coated plate in serum-free conditions consisting of DMEM/F12 (Gibco, 11320-33) and Neurobasal media (Gibco, 21103049) supplemented with N2 (Gibco, 17502-048), B27 (Gibco, 17504-044), BSA (Gibco, 15260-037), penicillin/streptomycin, 1,000 U/ml ESGRO LIF (Millipore, ESG1107), 3 μM CHIR 99021 (Stemgent, 04-0004), 1 μM PD0325901 (Stemgent, 04-0006), 1.5 × 10−4 M MTG (Sigma), and 2 mM L-glutamine (Gibco). Medium was changed every day. For SN-mediated cell differentiation, ESCs were dissociated with Accutase and plated for embryoid body (EB) formation (day 0) at 40,000 cells/ml in serum free differentiation (SFD) medium composed of 75% Iscove’s modified Dulbecco’s medium (Invitrogen) and 25% Ham’s F12 medium (Invitrogen) supplemented with N2, B27, penicillin/streptomycin, 0.05% BSA, 2 mM glutamine, 0.5 mM ascorbic acid (Sigma) and 0.1 mM β-mercaptoethanol (Gadue et al., 2006). After 48 hr, EBs were dissociated with Accutase and reaggregated in SFD media with the addition of human ACTIVIN-A (8 ng/ml), BMP4 (0.5 ng/ml), and VEGF (5 ng/ml) for another 48 hr. All cytokines were purchased from R&D Systems. For experiments using the double reporter line, at day 4, EBs were dissociated with Accutase and replated at 58,000 cells/cm2 in 0.1% gelatin-coated six-well plates with cardiomyocyte media containing RPMI (Corning cellgro, 10-041CV) with N2, B27 minus vitamin A (Life Technologies, 12587-010), penicillin/streptomycin, 2 mM glutamine, 0.5 mM ascorbic acid, and 0.1 mM β-mercaptoethanol. The next day, differentiated cells were treated with 100 μM SN for 5 days. Masterplex ReaderFit software was used to generate the efficacy curves (EC50).
High-Content Screening
CCS:Lacz ESCs were trypsinized and plated in SFD medium. After 48 hr, the EBs were dissociated with Accutase and reaggregated in SFD medium with the addition of human ACTIVIN-A (8 ng/ml), BMP4 (0.5 ng/ml), and VEGF (5 ng/ml). At day 5, EBs were dissociated with Accutase and replated at 5,000 cells per well in 0.1% gelatin-coated 384-well plates with cardiomyocyte media. The next day, cells were treated with compounds from chemical libraries in duplicate at either of two concentrations (10 μM or 1 μM). The libraries include compounds from Sigma LOPAC (1,280 pharmacologically active compounds, MicroSource US-Drug collection (2,320 compounds, including known drugs, experimental bioactivities, and pure natural products, and the Prestwick Chemical library (1,200 compounds, including 100% FDA-approved drugs). Cells were harvested at day 11 to measure β-galactosidase activity, using the Beta-Glo assay substrate (Promega, E4740). The effect of compound on β-galactosidase activity was quantified by calculating the fold induction, determined using the ratio A/B, where A represents the activity in the presence of compound and B represents the activity in wells with only DMSO added. Hit compounds were defined as those with A/B higher than 2.5. Hit compounds were picked from the original library and validated in three independent experiments using the same protocol.
Real-Time qPCR
Total RNA from FACS-sorted cells was purified using the Absolutely RNA Nanoprep kit (Agilent Technologies, 400753), quantified with a NanoDrop spectrophotometer (Thermo Scientific) and reverse transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems, 4374966). Real-time PCR was performed with a LightCycler 480 (Roche) instrument with LightCycler DNA master SYBR Green I reagents. Differences between samples and controls were calculated based on the 2−ΔΔCT method (Livak and Schmittgen, 2001) and normalized to GAPDH. Statistical significance was determined using a two-tailed Student’s t test (p < 0.05). Primer sequences are listed in Table S3.
Flow Cytometry and Immunohistochemistry
ESCs harboring the Cntn2:egfp and αMhc:mcherry reporter genes have been described previously (Maass et al., 2015) and were differentiated into cardiac and PC-like cells as described above. At day 25, cells were dissociated with 1 mg/ml type II collagenase (Life Technologies, 17101-015) and 20 U/μl DNase I (Millipore, 260913) for 40 min at 37°C. Cells were centrifuged at 1,400 rpm for 4 min and resuspended in FACS buffer containing DMEM without phenol red (Life Technologies, 21063-029), 1 mM EDTA, 25 mM HEPES, and 5% fetal bovine serum (FBS). Cells were collected using a BD FACSARIA cell sorter. For the flow analysis, approximately 50,000 single live-cell events were recorded and analyzed per sample on a C6 flow cytometer (Accuri) and analyzed using FCS express (De novo Software). For immunostaining, cells were fixed with 4% paraformaldehyde at room temperature for 10 min and washed three times with 1× PBS. Cells were then blocked with 5% horse serum and 0.3% Triton X-100 and incubated with antibodies against HCN4 (1:500, mouse; Abcam, ab85023), GFP (1:2,000, goat; R&D system, AF4240), TNNT2 (1:500, mouse, Thermo Scientific, MA5-12960), αMHC (1: 500, mouse, Abcam, ab15), Connexin43 (1:200, rabbit, ab11370), or TUBB3 (1:1,000, rabbit; Covance, MRB-435) followed by Alexa-488-, Alexa-Fluor-555-, and Alexa-Fluor-647-conjugated donkey secondary antibodies against mouse, goat or Rabbit (1:500, Invitrogen). Nuclei were counterstained with DAPI.
Electrophysiology
Cells were dissociated and plated onto gelatin-coated coverslips and then transferred into a recording chamber within 48 to 72 hr for whole-cell patch clamp studies. Cells were superfused with Tyrode’s solution containing (mM) NaCl 137.7, KCl 5.4, NaOH 2.3, CaCl21.8, MgCl2 1, Glucose 10, and HEPES 10 (pH adjusted to 7.4 with NaOH) at room temperature. Electrodes were filled with 50 mM KCl, 80 mM K-aspartic acid, 1 mM MgCl2, 10 mM EGTA, 10 mM HEPES, and 3 mM Na2-ATP (pH adjusted to 7.2 with KOH). The resistances of the electrodes were between 2 and 3 MΩ. Only cells with gigaseal were used to collect data. Stimulated action potentials were triggered by minimum positive pulses with 1-Hz frequency with current clamp mode. Signals were recorded by an amplifier (MultiClamp 700B, Axon Instruments) and a digitizer (Model DIGIDATA 1440A, Axon Instruments) connected to a PC. Data acquisition and analysis were performed using CLAMPEX 10.2 and CLAMFIT 10.2 software (Axon instruments), respectively.
RNA Sequencing and Data Analysis
RNA was prepared from sorted cells with Absolutely RNA Nanoprep kit. The cDNA libraries were generated using TruSeq RNA Sample Preparation (Illumina). Each library was sequenced using 50-bp single reads in the HiSeq2500 format (Illumina). Raw data were normalized with negative cells as a fold change. Gene lists were analyzed with the Cluster software (Eisen et al., 1998). Hierarchical clustering involved Pearson correlation for population comparisons and Euclidian distance calculations for gene level clustering. Data were visualized with TreeView software (Eisen et al., 1998). Global CCS and cardiomyocyte gene expression data were obtained from a published database (GSE60987) (Kim et al., 2014). False discovery rate (FDR) q values < 0.25 or nominal (NOM) p values < 0.05 were considered significant.
cAMP Activity Determination
Day 4 EBs were dissociated and replated in 96-well plates. The next day, differentiated cells were treated with activators, inhibitors, or DMSO as control. Cells were harvested after 24-hr treatment. The levels of cAMP activity were determined using the cAMP-Glo Kit (Promega, V1501), according to the manufacturer’s instruction, and using the EnVision automated microplate reader system (Perkin Elmer).
Statistics
A two-tailed Student’s t test was used to calculate statistical significance.
Author Contributions
S.-Y.T. conceived and designed the study, carried out experiments, collected and analyzed data, and wrote the manuscript; K.M. provided essential reagents prior to publication; J.L. carried out experiments and collected data; and G.I.F., S.C., and T.E. conceived and designed the study, analyzed data, and wrote the manuscript.
Acknowledgments
S.C. is a New York Stem Cell Foundation-Roberson Investigator. We are grateful for technical support and advice provided by Harold S. Ralph in the Cell Screening Core Facility, Jenny Xiang in the Genomics Resources Core Facility, and staff at the Flow Cytometry Core Facilities at Weill Cornell Medical College. We thank members of the Evans, Chen, and Fishman laboratories for advice and comments throughout the course of this work. This work was supported by a contract from the Empire State Stem Cell Research Program (NYSTEM, #C028115), the Korein Foundation, the B. and R. Knapp Foundation, and the Weisfeld Family Program in Cardiovascular Regenerative Medicine at NYU School of Medicine. Additional support was provided by the NHLBI through grants HL105983 (G.I.F.) and HL111400 (T.E.).
Published: May 28, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Contributor Information
Glenn I. Fishman, Email: glenn.fishman@nyumc.org.
Shuibing Chen, Email: shc2034@med.cornell.edu.
Todd Evans, Email: tre2003@med.cornell.edu.
Accession Numbers
The accession number for the primary data reported in this paper is GEO: GSE68025.
Supplemental Information
As shown by live imaging, αMhc:mCherry+ cells can be seen at differentiation day 12. The first faint Cntn2:eGFP+ cells can initially be seen at this time and the signal co-localizes with αMhc:mCherry+ cells at 12 days and gradually increases in GFP intensity over 20 hr.
References
- Biel M., Wahl-Schott C., Michalakis S., Zong X. Hyperpolarization-activated cation channels: from genes to function. Physiol. Rev. 2009;89:847–885. doi: 10.1152/physrev.00029.2008. [DOI] [PubMed] [Google Scholar]
- Burgoyne R.D., Cambray-Deakin M.A., Lewis S.A., Sarkar S., Cowan N.J. Differential distribution of beta-tubulin isotypes in cerebellum. EMBO J. 1988;7:2311–2319. doi: 10.1002/j.1460-2075.1988.tb03074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G., Litchenberg W.H., Cole G.J., Mikawa T., Thompson R.P., Gourdie R.G. Development of the cardiac conduction system involves recruitment within a multipotent cardiomyogenic lineage. Development. 1999;126:5041–5049. doi: 10.1242/dev.126.22.5041. [DOI] [PubMed] [Google Scholar]
- Cho H.C., Marbán E. Biological therapies for cardiac arrhythmias: can genes and cells replace drugs and devices? Circ. Res. 2010;106:674–685. doi: 10.1161/CIRCRESAHA.109.212936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFrancesco D. The cardiac hyperpolarizing-activated current, if. Origins and developments. Prog. Biophys. Mol. Biol. 1985;46:163–183. doi: 10.1016/0079-6107(85)90008-2. [DOI] [PubMed] [Google Scholar]
- Edwards A.V., Davis D.L., Juraszek A.L., Wessels A., Burch J.B. Transcriptional regulation in the mouse atrioventricular conduction system. Novartis Found. Symp. 2003;250:177–189. discussion 189–193, 276–279. [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain G.L., Quandt F.N., Bastian B.L., Gerschenfeld H.M. Contribution of a caesium-sensitive conductance increase to the rod photoresponse. Nature. 1978;272:466–469. doi: 10.1038/272467a0. [DOI] [PubMed] [Google Scholar]
- Friederich J.A., Butterworth J.F., 4th Sodium nitroprusside: twenty years and counting. Anesth. Analg. 1995;81:152–162. doi: 10.1097/00000539-199507000-00031. [DOI] [PubMed] [Google Scholar]
- Furley A.J., Morton S.B., Manalo D., Karagogeos D., Dodd J., Jessell T.M. The axonal glycoprotein TAG-1 is an immunoglobulin superfamily member with neurite outgrowth-promoting activity. Cell. 1990;61:157–170. doi: 10.1016/0092-8674(90)90223-2. [DOI] [PubMed] [Google Scholar]
- Gadue P., Huber T.L., Paddison P.J., Keller G.M. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourdie R.G., Mima T., Thompson R.P., Mikawa T. Terminal diversification of the myocyte lineage generates Purkinje fibers of the cardiac conduction system. Development. 1995;121:1423–1431. doi: 10.1242/dev.121.5.1423. [DOI] [PubMed] [Google Scholar]
- Hashem S.I., Claycomb W.C. Genetic isolation of stem cell-derived pacemaker-nodal cardiac myocytes. Mol. Cell. Biochem. 2013;383:161–171. doi: 10.1007/s11010-013-1764-x. [DOI] [PubMed] [Google Scholar]
- Holzmeister J., Leclercq C. Implantable cardioverter defibrillators and cardiac resynchronisation therapy. Lancet. 2011;378:722–730. doi: 10.1016/S0140-6736(11)61228-2. [DOI] [PubMed] [Google Scholar]
- Hoogaars W.M., Engel A., Brons J.F., Verkerk A.O., de Lange F.J., Wong L.Y., Bakker M.L., Clout D.E., Wakker V., Barnett P. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Tsoy I., Park M.K., Lee Y.S., Lee J.H., Seo H.G., Chang K.C. Iron released by sodium nitroprusside contributes to heme oxygenase-1 induction via the cAMP-protein kinase A-mitogen-activated protein kinase pathway in RAW 264.7 cells. Mol. Pharmacol. 2006;69:1633–1640. doi: 10.1124/mol.105.020487. [DOI] [PubMed] [Google Scholar]
- Kim E.E., Shekhar A., Lu J., Lin X., Liu F.Y., Zhang J., Delmar M., Fishman G.I. PCP4 regulates Purkinje cell excitability and cardiac rhythmicity. J. Clin. Invest. 2014;124:5027–5036. doi: 10.1172/JCI77495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita-Matsuo H., Barcova M., Prigozhina N., Salomonis N., Wei K., Jacot J.G., Nelson B., Spiering S., Haverslag R., Kim C. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS ONE. 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleger A., Seufferlein T., Malan D., Tischendorf M., Storch A., Wolheim A., Latz S., Protze S., Porzner M., Proepper C. Modulation of calcium-activated potassium channels induces cardiogenesis of pluripotent stem cells and enrichment of pacemaker-like cells. Circulation. 2010;122:1823–1836. doi: 10.1161/CIRCULATIONAHA.110.971721. [DOI] [PubMed] [Google Scholar]
- Laflamme M.A., Murry C.E. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maass K., Shekhar A., Lu J., Kang G., See F., Kim E.E., Delgado C., Shen S., Cohen L., Fishman G.I. Isolation and characterization of embryonic stem cell-derived cardiac purkinje cells. Stem Cells. 2015;33:1102–1112. doi: 10.1002/stem.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa K., Bahrudin U., Miake J., Igawa O., Kurata Y., Nakayama Y., Shirayoshi Y., Hisatome I. Identification, isolation and characterization of HCN4-positive pacemaking cells derived from murine embryonic stem cells during cardiac differentiation. Pacing Clin. Electrophysiol. 2010;33:290–303. doi: 10.1111/j.1540-8159.2009.02614.x. [DOI] [PubMed] [Google Scholar]
- Moskowitz I.P., Pizard A., Patel V.V., Bruneau B.G., Kim J.B., Kupershmidt S., Roden D., Berul C.I., Seidman C.E., Seidman J.G. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development. 2004;131:4107–4116. doi: 10.1242/dev.01265. [DOI] [PubMed] [Google Scholar]
- Moskowitz I.P., Kim J.B., Moore M.L., Wolf C.M., Peterson M.A., Shendure J., Nobrega M.A., Yokota Y., Berul C., Izumo S. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Pallante B.A., Giovannone S., Fang-Yu L., Zhang J., Liu N., Kang G., Dun W., Boyden P.A., Fishman G.I. Contactin-2 expression in the cardiac Purkinje fiber network. Circ Arrhythm Electrophysiol. 2010;3:186–194. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polte T., Schröder H. Cyclic AMP mediates endothelial protection by nitric oxide. Biochem. Biophys. Res. Commun. 1998;251:460–465. doi: 10.1006/bbrc.1998.9486. [DOI] [PubMed] [Google Scholar]
- Rentschler S., Vaidya D.M., Tamaddon H., Degenhardt K., Sassoon D., Morley G.E., Jalife J., Fishman G.I. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128:1785–1792. doi: 10.1242/dev.128.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen M.R., Brink P.R., Cohen I.S., Robinson R.B. Genes, stem cells and biological pacemakers. Cardiovasc. Res. 2004;64:12–23. doi: 10.1016/j.cardiores.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Ruhparwar A., Kallenbach K., Klein G., Bara C., Ghodsizad A., Sigg D.C., Karck M., Haverich A., Niehaus M. Adenylate-cyclase VI transforms ventricular cardiomyocytes into biological pacemaker cells. Tissue Eng. Part A. 2010;16:1867–1872. doi: 10.1089/ten.TEA.2009.0537. [DOI] [PubMed] [Google Scholar]
- Saito M., Sasaki T., Matsuoka H. Vitamin B(12) promotes Cx40 and HCN4 gene expression at an early stage of cardiomyocyte differentiation. Exp. Anim. 2009;58:57–60. doi: 10.1538/expanim.58.57. [DOI] [PubMed] [Google Scholar]
- Sastry A., Arnold E., Gurji H., Iwasa A., Bui H., Hassankhani A., Patel H.H., Feramisco J.R., Roth D.M., Lai N.C. Cardiac-directed expression of adenylyl cyclase VI facilitates atrioventricular nodal conduction. J. Am. Coll. Cardiol. 2006;48:559–565. doi: 10.1016/j.jacc.2006.01.082. [DOI] [PubMed] [Google Scholar]
- Scavone A., Capilupo D., Mazzocchi N., Crespi A., Zoia S., Campostrini G., Bucchi A., Milanesi R., Baruscotti M., Benedetti S. Embryonic stem cell-derived CD166+ precursors develop into fully functional sinoatrial-like cells. Circ. Res. 2013;113:389–398. doi: 10.1161/CIRCRESAHA.113.301283. [DOI] [PubMed] [Google Scholar]
- Spooner P.M., Albert C., Benjamin E.J., Boineau R., Elston R.C., George A.L., Jr., Jouven X., Kuller L.H., MacCluer J.W., Marbán E. Sudden cardiac death, genes, and arrhythmogenesis : consideration of new population and mechanistic approaches from a national heart, lung, and blood institute workshop, part I. Circulation. 2001;103:2361–2364. doi: 10.1161/01.cir.103.19.2361. [DOI] [PubMed] [Google Scholar]
- Stroud D.M., Darrow B.J., Kim S.D., Zhang J., Jongbloed M.R., Rentschler S., Moskowitz I.P., Seidman J., Fishman G.I. Complex genomic rearrangement in CCS-LacZ transgenic mice. Genesis. 2007;45:76–82. doi: 10.1002/dvg.20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan R., O’Connell R.P., Deo M., Milstein M.L., Furspan P., Herron T.J., Pandit S.V., Musa H., Berenfeld O., Jalife J., Anumonwo J.M. The ionic bases of the action potential in isolated mouse cardiac Purkinje cell. Heart Rhythm. 2013;10:80–87. doi: 10.1016/j.hrthm.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.M., Claycomb W.C. Embryonic stem cells form an organized, functional cardiac conduction system in vitro. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H670–H679. doi: 10.1152/ajpheart.00841.2004. [DOI] [PubMed] [Google Scholar]
- Wiese C., Grieskamp T., Airik R., Mommersteeg M.T., Gardiwal A., de Gier-de Vries C., Schuster-Gossler K., Moorman A.F., Kispert A., Christoffels V.M. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Wiese C., Nikolova T., Zahanich I., Sulzbacher S., Fuchs J., Yamanaka S., Graf E., Ravens U., Boheler K.R., Wobus A.M. Differentiation induction of mouse embryonic stem cells into sinus node-like cells by suramin. Int. J. Cardiol. 2011;147:95–111. doi: 10.1016/j.ijcard.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf C.M., Berul C.I. Inherited conduction system abnormalities—one group of diseases, many genes. J. Cardiovasc. Electrophysiol. 2006;17:446–455. doi: 10.1111/j.1540-8167.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- Woods C.E., Olgin J. Atrial fibrillation therapy now and in the future: drugs, biologicals, and ablation. Circ. Res. 2014;114:1532–1546. doi: 10.1161/CIRCRESAHA.114.302362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano S., Miake J., Mizuta E., Manabe K., Bahrudin U., Morikawa K., Arakawa K., Sasaki N., Igawa O., Shigemasa C. Changes of HCN gene expression and I(f) currents in Nkx2.5-positive cardiomyocytes derived from murine embryonic stem cells during differentiation. Biomed. Res. 2008;29:195–203. doi: 10.2220/biomedres.29.195. [DOI] [PubMed] [Google Scholar]
- Zaritsky J.J., Redell J.B., Tempel B.L., Schwarz T.L. The consequences of disrupting cardiac inwardly rectifying K(+) current (I(K1)) as revealed by the targeted deletion of the murine Kir2.1 and Kir2.2 genes. J. Physiol. 2001;533:697–710. doi: 10.1111/j.1469-7793.2001.t01-1-00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As shown by live imaging, αMhc:mCherry+ cells can be seen at differentiation day 12. The first faint Cntn2:eGFP+ cells can initially be seen at this time and the signal co-localizes with αMhc:mCherry+ cells at 12 days and gradually increases in GFP intensity over 20 hr.


