Summary
Reprogramming of somatic cells produces induced pluripotent stem cells (iPSCs) that are invaluable resources for biomedical research. Here, we extended the previous transcriptome studies by performing RNA-seq on cells defined by a combination of multiple cellular surface markers. We found that transcriptome changes during early reprogramming occur independently from the opening of closed chromatin by OCT4, SOX2, KLF4, and MYC (OSKM). Furthermore, our data identify multiple spliced forms of genes uniquely expressed at each progressive stage of reprogramming. In particular, we found a pluripotency-specific spliced form of CCNE1 that is specific to human and significantly enhances reprogramming. In addition, single nucleotide polymorphism (SNP) expression analysis reveals that monoallelic gene expression is induced in the intermediate stages of reprogramming, while biallelic expression is recovered upon completion of reprogramming. Our transcriptome data provide unique opportunities in understanding human iPSC reprogramming.
Highlights
-
•
Initial transcriptional change relies on histone modifications in fibroblast
-
•
Allele-specific gene expression is manifested during reprogramming
-
•
A large number of spliced forms of genes are identified during reprogramming
-
•
Pluripotent-specific splicing of CCNE1 (pCCNE1) enhances reprogramming
In this article, Park and colleagues provide robust transcriptome data acquired in cells undergoing human iPSC reprogramming. Significant changes in cell signaling pathways, allelic-specific expression, and an uncharacterized splicing of CCNE1 (pCCNE1) were identified during the progressive cell fate change of fibroblasts to iPSCs. The data will broaden the understanding of the reprogramming process and human-specific gene regulation.
Introduction
Induced pluripotent stem cells (iPSCs) have similar properties as embryonic stem cells (ESCs), such as self-renewal and differentiation capacity (Park et al., 2008c; Takahashi and Yamanaka, 2006). Reprogramming technique offers tremendous potential for disease modeling, cell-based therapy, and drug screening (Park et al., 2008a). Although the reprogramming process is quite robust and applicable to various types of adult differentiated cells, only a small fraction of donor cells reaches a fully pluripotent state, while the majority are refractory to reprogramming. Imperfect reprograming may carry somatic memory and may contribute to cancer development (Ohnishi et al., 2014). Therefore, efficient selection and generation of bona fide iPSCs are essential for safe uses in regenerative medicine.
Serial live cell imaging is one of the tools to distinguish bona fide human iPSCs (hiPSCs) from partially reprogrammed cells. Previously, we identified three distinct types of expandable hESC-like colonies during reprogramming via expression patterns of virus-derived GFP, fibroblast marker CD13 (ANPEP), and two pluripotent markers SSEA4 and TRA160 (Chan et al., 2009). Type I cells are defined by continuous expression reprogramming genes (CD13−GFP+SSEA4−TRA160−). Type II cells express pluripotency marker SSEA4 and continue expressing reprogramming factors (CD13−GFP+SSEA4+TRA160−). Type III cells show expression of TRA160 as well as SSEA4 (CD13−GFP−SSEA4+TRA160+). Among these types of colonies, only type III has similar molecular phenotypes with hESCs and become bona fide hiPSCs. Type I and type II cells are partially reprogrammed cells and display negative nuclear NANOG staining, low expression of several pluripotent genes (e.g., DNMT3B and REX1), and a distinct epigenetic state from type III cells and hESCs. Type I cells remain in their incomplete reprogramed state, while a small population of type II cells may still convert to type III cells and complete hiPSC reprogramming.
Reprogramming pathways have been extensively studied. Mesenchymal-to-epithelial transition (MET) occurs in the initial phase of reprogramming and is synergistically activated by OCT4, SOX2, KLF4, and MYC (OSKM) and BMP signaling, but is blocked by the transforming growth factor β (TGF-β) pathway (Li et al., 2010; Samavarchi-Tehrani et al., 2010). Despite the active function of BMP in the initial reprogramming, BMP proteins prevent the transition of pre-miPSCs to fully reprogrammed miPSCs by maintaining H3K9 methylation (Chen et al., 2013). In contrast, ACTIVIN/NODAL signaling pathway, which is a branch of TGF-β signaling, is essential for mESC self-renewal (Ogawa et al., 2007). WNT ligands and a downstream component of WNT signaling pathway, β-catenin, are required to prevent differentiation and maintain self-renewal in mESCs (Lyashenko et al., 2011). Whereas the transcriptional repressor TCF3 inhibits mESC self-renewal, an interaction with β-catenin followed by WNT3A stimulation activates the expression of self-renewal genes by blocking the TCF3 repressive activity (Yi et al., 2011). A recent study further defined the role of WNT, revealing that this pathway is a negative regulator in the early stages, but switches to a positive regulator in the late stage of mouse reprogramming (Ho et al., 2013).
Transcription profiling during reprogramming has provided critical insights into understanding reprogramming. Microarray-based transcriptome analysis in miPSCs and partially reprogrammed murine cell populations sorted by a fibroblast marker (THY1) and two pluripotent markers (SSEA1 and Oct4-GFP) revealed that the reprogramming process is composed of two main transcriptional waves (Polo et al., 2012). The first wave is driven by Myc and Klf4 and characterized by the loss of fibroblast identity and a gain in cell proliferation. The second wave is controlled by Oct4, Sox2, and Klf4 and is associated with changes in DNA methylation that facilitate stable pluripotency. A microarray and single-cell qPCR study of cell populations sorted by virus-driven EGFP and TRA160 in hiPSC reprogramming, showed that TRA160+ cell populations at late time points (approximately day 28) exhibit more similar gene expression patterns to hESCs and less heterogeneous than those at early time points (approximately day 11) (Tanabe et al., 2013). However, most of the nascent TRA160+ cells fail to complete reprogramming. These recent reports indicate that transcriptional and signaling regulatory networks are different among intermediate steps.
Here, we set out to investigate the progressive steps of hiPSC reprogramming by Phi29 DNA polymerase-based mRNA-sequencing (Phi29-mRNA amplification [PMA] RNA-seq) that enables us to monitor transcriptomes in scarce intermediate cell populations (Pan et al., 2013). We identified unique pluripotency-specified spliced transcripts and determined a surprising function of a spliced form of CCNE (pCCNE1) in improving the reprogramming efficiency. We also found that the actively reprogramming intermediate stage cells acquire a unique ASE pattern, which is erased when reprogramming is completed. Overall, our data analyses allowed us to further dissect the mechanism of hiPSC reprogramming.
Results
Strategy of Transcriptome Profiling from Partially Reprogrammed Cell States
In order to facilitate isolating cells undergoing reprogramming, we initiated reprogramming in human primary fibroblasts with pMSCV-IRES-GFP-based retroviral vectors expressing OSKM (Park et al., 2008b). Cells were harvested at day 3 and weeks 1, 2, 3 and 4 after the viral infection (Figure S1A). The intermediate reprogramed cells from weeks 1 to 4 were further separated by fluorescence-activated cell sorting (FACS) using antibodies for CD13, SSEA4, and TRA160 or GFP expression. At week 1, the majority of cells express virus-derived GFP (Figure S1B), and around 96.9% of those GFP+ cells expressed CD13. Double-positive cells (GFP+CD13+) also made up the majority of week 2 cell populations (31.3%), but the ratio of GFP+CD13− cells was greatly increased (20.9%). We observed that 2.7% (GFP+CD13− SSEA4+ and GFP−CD13− SSEA4+) of cells at week 2 showed SSEA4 expression with loss of CD13 expression. At weeks 3 and 4, the major cell population consisted of GFP+SSEA4−TRA160− cells (70.0% and 17.5%, respectively), but around 4%–6% of cells displayed expression of two pluripotent markers without GFP expression (GFP−SSEA4+TRA160+). At week 4, colonies showing hESC-like morphology with CD13−GFP−SSEA4+TRA160+ cell surface markers were picked for expansion and here on referred to as established iPSCs (grouped together with ESCs in subsequent analyses). PMA RNA-seq was performed in 18 intermediate cell populations, three replicates of parental fibroblasts, fibroblasts at day 3 post-OSKM induction, as well as ESCs and two types of established iPSCs (Pan et al., 2013).
Initial Gene Regulation by OSKM Overexpression in hiPSC Reprogramming
To examine genes immediately regulated by OSKM induction, we compared the transcriptome profile in cells 3 days post-ectopic OSKM overexpression with that of parental fibroblast cells (Figure 1A). Gene Ontology (GO) analysis showed that upregulated genes at day 3 are related to “type I interferon signaling pathway” and “histone modification” (Figure 1B). These genes include EHMT1, EZH2 (Onder et al., 2012), HMGA1 (Shah et al., 2012), MED12 (Chia et al., 2010), RARG (Wang et al., 2011), and TAF11 (Maston et al., 2012), which are highly expressed in hESCs and are required for self-renewal, maintenance of pluripotency, or hiPSC reprogramming. Downregulated genes are involved with “cell development” and “TGF-β signaling pathway.” Inhibition of the TGF-β signaling pathway has been characterized and previously shown to enhance iPSC reprogramming (Ichida et al., 2009). These initial responses to OSKM are also detected by reprogramming with electroporation of episomal vectors (Figure S1C). Since the type I interferon pathway is also triggered by the empty vector with infection or electroporation, the induction of this pathway seems to be a general cellular response to foreign viral DNA and not OSKM per se, as both the pMSCV construct and episomal plasmids have been assembled with viral elements (retrovirus and Epstein-Barr virus, respectively). Thus, our data support that the major role of OSKM in the early phase of reprogramming is the activation of reprogramming-related histone remodelers and transcription factors and the suppression of signaling pathways interfering with iPSC reprogramming. This early plasticity, also observed in our 3-day RNA-Seq data, can be utilized to direct differentiation to any lineage of choice (Efe et al., 2011).
Figure 1.
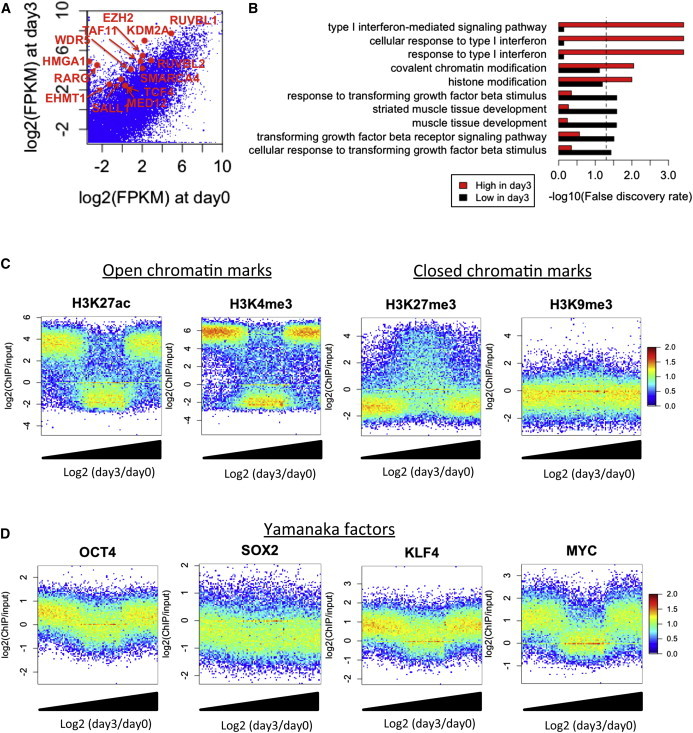
Initial Gene Regulation by OSKM
(A) Comparison of gene expression in OSKM-induced cells between days 0 and 3. Differentially expressed genes (>4-fold) related to “histone modification (GO: 0016570)” or “transcription factor binding (GO: 0008134)” are shown by red dots.
(B) GO analysis of upregulated and downregulated genes at day3. Dashed line represents 0.05 FDR.
(C and D) Comparison of (C) histone modification and (D) OSKM binding level in fibroblast stage with gene expression changes at day3. The x axis represents the rank of genes sorted by increasing order of log2(day 3/day 0) values. The y axis represents log2(ChIP/input). Colors represent log10(count).
See also Figure S1.
We next asked whether chromatin signatures in the parental fibroblasts and the initial binding of OSKM at promoters determine the genes regulated in the initial phase of reprogramming. To this end, the upregulated and downregulated genes at day 3 were compared with public ChIP-seq studies for histone modifications (Bernstein et al., 2010) and OSKM (Soufi et al., 2012) in fibroblast cells. We did not observe a distinct correlation of the histone modification level and initial OSKM binding between upregulated and downregulated genes at day 3. However, both upregulated and downregulated genes at day 3 showed significantly higher open chromatin marks H3K4me3 and H3K27ac and lower closed chromatin mark H3K27me3 than non-regulated genes (Figure 1C). In addition, OCT4, KLF4, and MYC, but not SOX2, are significantly enriched in both initially regulated promoters (Figure 1D), indicating that genes within pre-existing open chromatin regions are initially regulated by OKM, which act as both activators and repressors.
Transcriptome Analysis Revealed Three Representative Intermediate States during hiPSC Reprogramming
Consistent with our previous classification (Chan et al., 2009), principle component analysis (PCA) segregates the partially reprogrammed cell populations into three distinct stages (types I, II, and III) as well as fibroblast-like and ESC/iPSC stage (Figure 2A). Parental fibroblasts, day 3 reprogrammed cells, and CD13+GFP+ cells at weeks 1 and 2 were grouped into the fibroblast-like stage. Typical type I cells, grouped as type I stage, represented by CD13−GFP+SSEA4− at weeks 1, 2, and 4, are distinguishable from the fibroblast-like stage, and close to CD13+GFP+SSEA4+, CD13−GFP+SSEA4+, or CD13−GFP−SSEA4+ at week 2, suggesting that repression of the fibroblast phenotype (transition from CD13+ to CD13−) or induction of a pluripotent phenotype (SSEA4− to SSEA4+) represents the exit from the fibroblast-like stage. Type I cells are the closest to the fibroblast-like stage and neighbor type II and III stages of cells, suggesting that the fibroblast-to-type I transition is the first barrier in the path to iPSCs. Type II stage represents GFP+SSEA4+TRA160− cell populations and resides closer to type I stage than type III. Type II is the most distant stage from fibroblasts and ESC/iPSCs. Type III stage is composed of GFP−SSEA4+TRA160+ cells and shows the most similar transcriptional patterns with ESCs and iPSCs. Despite the repression of CD13 from the fibroblast-like stage, the expression levels of several other fibroblast markers, such as COL1A1 and COL1A2, are higher in types I and II than ESC/iPSCs. Meanwhile, the expression of these genes in type III cells is as low as that of ESC/iPSCs, indicating that the fibroblast signature still exists in types I and II stage (Figure S1D). GFP−SSEA4+TRA160− cell populations at weeks 3 and 4 are located between type II and type III stages and are hypothesized to be in the course of transition from types II to III. Between type III and ESC/iPSCs stage, the expression levels of OSKM and the other pluripotency regulators (e.g., NANOG) were not significantly different (Table S1). Around 900 genes show significantly higher expression in ESC/iPSC stage compared with type III (Figure S1E) and are overrepresented as “chromatin modifications” and “transcription cofactor activity” (Figure S1F).
Figure 2.
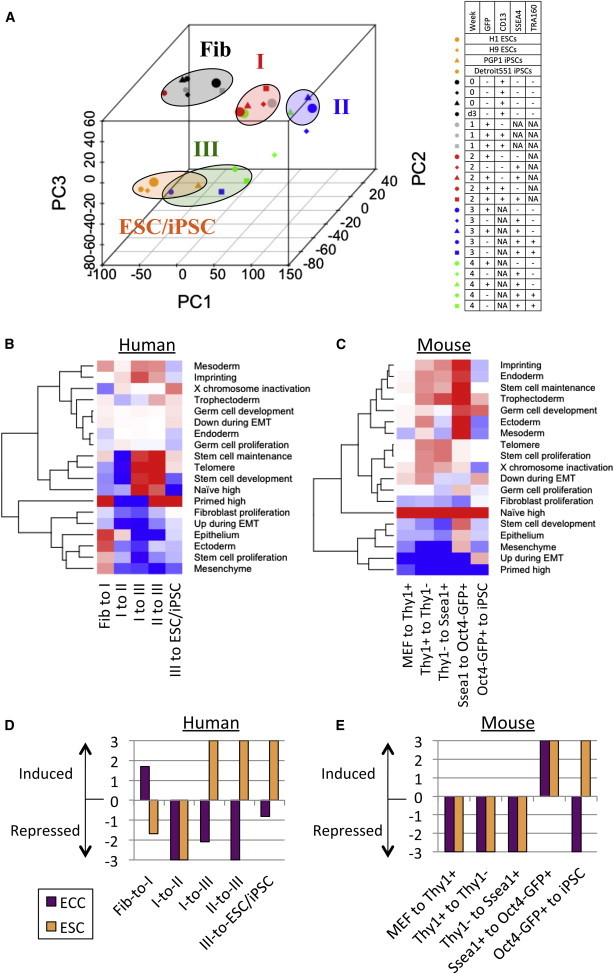
Characterization of Intermediate Stages in hiPSC Reprogramming
(A) PCA classification of the human intermediate states.
(B and C) GSEA of stem cell functions (B) between distinct human intermediate stages and (C) mouse intermediate stages. Gene sets induced or repressed in the transition between two stages (−log10(FDR)) are shown by red and blue color, respectively.
(D and E) GSEA of ECC and ESC-specific genes in (D) human and (E) mouse.
See also Figure S1.
Next, our transcriptome data were compared with gene signatures of unsorted and sorted populations (GFP+TRA160−, or TRA160+) from the published work (Tanabe et al., 2013) by gene set enrichment analysis (GSEA) (Table S2A). All of these signatures are significantly induced in the transition from fibroblast-like to type I stage and also are upregulated in later stages (Figure S1G). Gene signatures at mature stages (TRA160+ cells and iPSCs) are significantly enriched in the I-to-III and II-to-III transitions (false discover rate [FDR] < 0.001), but not in the I-to-II, supporting our observations that type III is closer to ESC/iPSC. In the I-to-II transition, only the gene signature at middle time point (day 11) is significantly enriched (FDR < 0.001). The iPSC signature is also induced in III-to-ESC/iPSC transition (FDR = 0.001), suggesting that while close to ESC/iPSC, type III cells have not fully completed reprogramming.
Population-based transcriptome analysis provides a more robust quantification of gene expression and has relatively low technical noise and high reproducibility (Marinov et al., 2014). Although it is very useful to flesh out the characteristics of the whole population, we cannot gauge the biological variation between the cells comprising that population. In order to investigate the heterogeneity of the intermediates, we compared our data with single-cell datasets obtained from partially reprogrammed cells (Chung et al., 2014). Consistently, the majority of double-positive cells (SSEA4+TRA160+) and none of SSEA4+TRA160− and GFP+ cells were classified into type III group (Figures S1H and S1I). While more than 75% of type II cells are SSEA4+TRA160−, more than 60% of type I cells are GFP+, indicating that the sorted-cell populations display heterogeneity, but mainly occupy specific intermediate stages. Overall, our transcriptome data are highly reliable and allow us to understand gene regulation changes during hiPSC reprogramming.
Primed and Naive-State Signatures Are Induced during iPSC Reprogramming
Despite many previous efforts to induce a naive-state in hESCs and hiPSCs (Takashima et al., 2014; Theunissen et al., 2014), it is still unclear whether or when OSKM induction is responsible for naive- and primed-state properties. To address the ground state in intermediate reprogramming stages, we analyzed the enrichment of genes specifically expressed in naive or primed ESCs (Figure 2B; Table S2B). GSEA revealed that primed-state signatures were significantly induced in fibroblast-to-I (FDR = 0.001) and type III-to-ESC/iPSC transition (FDR = 0.001). In contrast, naive-state signatures were significantly enriched in I-to-III (FDR = 0.001) and II-to-III transitions (FDR = 0.017). Significant repression of the primed-state was observed in I-to-II (FDR = 0.001) and I-to-III transitions (FDR = 0.001). These results indicate that type I and ESC/iPSC are biased to the primed state, whereas type III is to naive state. Type II is represented by a large depletion of primed-state signatures and no induction of naive-state signatures. Unlike dynamic changes of naive and primed signatures in human, murine iPSC reprogramming showed across-the-board increase of naive-specific (FDR < 0.001) and decrease of primed-specific genes (FDR < 0.017) in all intermediate stages (Figure 2C) (Polo et al., 2012).
We further addressed the expression changes in genes related to stem cell functions (Figure 2B). Genes related to stem cell maintenance and development and telomere maintenance are significantly induced in I-to-III and II-to-III transitions (FDR < 0.005). These gene sets are significantly depleted in I-to-II transition (FDR < 0.002), indicating that stem cell properties are gained with naive-state induction in type III. Gene sets involved in fibroblast proliferation are significantly suppressed in I-to-II and I-to-III transitions, confirming that type I stage still has fibroblast features. We observed a significant reduction of EMT-upregulated genes in MEF-to-ThyI+ transition in mouse (FDR = 0.001) (Figure 2C). On the other hand, we found a significant induction of epithelium developmental genes in fibroblast-to-I transition (FDR = 0.005) and a reduction of EMT-upregulated genes in I-to-II and I-to-III transitions (FDR = 0.003 and 0.001, respectively) in hiPSC reprogramming. This suggests that MET is required in both early and intermediate phases and promotes the exit of human reprogramed cells from the type I stage. Consistent with our previous finding that human female fibroblasts reactivate their inactive X chromosome during hiPSC reprogramming (Kim et al., 2014b), X-chromosome inactivation (XCI)-related genes are significantly repressed in fibroblast-to-I (FDR = 0.047) and are induced in III-to-ESC/iPSC stage (FDR = 0.042).
Cells in Type I Stage Present the Tumorigenic Potential
Since somatic reprogramming is induced by multiple oncogenic factors, the tumorigenic potential of iPSCs is a major concern for using iPSCs in cell therapy. To examine the tumorigenicity of each intermediate stage of reprogramming, we performed GSEA of cancer-related genes (Figure 2D). Since many oncogenes overlap with pluripotent genes, differentially expressed genes between ESCs and embryonic carcinoma cells (ECCs), a malignant counterpart of ESCs, were used as a cancer-related gene set (Table S2D) (Chang et al., 2010; Sperger et al., 2003). In hiPSC reprogramming, we observed that ECC-specific genes are significantly enriched in fibroblast-to-I transition (Figure 2D; FDR = 0.019). Interestingly, ECC-specific genes are significantly depleted in I-to-II, I-to-III, and II-to-III transitions (FDR = 0.001, 0.007, and 0.001, respectively). Additionally, a significant induction of ESC-specific genes was observed in I-to-III and II-to-III transitions (FDR = 0.001 and 0.001, respectively), indicating that type I is more tumorigenic than the other intermediate stages. This is consistent with our previous report demonstrating the formation of poorly differentiated teratomas from type I cells when injected into immunodeficient mice (Chan et al., 2009). In mouse, ECC-specific genes are significantly induced at Oct4-GFP+ stage (FDR = 0.001), but are reduced at mature iPSCs (FDR = 0.001) (Figure 2E). Our results show that tumorigenic potential was induced at the early and late stage of iPSC reprogramming in human and mouse, respectively.
Unique Alternative Splicing in Reprogramming
Alternative splicing (AS) is a key event to generate multiple isoforms and functional diversity in proteins. ESC/iPSC- or type III-specific isoforms are hypothesized to modulate the regulation of pluripotency and self-renewal. To identify stage-specific AS events, we compared spliced read alignments among different reprogramming stages (Figure S2A). A total of 636,803 junctions were aligned by our RNA-seq libraries, and about 24.6% of them were matched with splicing sites of RefSeq genes; 47.7% of them were not matched with RefSeq splicing sites, but were observed within RefSeq gene bodies. Spliced junctions within RefSeq genes were further filtered by (1) stage specificity score, (2) gene expression level, and (3) normalized counts of reads spanning the junction (see Experimental Procedures). Finally, a total of 2,342 (0.367%) splice junctions in 774 genes were identified as stage-specific AS candidates (Figure 3A). These candidates include spliced junctions in known differentiated cell- or ESC-specific isoforms of FOXP1 and MBD2 (Gabut et al., 2011; Lu et al., 2014) (Figures S2B and S2C).
Figure 3.
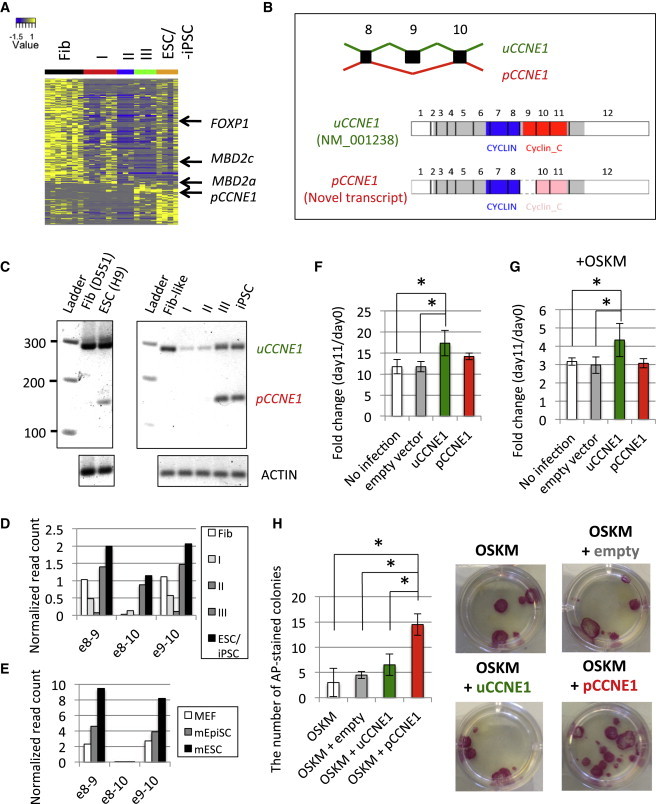
Alternative Spliced Forms of Genes Specific to Each Stage of Reprogramming
(A) Differential expression patterns of splice junctions. Colors represent the normalized read count mapped to each splice junction.
(B) Schematic representation of functional domains of splicing isoforms of CCNE1. Gray, blue, and red rectangles represent open reading frame, CYCLIN, and Cyclin_C domain, respectively. Pink rectangles represent the truncated Cyclin_C domain resulting from exon 9 skipping.
(C) RT-PCR assay using primers targeting exons 8 and 10. (Left) is derived from parental fibroblasts and H9 ESCs. (Right) is derived from sorted intermediate populations: Fib-like (w1 CD13+ GFP+), type I (w2 CD13+ GFP+ SSEA4+), II (w4 GFP+ SSEA4+ TRA160+), III (w4 GFP− SSEA4+ TRA160+), and iPSC.
(D and E) Exon 9 skipping of CCNE1 in (D) human and (E) mouse somatic and pluripotent stem cells.
(F and G) Effect of CCNE1 variants on cell growth rate. Fold change of cell count at day 11 to that at day 0 was calculated (F) without and (G) with OSKM induction (∗p < 0.05 by one-side t test, three biological replicates). Error bars represent SD.
(H) Positive regulation of hiPSC reprogramming by pCCNE1 overexpression. (Right) represents representative AP+ colonies in 12-well plate induced by overexpression of empty vector, uCCNE1, or pCCNE1 with reprogramming factors OSKM (three biological replicates). Error bars represent SD.
See also Figures S2 and S3.
In this study, we focused on the function of a previously uncharacterized variant from the CCNE1 gene. This variant excludes a highly conserved exon 9 of CCNE1 (Figure S3A), leading to the modification of Cyclin C-terminal (Cyclin_C) domain (Figure 3B). RT-PCR assay confirmed that the exclusion of exon 9 is observed only in pluripotent-cell stages (type III and ESC/iPSC) (pCCNE1, pluripotent CCNE1) (Figure 3C). In contrast, the known isoform of CCNE1 (NM_001238) is ubiquitously expressed from fibroblasts to ESC/iPSC stage (uCCNE1, ubiquitous CCNE1). Since pCCNE1 is also detectable in reprogramming with somatic cell nuclear transfer, Sendai virus (Figure S3B), episomal vectors (Figure S3C) and polycistronic OSKM lentivirus (Figures S3D and S3E), its induction does not depend on reprogramming methods. Whereas ESC-specific isoforms of Foxp1 and Mbd2 were also observed in mESCs (Figures S2B and S2C), exon 9 skipping of Ccne1 was not detected in mouse embryonic fibroblasts (MEFs), epiblast stem cells (EpiSCs) and ESCs (Figures 3D and 3E), indicating that pCCNE1 is a human-specific transcript variant.
Despite the high levels of uCCNE1 and pCCNE1 in type III stage and ESCs/iPSCs (Figures 3D and S3B–S3E), neither isoform is considerably expressed in fibroblasts after individual or combinatorial OSKM overexpression (Figures S3F and S3G). However, pCCNE1 expression is significantly increased by uCCNE1 overexpression (p = 2.21e-4), whereas pCCNE1 does not affect uCCNE1 transcription (p = 0.077). These results suggest that the stem cell-specific splicing of CCNE1 is not an initial target of OSKM; instead, it is most likely controlled by a higher amount of uCCNE1 and the transcriptional and signaling networks of pluripotency established in mature hiPSCs (Figure S3H).
Given the specificity of pCCNE1 expression in the pluripotent stage, we next asked about the functional differences between pCCNE1 and uCCNE1. Consistent with our knowledge that CCNE1 is involved in the cell cycle (Honda et al., 2005), overexpression of uCCNE1 significantly accelerates cell proliferation (p = 0.033 by one-side t test; Figure 3F). In contrast, pCCNE1 displays little effect on cell-cycle progression (p = 0.058). Furthermore, pCCNE1 cannot enhance cell proliferation even after OSKM induction (p = 0.312; Figure 3G), indicating that pCCNE1 loses its (if any) functional role in the cell-cycle progression during reprogramming. Interestingly, overexpression of pCCNE1, but not uCCNE1, with OSKM significantly increased the efficiency of hiPSC reprogramming by 4-fold more than OSKM alone (p = 0.022) or empty vector + OSKM (p = 0.022) (Figure 3H), as quantified by alkaline phosphatase (AP) staining. We validated our reprogramming data by double staining iPSCs with pluripotency markers SSEA4 and TRA160 (Figure S3I). Taken together, our results indicate that pCCNE1 is a newly identified pluripotent spliced form utilized by somatic cells to acquire pluripotency in a cell cycle-independent manner.
Monoallelic Gene Expression Is Uniquely Induced in Reprogramming
Allele-specific expression (ASE) is one of the gene regulatory systems that increase gene variations in a cell. A major change in ASE is known to occur during the pre-implantation development following maternal mRNA loss and paternal genome activation. Zygotic gene activation is induced at four- to eight-cell transition in humans and at one- to two-cell transition in mice (Xue et al., 2013), whereas in the blastocyst, the majority of genes are expressed biallelically. ESCs and differentiated cells display around 65%–80% of biallelic gene expression (Eckersley-Maslin et al., 2014). Despite much interest in its regulation, the ASE change during hiPSC reprogramming has been poorly understood due to the absence of advanced molecular tools. Thus, we measured the heterozygous single nucleotide polymorphism (SNP) expressions in each cell population isolated during reprogramming and calculated ASE ratios (reference:alternative allele expression ratios) for 105 SNPs observed within genes expressed in parental fibroblasts, intermediate stages, and established iPSCs; 68 of 105 SNPs were known SNPs registered in dbSNP Build 132 (Figure 4A). ASE ratios showing symmetric distribution with the highest peak at 0.5 were observed in parental fibroblasts, cell populations expressing fibroblast marker CD13 (GFP+CD13+), and iPSCs (Figure 4B), consistent with our previous report (Lee et al., 2009). This indicates that most genes are expressed from both alleles, or cells expressing either allele are equally mixed in these populations. On the other hand, in types I, II, and III-stage cell populations, ASE ratios in several SNPs were increased and decreased closer to 1 or 0, respectively, indicating that either allele is preferentially expressed during hiPSC reprogramming. The bias level of allelic preference is significantly higher in types I, II, and III than the fibroblast stage (Figure 4C; p = 4.14e-3, 4.29e-2, and 6.50e-4, respectively). This ASE bias was also observed in polycistronic vector-based reprogramming, indicating that the occurrence of ASE is not a corollary to individually expressed transgenes (Figure S3J).
Figure 4.
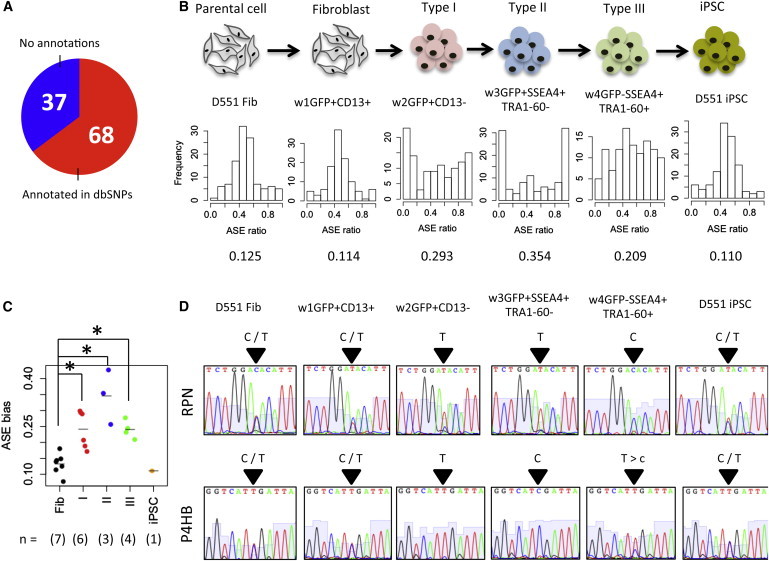
ASE Occurs in Intermediate Stages of hiPSC Reprogramming
(A) Overlap of 105 detected SNPs with dbSNP.
(B) Histograms of ASE ratios in six representative cell populations. Value below histogram represents ASE bias.
(C) Comparison of average ASE bias among different intermediate stages (∗p < 0.05 by one-sided t test). The number in parentheses denotes the number of populations in each class.
(D) Confirmation of ASE patterns of RPN and P4HB by Sanger sequencing.
See also Figure S3.
To validate ASE during iPSC reprogramming, we selected two SNPs in the RPN and P4HB genes and analyzed the SNP expression by Sanger sequencing (Figure 4D). These genes were expressed from both alleles (C and T) in parental D551 fibroblast, fibroblast-stage cell population, and iPSCs, while either allele (C or T) was predominantly or preferentially expressed in types I, II, and III. These results indicate that ASE occurs in the intermediate stages and that biallelic expression is restored when cells complete iPSC reprogramming.
Biphasic Change of Signaling Pathways
To gain insight into the mechanisms of signaling pathways in iPSC reprogramming, we analyzed their enrichment at each intermediate stage (Figure 5A; Table S2C). Type I-to-II transition was well represented by the reduction of most signaling pathways, while type II-to-III transition was characterized by the induction of NOTCH and WNT (FDR < 0.042; Figure 5B). Signaling pathways normally reduced or blocked in iPSC reprogramming (p53, neurotrophin, and MAPK) were indeed significantly repressed in I-to-II, I-to-III, and II-to-III transitions (FDR < 0.009) (Hong et al., 2009; Ishizuka et al., 2014; Levenberg et al., 2005). No significant induction or repression of any signaling pathways was observed in fibroblast-to-I and III-to-ESC/iPSC transition.
Figure 5.
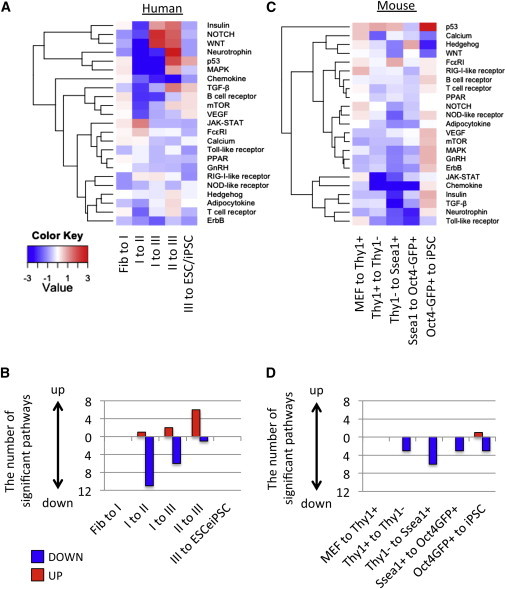
Biphasic Change of Signaling Pathways during hiPSC Reprogramming
(A and B) GSEA of signaling pathways (A) between distinct human intermediate stages and (B) between distinct mouse intermediate stages.
(C and D) The count of significantly upregulated (red) or downregulated (blue) pathways in (C) human and (D) mouse iPSC reprogramming.
See also Figure S4.
NOTCH signaling is one of the pathways that display a biphasic change. By adding NOTCH inhibitor DAPT or activator DLL4 ligand at specific periods of reprogramming (Figure S4A), we found that NOTCH inhibition at an early time point and activation at a late time point is more efficient than vice versa in enhancing reprogramming (Figures S4B and S4C). These data suggest that biphasic change of signaling pathway is an important consideration to improve the efficiency of iPSC reprogramming.
Conversely, we found no significant induction in most of signaling pathways between intermediate cells during murine iPSC reprogramming (Figures 5C and 5D). Only the P53 signaling pathway was significantly upregulated in Oct4-GFP+-to-iPSC transition (FDR = 0.001). These results suggest distinct signaling mechanisms during iPSC reprogramming between human and mouse or, alternatively, that hiPSC reprogramming is more sensitive to signaling pathways.
Type III and ESC/iPSC Signatures Are Co-regulated by Multiple Pluripotent Transcription Factors
Developmental genes have high factor loadings (FLs), while genes associated with the cell cycle and stem cell development have low FLs in principle component (PC) 2 and 3 (Figure S5A). Using FLs in PC1-3, we classified genes into three groups that are highly expressed in fibroblast type I (957 genes), type II (123 genes), and III-ESC/iPSC (511 genes) (Figure 6A; Table S3). The fibroblast type I group includes many fibroblast-specific markers such as CD13, COL1A1, COL1A2, and S100A4. In contrast, type III-ESC/iPSC group contains known pluripotency genes such as LIN28A, NANOG, PRDM14, ZFP42 (REX1), and DNMT3B. The type II group includes genes that both promote (OGT and PAF1) and block pluripotency and self-renewal (LEFTY2) (Ding et al., 2009; Jang et al., 2012; Kim et al., 2014a).
Figure 6.
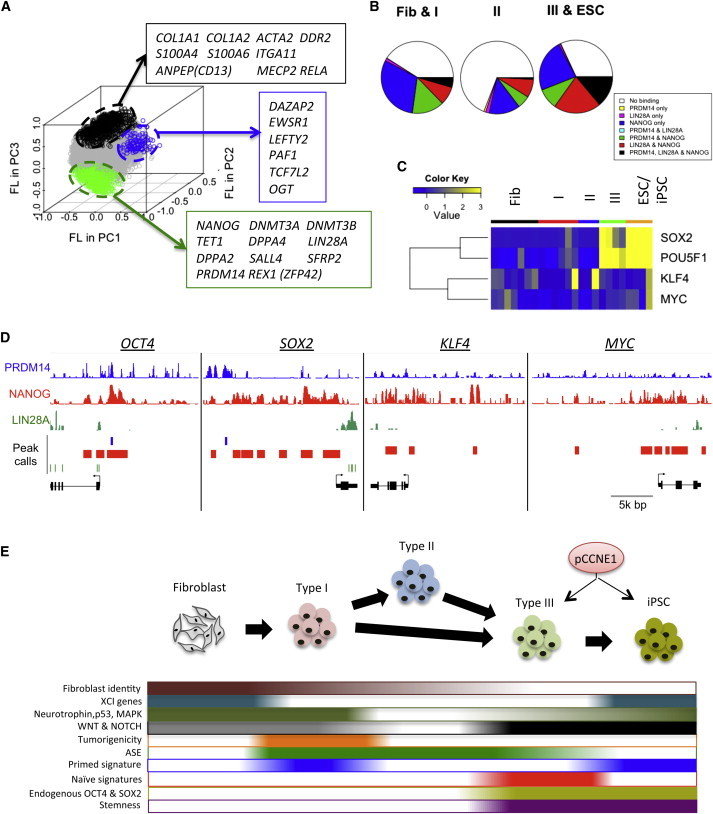
Transcriptional Regulation of Type III and ESC/iPSC Signatures by Multiple Pluripotent Factors
(A) Genes preferentially expressed in fibroblasts and type I, type II, and type III and ESC/iPSC. FLs in PC1–3 of each gene are plotted.
(B) Ratios of NANOG, PRDM14, and LIN28A target genes in fibroblast and type I, type II, and type III and ESC/iPSC gene sets.
(C) Endogenous OSKM expression patterns during hiPSC reprogramming. Relative expression to average was shown by color range blue (low expression) to yellow (high expression).
(D) NANOG, PRDM14, and LIN28A binding patterns in OSKM loci.
(E) Model of reprogramming milestones.
See also Figure S5.
To understand the regulatory mechanism of type III and iPSC gene signatures, we analyzed genes targeted by three main pluripotency regulatory factors (NANOG, PRDM14, and LIN28A) enriched in type III/ESC/iPSCs by using publicly available ChIP-seq and CLIP-seq datasets (Chia et al., 2010; Kunarso et al., 2010; Wilbert et al., 2012). Whereas NANOG binds more than 15,000 gene loci, PRDM14 and LIN28A targets comprise around 5,000 genes (Figure S5B). In addition, more than 95% of LIN28A and PRDM14 targets were co-targeted by NANOG. NANOG targets were significantly enriched in the fibroblast type I (p = 1.20e-12 by hypergeometric test) and type III-ESC/iPSC groups (p = 6.26e-3), but not in type II (p = 0.999) (Figures 6B and S5C). However, unique targets of NANOG are only significantly enriched in the fibroblast type I group (p = 2.86e-5), but not in type II (p = 0.983) and type III-ESC/iPSC groups (p = 0.871), suggesting that the gene regulation of type III-ESC/iPSC group is mediated by co-regulation of NANOG and the other pluripotent factors.
We found that endogenous OCT4 and SOX2 RNA expressions are only induced in type III and ESC/iPSCs (Figure 6C; Table S4). Since endogenous Oct4, Sox2, and Klf4 are induced in iPSCs and ESCs (Figure S5D), human and mouse employ distinct regulatory mechanisms to establish iPSCs. Co-targets of OCT4 and SOX2 were significantly enriched in type III-ESC/iPSC group (Figure S5E; p = 3.59e-14). These results indicate that the activation of endogenous OCT4 and SOX2 is correlated with the induction of type III and ESC/iPSC gene signatures in human. In addition, we found that whereas MYC and KLF4 are targeted by NANOG only, OCT4 and SOX2 are co-targeted by NANOG, PRDM14, and LIN28A (Figure 6D), supporting our hypothesis that co-regulation of multiple pluripotent transcription factors is required to regulate type III and iPSC gene signatures.
Discussion
Dissecting the transcriptional landscape of reprogramming represents one of the most straightforward ways to understand cell fate change. Most previous studies performed gene expression profiling in whole population of cells undergoing reprogramming. Only recently, the Yamanaka group described the transcriptome changes during human somatic cell reprogramming by microarray analysis of TRA160 sorted cells (Tanabe et al., 2013). Here, we used RNA-Seq to perform extensive transcriptome analyses of somatic cells undergoing reprogramming based on more elaborate combinatorial staining with CD13, SSEA4, and TRA160 and retroviral GFP.
By analyzing cells 3 days post-reprogramming factor induction, we demonstrated that the earliest gene expression response is independent of chromatin changes induced by OSKM. Although a previous study demonstrated that as pioneer regulators OCT4, SOX2, and KLF4 bind to the closed chromatin regions and initiate chromatin rearrangements (Soufi et al., 2012), our results showed that genes located at the closed chromatin regions do not show large transcriptional differences at day 3. Our observation suggests that 3 days is too short a time to remodel the fibroblast closed chromatin structure by OSK and that the initial gene regulation is mainly controlled by OKM transcriptional regulatory function.
Current transcriptome analysis by RNA-seq identified a large number of splicing variants of genes expressed at progressive stages of reprogramming, in addition to parental fibroblasts and iPSCs. In particular, we found that CCNE1 expresses human-specific pluripotent splicing variant pCCNE1 only when cells acquire pluripotency. One of the known functions of CCNE1 involves promoting the entry of G1 to S phase by binding to phospho-cyclin-dependent kinase 2 (pCDK2). Overexpression of a full-length uCCNE1 was not effective in promoting reprogramming, while pCCNE1 improved reprogramming without influencing cell-cycle progression. These data suggest that pCCNE1 possesses a pluripotency-specific function different from the cell-cycle-related general function of uCCNE1. The pCCNE1 isoform lacks exon 9, which is composed of two α helices and a loop (Figure S3A), and may thus play a role independently of its interaction with pCDK (Honda et al., 2005) and its localization at the centrosome (Matsumoto and Maller, 2004). In addition to pCCNE1, a large number of spliced forms of previously uncharacterized genes were identified in our analysis, and our data will be a very useful resource to dissect the regulation of gene splicing during reprogramming and function of genes uniquely spliced at pluripotency.
We found that the transitions of type I to types II and III are accompanied by dramatic changes in multiple signal transduction pathways. Interestingly, the P53 pathway was enriched in type III to ESC/iPSC in human and Oct4-GFP+ to iPSCs in mouse. Initially this finding seems somewhat contradictory, as P53 downregulation has been consistently shown to enhance the reprogramming process. However, at least in the human data, we found enrichment of cell-cycle-related genes, stress response, and DNA repair at later reprogramming stages. Since iPSCs have somatic mutations independently of derivation method as well as chromosomal aberrations of parental origin and from early and late passages (Gore et al., 2011; Johannesson et al., 2014), upregulation of P53 pathway could be a response to counter these genetic changes and maintain DNA integrity. Thus, although the purpose of late P53 induction is unclear at present, our data and previous studies point to one or more combinations of a faster cell cycle, reprogramming itself, original parental aberrations, and culture conditions. Similarly, we identified the biphasic repression and induction of the NOTCH signaling pathway, consistent with a recent report (Ichida et al., 2014). We further validated that activation of NOTCH pathway at a late time point increases reprogramming efficiency. We provide valuable information on the distinct function of signaling factors during different stages of reprogramming in order to more efficiently generate iPSCs.
Overall, our robust transcriptome data in cells undergoing hiPSC reprogramming showed dramatic changes in cell signaling pathways, human-specific AS, and ASE during the progressive cell fate change of fibroblasts to iPSCs (Figure 6E). The data will broaden the knowledge of the reprogramming process and human-specific gene regulation.
Experimental Procedures
Cell Culture
Normal primary fibroblast Detroit 551 were purchased from American Type Culture Collection (CCL-110) and maintained in DMEM high glucose (GIBCO) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. Human ESCs and iPSCs were cultured on irradiated murine embryonic feeder cells in medium containing DMEM/F12, 20% knockout serum replacement, and 4 ng/ml basic fibroblast growth factor (bFGF).
iPSC Reprogramming and Cell Sorting
The reprogramming procedure was conducted as previously described (Park et al., 2008b). Detroit 551 cells were seeded at 100,000 cells/well of a six-well plate 1 day prior to infection. A retrovirus cocktail containing OSKM was added to each well at MOI 5. On day 5 post-infection, the cells were trypsinized and transferred to 10-cm culture dishes containing MEFs. Prior to sorting, the cells were detached using accutase, washed, and incubated in 20% FBS in 1× PBS with the following antibodies according to manufacturer’s recommended dilutions: anti-human CD13 (BD catalog number 555394), anti-human/mouse SSEA4 (R&D catalog number FAB1435A), anti-human TRA160 (BD catalog number 560193). Sorting was conducted using a BD FACSAria cell sorter. Then the cells were pelleted and quickly frozen in liquid nitrogen or sorted directly in RLT + 2-mercaptoethanol lysis buffer (QIAGEN).
PMA RNA-Seq Library Construction and Illumina Sequencing
PMA RNA-seq library was prepared as previously described (Pan et al., 2013). Reads mapped to hg19 human genome were used for subsequent analyses. The details are given in Supplemental Experimental Procedures. All public data used in this study were summarized in Table S5.
Gene Expression Analysis
RNA was isolated using an RNeasy minikit (QIAGEN) and used for reverse transcription with iScript (BioRad) according to the manufacturer’s protocol with primer sets in Table S6.
Author Contributions
Y.T. performed all bioinformatics analysis. E.H. planned and conducted most of the experiments. Y.T., J.S., Y.X., K.-Y.K, and K.H. performed some of the experiments. E.H., Y.L., M.Z., X.P., S.M.W., G.E., and M.S. were involved in designing, generating, and performing PMA RNA-seq. I.-H.P. conceived and coordinated the project. Y.T., E.H., J.S. and I.-H.P. wrote the manuscript.
Acknowledgments
I.-H. P. was partly supported by NIH (GM0099130-01, GM111667-01), CSCRF (12-SCB-YALE-11, 13-SCB-YALE-06), KRIBB/KRCF research initiative program (NAP-09-3), and CTSA Grant UL1 RR025750 from the National Center for Advancing Translational Science (NCATS), a component of the NIH, and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH. Computation time was provided by Yale University Biomedical High Performance Computing Center.
Published: May 21, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Accession Numbers
The accession number for the pCCNE1 reported in this paper is GenBank: KR134287. All data are deposited to GEO with accession number GEO: GSE67915.
Supplemental Information
(A) Tanabe et al. (2013) datasets, (B) stem cell functions, (C) signaling pathways, and (D) cancer-related genes.
References
- Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.M., Ratanasirintrawoot S., Park I.H., Manos P.D., Loh Y.H., Huo H., Miller J.D., Hartung O., Rho J., Ince T.A. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Chang G., Miao Y.L., Zhang Y., Liu S., Kou Z., Ding J., Chen D.Y., Sun Q.Y., Gao S. Linking incomplete reprogramming to the improved pluripotency of murine embryonal carcinoma cell-derived pluripotent stem cells. PLoS ONE. 2010;5:e10320. doi: 10.1371/journal.pone.0010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Chen J., Wu Y. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Chia N.Y., Chan Y.S., Feng B., Lu X., Orlov Y.L., Moreau D., Kumar P., Yang L., Jiang J., Lau M.S. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Chung K.M., Kolling F.W., 4th, Gajdosik M.D., Burger S., Russell A.C., Nelson C.E. Single cell analysis reveals the stochastic phase of reprogramming to pluripotency is an ordered probabilistic process. PLoS ONE. 2014;9:e95304. doi: 10.1371/journal.pone.0095304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Paszkowski-Rogacz M., Nitzsche A., Slabicki M.M., Heninger A.K., de Vries I., Kittler R., Junqueira M., Shevchenko A., Schulz H. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin M.A., Thybert D., Bergmann J.H., Marioni J.C., Flicek P., Spector D.L. Random monoallelic gene expression increases upon embryonic stem cell differentiation. Dev. Cell. 2014;28:351–365. doi: 10.1016/j.devcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe J.A., Hilcove S., Kim J., Zhou H., Ouyang K., Wang G., Chen J., Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- Gabut M., Samavarchi-Tehrani P., Wang X., Slobodeniuc V., O’Hanlon D., Sung H.K., Alvarez M., Talukder S., Pan Q., Mazzoni E.O. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Gore A., Li Z., Fung H.L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho R., Papp B., Hoffman J.A., Merrill B.J., Plath K. Stage-specific regulation of reprogramming to induced pluripotent stem cells by Wnt signaling and T cell factor proteins. Cell Rep. 2013;3:2113–2126. doi: 10.1016/j.celrep.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Lowe E.D., Dubinina E., Skamnaki V., Cook A., Brown N.R., Johnson L.N. The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2-independent roles. EMBO J. 2005;24:452–463. doi: 10.1038/sj.emboj.7600554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida J.K., Blanchard J., Lam K., Son E.Y., Chung J.E., Egli D., Loh K.M., Carter A.C., Di Giorgio F.P., Koszka K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida J.K., Tcw J., Williams L.A., Carter A.C., Shi Y., Moura M.T., Ziller M., Singh S., Amabile G., Bock C. Notch inhibition allows oncogene-independent generation of iPS cells. Nat. Chem. Biol. 2014;10:632–639. doi: 10.1038/nchembio.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T., Goshima H., Ozawa A., Watanabe Y. Involvement of β-adrenoceptors in the differentiation of human induced pluripotent stem cells into mesodermal progenitor cells. Eur. J. Pharmacol. 2014;740:28–34. doi: 10.1016/j.ejphar.2014.06.056. [DOI] [PubMed] [Google Scholar]
- Jang H., Kim T.W., Yoon S., Choi S.Y., Kang T.W., Kim S.Y., Kwon Y.W., Cho E.J., Youn H.D. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012;11:62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Johannesson B., Sagi I., Gore A., Paull D., Yamada M., Golan-Lev T., Li Z., LeDuc C., Shen Y., Stern S. Comparable frequencies of coding mutations and loss of imprinting in human pluripotent cells derived by nuclear transfer and defined factors. Cell Stem Cell. 2014;15:634–642. doi: 10.1016/j.stem.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Kim D.K., Cha Y., Ahn H.J., Kim G., Park K.S. Lefty1 and lefty2 control the balance between self-renewal and pluripotent differentiation of mouse embryonic stem cells. Stem Cells Dev. 2014;23:457–466. doi: 10.1089/scd.2013.0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.Y., Hysolli E., Tanaka Y., Wang B., Jung Y.W., Pan X., Weissman S.M., Park I.H. X Chromosome of female cells shows dynamic changes in status during human somatic cell reprogramming. Stem Cell Reports. 2014;2:896–909. doi: 10.1016/j.stemcr.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G., Chia N.Y., Jeyakani J., Hwang C., Lu X., Chan Y.S., Ng H.H., Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Park I.H., Gao Y., Li J.B., Li Z., Daley G.Q., Zhang K., Church G.M. A robust approach to identifying tissue-specific gene expression regulatory variants using personalized human induced pluripotent stem cells. PLoS Genet. 2009;5:e1000718. doi: 10.1371/journal.pgen.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenberg S., Burdick J.A., Kraehenbuehl T., Langer R. Neurotrophin-induced differentiation of human embryonic stem cells on three-dimensional polymeric scaffolds. Tissue Eng. 2005;11:506–512. doi: 10.1089/ten.2005.11.506. [DOI] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lu Y., Loh Y.H., Li H., Cesana M., Ficarro S.B., Parikh J.R., Salomonis N., Toh C.X., Andreadis S.T., Luckey C.J. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell. 2014;15:92–101. doi: 10.1016/j.stem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyashenko N., Winter M., Migliorini D., Biechele T., Moon R.T., Hartmann C. Differential requirement for the dual functions of β-catenin in embryonic stem cell self-renewal and germ layer formation. Nat. Cell Biol. 2011;13:753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinov G.K., Williams B.A., McCue K., Schroth G.P., Gertz J., Myers R.M., Wold B.J. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24:496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston G.A., Zhu L.J., Chamberlain L., Lin L., Fang M., Green M.R. Non-canonical TAF complexes regulate active promoters in human embryonic stem cells. eLife. 2012;1:e00068. doi: 10.7554/eLife.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y., Maller J.L. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 2004;306:885–888. doi: 10.1126/science.1103544. [DOI] [PubMed] [Google Scholar]
- Ogawa K., Saito A., Matsui H., Suzuki H., Ohtsuka S., Shimosato D., Morishita Y., Watabe T., Niwa H., Miyazono K. Activin-Nodal signaling is involved in propagation of mouse embryonic stem cells. J. Cell Sci. 2007;120:55–65. doi: 10.1242/jcs.03296. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Semi K., Yamamoto T., Shimizu M., Tanaka A., Mitsunaga K., Okita K., Osafune K., Arioka Y., Maeda T. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Durrett R.E., Zhu H., Tanaka Y., Li Y., Zi X., Marjani S.L., Euskirchen G., Ma C., Lamotte R.H. Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proc. Natl. Acad. Sci. USA. 2013;110:594–599. doi: 10.1073/pnas.1217322109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M.W., Cowan C., Hochedlinger K., Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H., Lerou P.H., Zhao R., Huo H., Daley G.Q. Generation of human-induced pluripotent stem cells. Nat. Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Shah S.N., Kerr C., Cope L., Zambidis E., Liu C., Hillion J., Belton A., Huso D.L., Resar L.M. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS ONE. 2012;7:e48533. doi: 10.1371/journal.pone.0048533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G., Zaret K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperger J.M., Chen X., Draper J.S., Antosiewicz J.E., Chon C.H., Jones S.B., Brooks J.D., Andrews P.W., Brown P.O., Thomson J.A. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K., Nakamura M., Narita M., Takahashi K., Yamanaka S. Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc. Natl. Acad. Sci. USA. 2013;110:12172–12179. doi: 10.1073/pnas.1310291110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang J., Liu H., Lu D., Chen X., Zenonos Z., Campos L.S., Rad R., Guo G., Zhang S. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc. Natl. Acad. Sci. USA. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert M.L., Huelga S.C., Kapeli K., Stark T.J., Liang T.Y., Chen S.X., Yan B.Y., Nathanson J.L., Hutt K.R., Lovci M.T. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Huang K., Cai C., Cai L., Jiang C.Y., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y.E. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Pereira L., Hoffman J.A., Shy B.R., Yuen C.M., Liu D.R., Merrill B.J. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat. Cell Biol. 2011;13:762–770. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Tanabe et al. (2013) datasets, (B) stem cell functions, (C) signaling pathways, and (D) cancer-related genes.


