Summary
Human embryonic stem cell (hESC)-derived dopamine neurons are currently moving toward clinical use for Parkinson’s disease (PD). However, the timing and extent at which stem cell-derived neurons functionally integrate into existing host neural circuitry after transplantation remain largely unknown. In this study, we use modified rabies virus to trace afferent and efferent connectivity of transplanted hESC-derived neurons in a rat model of PD and report that grafted human neurons integrate into the host neural circuitry in an unexpectedly rapid and extensive manner. The pattern of connectivity resembled that of local endogenous neurons, while ectopic connections were not detected. Revealing circuit integration of human dopamine neurons substantiates their potential use in clinical trials. Additionally, our data present rabies-based tracing as a valuable and widely applicable tool for analyzing graft connectivity that can easily be adapted to analyze connectivity of a variety of different neuronal sources and subtypes in different disease models.
Graphical Abstract
Highlights
-
•
Monosynaptic tracing is used to uncover circuit integration of hESC-derived neurons
-
•
Afferent and efferent synaptic contacts of grafted hESC-derived neurons are revealed
-
•
Network integration in a rat model of Parkinson’s disease is stable for 6 months
In this study, Parmar and colleagues have applied monosynaptic tracing technology, using modified rabies vectors, to visualize the neuronal circuit networks formed between transplanted hESC-derived neurons and the host rat Parkinsonian brain. The afferent and efferent connectivity formed between grafted human neurons and the host rat brain has been visualized and mapped using this approach.
Introduction
Dopamine (DA) cell replacement for Parkinson’s disease (PD), using transplantation of human fetal ventral mesencephalic tissue to the striata of PD patients, has provided proof of principle that DA-neuron-rich grafts can survive long term and restore striatal DAergic function (Barker et al., 2013). The lack of available fetal tissue, as well as the logistic and ethical issues associated with the use of obtaining such tissue, has spurred the development of protocols for the generation of authentic and functional midbrain DA neurons from human embryonic stem cells (hESCs) (Kirkeby et al., 2012; Kriks et al., 2011). Pre-clinical validation of such hESC-derived neurons in a rat model of PD shows that they function with equal potency and efficacy to fetal DA neurons after transplantation (Grealish et al., 2014). With the overall goal of mapping how transplants of stem cell-derived human neurons integrate in the host brain in a preclinical rat model of PD, we used a method based on modified rabies virus (ΔG-rabies) that allows for the tracing of afferent and efferent connections of transplanted human neurons. ΔG-rabies is a deletion mutant virus in which the gene coding for glycoprotein (GP; necessary for transsynaptic spread) is replaced by the gene coding for a fluorescent protein, and the envelope is pseudotyped to only infect cells expressing the TVA receptor (Wickersham et al., 2007). Therefore, the initial infection can be targeted to any cell engineered to express the TVA receptor. If that cell is modified to also express GP, the ΔG-rabies can assemble into infectious particles in this cell and spread retrograde across one synapse. This system has previously been used to trace connectivity of endogenous neural circuitry (Miyamichi et al., 2011; Watabe-Uchida et al., 2012), as well as newly born neurons in the olfactory bulb and hippocampus (Deshpande et al., 2013; Vivar et al., 2012). Here, we use the system to trace connectivity of transplanted hESC-derived neurons in a rat model of PD.
Results
We used a polycistronic lentiviral tracing vector (Miyamichi et al., 2011) expressing a histone-tagged GFP; the TVA receptor, which allows for selective infection of ΔG-rabies; and rabies GP to allow for transsynaptic spread (Figure 1A). To control for unspecific labeling, we used a non-synaptic spreading of ΔG-rabies, a control vector containing GFP and TVA, but lacking GP (Figure 1A) in parallel.
Figure 1.
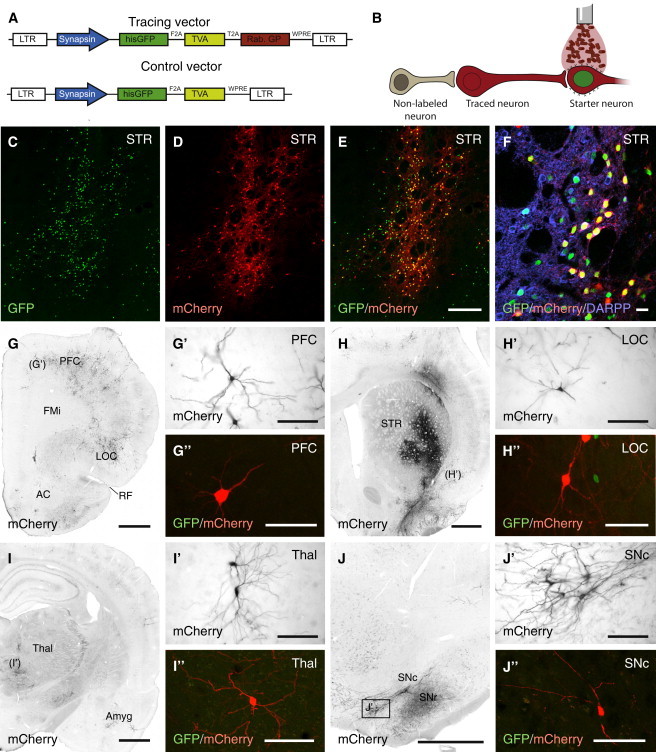
Overview of Monosynaptic Tracing Methodology
(A) Schematic representation of the lentivector constructs used in the study. The tracing vector labels cells with a histone-tagged GFP, the TVA receptor necessary for ΔG-rabies infection as well as the rabies GP that allows ΔG-rabies to transmit across synapses. The control vector lacks the GP and thus cannot transmit.
(B) Neurons that have been infected with the tracing construct are termed starter neurons. After ΔG-rabies infection, these cells turn mCherry+, and due to the presence of GP in the starter neuron, ΔG-rabies is transmitted retrogradely to label the traced neuron and cannot transmit further due to the lack of GP.
(C) Upon injection of the tracing lentivector to the rat striatum (n = 6), strong nuclear GFP+ expression is observed after 4 weeks.
(D and E) At 7 days after ΔG-rabies injection, a clear mCherry+ signal is observed co-localized with GFP+ nuclei.
(F) The main starter neuron population in this paradigm was DARPP32-expressing MSNs of the striatum.
(G–H″) Traced mCherry+/GFP− neurons could also be observed in rostral structures, such as the prefrontal and lateral orbital cortex and close to the injection site within the striatum.
(I–J″) In more caudal sections, traced neurons were found in regions known to project to the striatum, such as thalamus, amygdala, and substantia nigra pars compacta.
AC, anterior commissure; Amyg, amygdala; FMi, forceps minor; LOC, lateral orbital cortex; PFC, prefrontal cortex; RF, rhinal fissure; SNc, substantia nigra pars compacta; SNr, substantia nigra pars reticulata; STR, striatum; and Thal, thalamus. Scale bars represent 250 μm (C–E), 30 μm (F), 1 mm (G–J), 100 μm (G′–J′), 50 μm (G″ and H″), 100 μm (I″ and J″). See also Figure S1.
The targeted cells, here termed starter neurons, can be visualized by nuclear GFP expression (Figure 1B). Upon infection with the ΔG-rabies vector encoding mCherry, the targeted starter neurons expressing the TVA receptor can be infected by ΔG-rabies and easily identified by expression of GFP and mCherry (Figure 1B). These neurons are engineered to contain GP, and hence, ΔG-rabies can assemble into infectious particles in the starter cells. Any neuron that forms presynaptic contacts with the starter neuron, here called traced neuron, will be transduced with mCherry due to the selective transmission of rabies virus retrogradely across active synapses (Ugolini, 1995). The traced neurons can be easily identified from the starter neurons, as they express mCherry, but not GFP (Figure 1B). Further propagation of the ΔG-rabies vector does not occur as expression of the GP is restricted to the starter neuron and therefore only first-order synapses are traced (Etessami et al., 2000).
First, we traced endogenous striatal connections by injecting adult rats with either the tracing or control lentiviral vector into the striatum, and after 4 weeks, we injected the ΔG-rabies vector into the same site. When analyzed 1 week after ΔG-rabies injection, we found a large number of GFP+/mCherry+ starter neurons throughout the striatum (Figures 1C–1E). The vast majority of the GFP+ starter neurons co-expressed the medium spiny neuron (MSN) marker DARPP-32 (Figure 1F). Selective infection via the TVA receptor by ΔG-rabies vector was further confirmed by injecting ΔG-rabies into naive animals, where mCherry expression was never observed (not shown). mCherry+/GFP− neurons could also be found in the prefrontal, lateral orbital, and sensorimotor cortices (Figures 1G–1H″), as well as some scattered cells in thalamus, amygdala, and midbrain (Figures 1I–1J″). In animals that received the control lentiviral vector prior to injection of ΔG-rabies (i.e., where the targeted cells expressed TVA receptor but lacked GP), a large number of starter cells were observed expressing GFP and mCherry (Figure S1B). In these animals, however, we did not observe any tracing events (Figures S1A–S1H), confirming previous reports documenting that spread depends on expression of GP and occurs only via synaptically connected neurons (Etessami et al., 2000). The tracing observed is consistent with the known anatomy of striatal afferents (Gerfen and Wilson, 1996) and confirms the usefulness of the rabies-based tracing method for the visualization of striatal afferents throughout the brain.
To trace host-to-graft connectivity of transplanted hESC-derived neurons, we next generated hESC lines that stably expressed the tracing and control constructs (Figures 2A and S2A). The engineered hESCs were differentiated using a protocol that results in a high yield of authentic and functional ventral midbrain patterned DA neurons (Kirkeby et al., 2012). The midbrain-patterned cells (Figures S2C–S2F) were transplanted into a 6-hydroxydopamine (6-OHDA)-lesion model of PD, with athymic, nude rats as hosts (n = 8 per group). At 6 weeks post-transplantation, two rats per group were injected with ΔG-rabies within and adjacent to the graft core and perfused 7 days later. Remaining animals were left for their grafts to mature for 6 months, as detailed in the experimental timeline (Figure S2B). At this time point, the grafts were rich in DA neurons (Figures S2G and S2H). Based on quantifications of mCherry, GFP and tyrosine hydroxylase (TH) expression (Figure S2I), we could determine that 44.7% ± 15.7% of the starter neurons expressed TH. We also stained for serotonergic neurons and found only a small number (2.1% ± 0.4%) of the starter neurons expressed 5-HT (Figure S2J).
Figure 2.
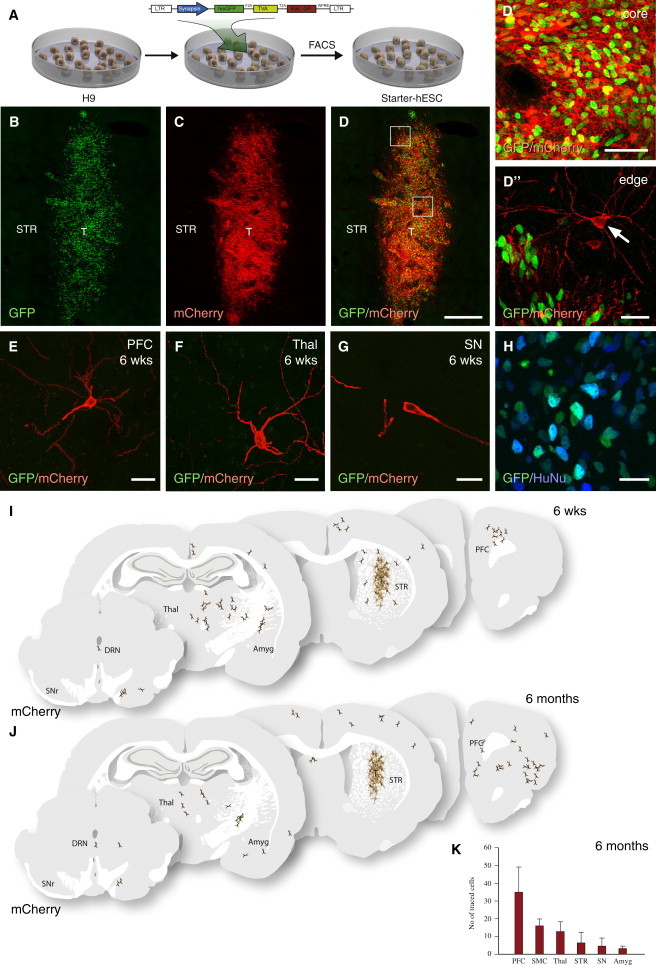
Host-to-Graft Connectivity of Intrastriatal Grafts of hESC-Derived Neurons
(A) Schematic illustration of generation of starter hESCs.
(B) Transplanted cells were detected based on GFP expression (n = 2 rats).
(C and D) ΔG-rabies selectively infected the transplanted tracer hESC-derived neurons.
(D′) Within the core of the transplant almost all GFP+ nuclei co-expressed mCherry.
(D″) mCherry+/GFP−-traced neurons were found in the host striatum at the graft edge.
(E–G) Traced host neurons could be detected in distal structures known to provide afferent inputs to striatum, such as the prefrontal cortex (E), thalamus (F), and substantia nigra (G).
(H) All GFP+ co-expressed the human-specific marker HuNu.
(I and J) Schematic illustration of the locations of the graft core, and distribution of mCherry+/GFP−-traced neurons in a brain 6 weeks (n = 2) and 6 months (n = 6) post-grafting.
(K) Quantifications of traced neurons in different host regions 6 months after transplantation (n = 5 rats, mean ± SEM).
Amyg, amygdala; DRN, dorsal raphe nucleus; PFC, prefrontal cortex; SNr, substantia nigra pars reticulata; SMC, sensory motor cortex; STR, striatum; T, transplant; Thal, thalamus. Scale bars represent 400 μm (B–D), 25 μm (D″, E, F, and H), and 50 μm (D′, J, G, and K). See also Figure S2.
Already 6 weeks after transplantation, grafts were readily detectable using GFP immunostaining (Figure 2B), and we observed that the majority of GFP+ grafted neurons were infected with ΔG-rabies (Figures 2C–2D′). Interestingly, mCherry+ neurons that did not co-express GFP (thus representing host cells that had made synaptic contacts with the transplanted neurons) could be observed adjacent to the graft core (Figure 2D″). In areas distal to the transplant, we observed mCherry+/GFP− host neurons within the prefrontal and sensorimotor cortices (Figures 2E and 2I), the thalamus (Figures 2F and 2I), and a few as far caudal as dorsal raphe nucleus and the substantia nigra, which is ≥5 mm away from graft core (Figures 2G and 2I).
At 6 months post-transplantation we confirmed that all starter cells (as detected by GFP expression) co-expressed human nuclei (HuNu; Figure 2H). At this time point, traced mCherry+/GFP− host neurons could be found in the striatum, close to the graft core, as well as in distal structures matching those observed at 6 weeks (Figure 2J). Quantifications showed that the majority of neurons connecting to the graft at this time point were located in the cortex, but connected neurons were also detected in thalamus, striatum, substantia nigra, and amygdala (Figure 2K; n = 5). The traced neurons in the prefrontal cortex were located mainly in layers III/V, displaying a classical pyramidal (Figure 3A) or basket cell (Figure 3B) morphology, and co-expressed the cortical marker TBR1 (Figure 3C). In the thalamus, the traced neurons were located in all anterior intralaminar nuclei (central medial, paracentral, and central lateral) (Figures 3D and 3F), which are the thalamic regions that normally project to the striatum (Van der Werf et al., 2002). These thalamic cells expressed Calbindin (CALB), but not Parvalbumin (PV) (Figures 3G and 3H). The traced host striatal cells connecting the grafted cells were distributed around the periphery of the graft and displayed a typical MSN morphology (Figures 2D″ and 3I) and expressed DARPP32 (Figure 3I). Thus, the connectivity observed after 6 weeks remained stable for up to 6 months post-transplantation, and the host brain regions that provide input to the grafted human neurons matched those that connected to endogenous striatal neurons.
Figure 3.
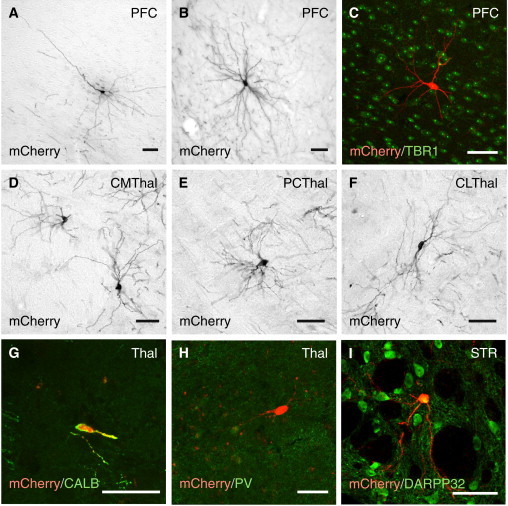
Characterization of Host-to-Graft Connectivity
(A and B) Traced cells in the prefrontal cortex (layers III/V) displayed (A) a classical pyramidal and (B) basket cell morphology.
(C) The traced host cells in cortex co-expressed the cortical marker TBR1.
(D–F) Representative images of traced cells in the three anterior intralaminar thalamic nuclei, which normally project to the striatum.
(G and H) Traced neurons in the thalamus co-express mCherry and CALB (G), but not PV (H).
(I) Locally traced cells in the vicinity of the transplant (n = 6) co-express mCherry and the MSN marker DARPP-32.
CLThal, central lateral thalamic nucleus; CMThal, central medial thalamic nucleus; PCThal, paracentral thalamic nucleus; PFC, prefrontal cortex; STR, Striatum; Thal, thalamus. Note that the far-red channel has been false color coded in green for easy visualization of co-labeling in (C), (G), (H), and (I). Scale bars represent 50 μm.
In the next set of experiments, we altered our experimental design to trace graft-to-host connectivity. Again, we used 6-OHDA lesioned nude rats, but this time the rats had been pre-injected with the tracing or control lentiviral vectors 4 weeks prior to transplantation. These virus injections were placed into two regions known to be densely innervated by transplanted human DA neurons: striatum (n = 6 for tracing and n = 4 for control) and prefrontal cortex (n = 5 for tracing and n = 4 for control). Thus, on the day of transplantation, the host brain contained GFP+ TVA-GP-expressing starter neurons and served as the recipient environment for the transplanted midbrain patterned WT hESCs (Figures S3A and S3B). Six months after transplantation, cells in the grafts had matured into functional DA neurons as confirmed by recovery in amphetamine-induced rotation (Figure S3C). At this time point, ΔG-rabies was injected at the same host brain regions that were initially injected with lentiviral vectors, resulting in selective infection of the modified host neurons via the TVA receptor. Subsequently, retrograde labeling of transplanted hESC-derived neurons that connected to host neurons in these regions should occur in animals pre-injected with tracing, but not control, vectors.
Local connectivity was assessed in animals pre-injected with lentiviral vectors into the striatum prior to grafting (Figure 4A). In these animals, a widespread mCherry expression in host striatum was observed (Figures 4B and 4C), and a distinct mCherry− graft core could be easily delineated (Figures 4B and 4C, dotted line demarcates graft-host border in B). In higher magnification, we observed traced mCherry+ neurons within the graft core of the animals pre-injected with the tracing vector (Figure 4D), but not in animals with the control vector (Figure 4E), indicating retrograde synaptic transfer of ΔG-rabies from starter neurons in the host striatum only in the presence of GP. In both groups, the grafted cells could be detected by the expression of the human-specific marker HuNu (blue cells in Figures 4F and S3D). The ΔG-rabies selective infection of the host starter neurons was confirmed by co-expression of GFP and mCherry (Figures 4G and S3E), whereas no HuNu+ human cells were found to co-express GFP (Figures 4G and S3E).
Figure 4.
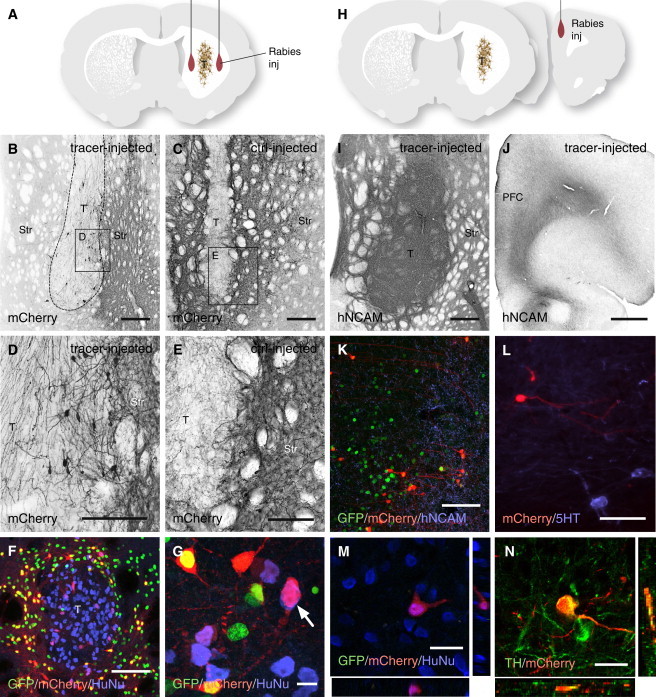
Graft-to-Host Connectivity of Intrastriatal Grafts of hESC-Derived Neurons
(A and H) Sites of injection of the ΔG-rabies vectors used to visualize graft-to-host connectivity.
(B and C) mCherry expressed in the host starter neurons was confined to the host striatum, while the transplant (T) was seemingly void of mCherry expression.
(D and E) In high-power magnification, mCherry+ neurons could be detected within the graft in the tracing group (n = 6; D), but never in the control group (n = 4; E).
(F and G) The HuNu+ graft was surrounded by GFP+ host nuclei, but none of the HuNu+ cells co-expressed GFP.
(I–K) Immunostaining for hNCAM showed that the grafts had established a dense innervation with the surrounding striatum (I), as well as the prefrontal cortex (J), including the site of prefrontal cortex that was targeted with the tracing lentivirus (K) (n = 5).
(L and M) The mCherry+ traced cells within the transplant core did not express 5HT (L), but were found to co-express HuNu (M) and TH (N), confirming that grafted human DA neurons had established synaptic contacts with neurons in the surrounding host striatum/prefrontal cortex.
PFC, prefrontal cortex; STR, striatum; T, transplant. Scale bars represent 1 mm (B, C, I, and J), 500 μm (D and E), 10 μm (G), 100 μm (K and F), 50 μm (L), and 25 μm (M and N). See also Figure S3.
Long distance connectivity was assessed in the animals pre-injected with lentiviral vectors into the prefrontal cortex prior to grafting (Figure 4H). The hESC-derived neurons had extended axons rostral innervating the prefrontal cortex (Figures 4I and 4J), where human neural cell adhesion molecule (hNCAM)-expressing axons could be found closely intermingled with mCherry+/GFP+ host starter neurons (Figure 4K). Analysis of the graft core, located in the striatum, 5 mm away from the site of the starter host neurons, revealed mCherry+/GFP−/HuNu+ neurons scattered within the graft core (Figure 4M). Phenotypic assessment of the traced mCherry+/GFP− cells showed that they co-expressed TH (Figure 4M), but not serotonin (5-HT) (Figure 4L), confirming that the grafted hESC-derived DA neurons had established synaptic contacts with the host neurons.
Discussion
We have used monosynaptic tracing based on modified rabies virus to analyze circuit integration of transplanted hESC-derived DA neurons in a rat model of PD. The technique allowed us to confirm the presence of graft-to-host and host-to-graft connectivity of transplanted human neurons, which is in line with previous studies of transplanted primary rodent DA neurons using electrophysiology, electron microscopy, or conventional tract-tracing methods (Fisher et al., 1991; Sørensen et al., 2005; Thompson et al., 2005; Tønnesen et al., 2011).
Our results revealed that the grafted hESC-derived DA neurons have a propensity for integrating in the host brain at a level exceeding what is commonly thought possible to achieve (Tønnesen and Kokaia, 2012). Additionally, it allowed for the precise identification of the source of host afferent monosynaptic inputs to the grafted cells, which has not been feasible until now due to limitations in existing technology. Our dataset shows that besides adjacent striatal cells, cortico-striatal input represents the most significant population of host afferents connecting with the grafted human neurons, but that also host neurons in the thalamus, dorsal raphe nucleus, amygdala, and substantia nigra innervate the grafted cells. This largely matches the endogenous striatal afferents mapped in this study and is in line with what has been previously described for transplanted striatal cells (Dunnett et al., 2000; Gerfen and Wilson, 1996). Interestingly, midbrain DA neurons normally receive afferent inputs from these cortical regions, as well as from striatal projection neurons (Watabe-Uchida et al., 2012), suggesting that the host brain may provide a regulatory control of the grafted DA neurons similar to that exerted by the intrinsic cortical and striatal afferents. One noteworthy and unexpected observation in this study is that the host afferents are formed as soon as 6 weeks after transplantation, at a time point much earlier than functional effects of transplants of human DA neurons normally can be detected.
Monosynaptic tracing using modified rabies virus has previously been successfully used to trace existing neuronal circuits in vivo (Wall et al., 2013; Watabe-Uchida et al., 2012) and also to map the integration of newborn neurons in the olfactory bulb and hippocampus (Deshpande et al., 2013; Vivar et al., 2012). Adapting it for tracing the connectivity of transplanted hESC-derived neurons, as done here, overcomes some limitations of existing tracing technology and opens up many new possibilities to analyze circuit integration after transplantation. First, it enables mapping in 3D of pre-synaptic inputs from host cells to grafted neurons in whole brains, which has not previously been possible because of a lack of adequate technology. Second, experiments in vitro and in vivo have confirmed the selective spread of ΔG-rabies only to synaptically coupled neurons (Wickersham et al., 2007), making it possible to distinguish between innervation and synaptic coupling. Third, the use of genetic targeting allows for analysis of integration of very defined cell populations. This makes the technology more refined than the use of conventional synaptic tracers, and it has already been used to re-define knowledge of neural circuits in vivo (Watabe-Uchida et al., 2012). One should keep in mind, though, that the technology is qualitative rather than quantitative and that it underrepresents the actual number of connected cells (Marshel et al., 2010).
Nevertheless, the technology offers new possibilities to qualitatively study functional integration of grafted neurons and serves as a valuable complement to electrophysiology, conventional tracers, and electron microscopy, as it allows for higher throughput analysis, mapping of both afferent and efferent connectivity, as well as visualization of connectivity from both local and distal brain structures.
Experimental Procedures
All data are presented as mean ± SEM.
Research Animals and Ethical Permissions
Adult (<180 g) athymic, nude female rats (n = 40) were purchased from Harlan Laboratories (Hsd:RH-Foxn1rnu). They were housed in individual ventilated cages, under a 12-hr light/dark cycle with ad libitum access to sterilized food and water. Female adult (225–250 g) Sprague Dawley rats (n = 12) were purchased from Charles River and were used to validate all viral vectors for these experiments; they were housed as described above but in standard caging.
All procedures were conducted in accordance with the European Union Directive (2010/63/EU) and were approved by the ethical committee for the use of laboratory animals at Lund University and the Swedish Department of Agriculture (Jordbruksverket).
Details of transplantation procedures are provided in detail in Supplemental Experimental Procedures.
Lentiviral Production
The constructs for the tracing and control vectors (as detailed in Figure 1A) were purchased from AddGene (IDs: 30195 and 30456, respectively). High-titer preparations of lentiviral particles were produced as previously described (Zufferey et al., 1997). Titers for the control vector were 3.1 × 108 U/ml and were used in vivo at a concentration of 20%. Titers for the tracing vector were 3.9 × 108 U/ml and were used in vivo at a concentration of 10%.
ΔG-Rabies Production
We produced our own pseudotyped rabies vectors according to Osakada and Callaway (2013) with minor adjustments. The protocol was stopped after step 60 as the virus was concentrated via ultracentrifugation only once and no sucrose cushion was used. Titering was performed using TVA-expressing HEK293T cells as defined in the protocol. Titers were 20–30 × 106 TU/ml, and for in vivo experiments were used at a dilution of 5%, as determined by testing different dilutions for a concentration that gave specific infection and tracing, in the absence of toxicity.
Differentiation of hESCs
Human ESCs H9 (WA09, passage 31–45) were differentiated to a ventral midbrain fate, using the protocol as described in detail in Supplemental Experimental Procedures, and generation of transgenic lines is also provided.
Microscopy and Quantifications
All bright-field images were captured using a Leica DMI6000B microscope, while all fluorescent images we acquired using a TCS SP8 laser-scanning confocal microscope. A full description of quantifications performed is provided in Supplemental Experimental Procedures.
Immunohistochemistry
All samples, cultured cells and brain tissue, were fixed in fresh 4% paraformaldehyde. A complete list of suppliers and concentrations of primary and secondary antibodies used is detailed in Supplemental Experimental Procedures.
Author Contributions
S.G., A.H., A.K., T.C., and M.J. performed and designed experiments. A.B., J.J., and M.P. designed experiments, guided preparation of the data and figures, and led data interpretation and analysis. S.G. and M.P. wrote the manuscript. All authors gave input for the manuscript.
Acknowledgments
We would like to thank Dr. Ed Callaway for help with establishing the technique and virus production and Bengt Mattsson, Michael Sparrenius, Ingar Nilsson, and Ulla Jarl for excellent technical assistance. This study was supported by grants from the European Community’s 7th Framework Programme through NeuroStemcellRepair (number 602278), the Strategic Research Area at Lund University Multipark (Multidisciplinary research in Parkinson’s disease), the Swedish Research Council (70862601/Bagadilico, K2012-99X-22324-01-5 and K2014-61X-20391-08-4), and the Swedish Parkinson Foundation (Parkinsonfonden). S.G. was supported by a postdoctoral stipend from the Swedish Brain Foundation (Hjärnfonden). The research leading to these results has received funding from the European Research Council under the European Union's 7th Framework Programme (FP/2007-2013)/ERC Grant Agreement number 309712.
Published: May 21, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information
References
- Barker R.A., Barrett J., Mason S.L., Björklund A. Fetal dopaminergic transplantation trials and the future of neural grafting in Parkinson’s disease. Lancet Neurol. 2013;12:84–91. doi: 10.1016/S1474-4422(12)70295-8. [DOI] [PubMed] [Google Scholar]
- Deshpande A., Bergami M., Ghanem A., Conzelmann K.K., Lepier A., Götz M., Berninger B. Retrograde monosynaptic tracing reveals the temporal evolution of inputs onto new neurons in the adult dentate gyrus and olfactory bulb. Proc. Natl. Acad. Sci. USA. 2013;110:E1152–E1161. doi: 10.1073/pnas.1218991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnett S.B., Nathwani F., Björklund A. The integration and function of striatal grafts. In: Björklund A., Dunnett S.B., editors. Progress in Brain Research. Elsevier; 2000. pp. 345–380. [DOI] [PubMed] [Google Scholar]
- Etessami R., Conzelmann K.K., Fadai-Ghotbi B., Natelson B., Tsiang H., Ceccaldi P.E. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J. Gen. Virol. 2000;81:2147–2153. doi: 10.1099/0022-1317-81-9-2147. [DOI] [PubMed] [Google Scholar]
- Fisher L.J., Young S.J., Tepper J.M., Groves P.M., Gage F.H. Electrophysiological characteristics of cells within mesencephalon suspension grafts. Neuroscience. 1991;40:109–122. doi: 10.1016/0306-4522(91)90178-q. [DOI] [PubMed] [Google Scholar]
- Gerfen C.R., Wilson C.J. The basal ganglia. In: Björklund A., Swanson L.W., Hökfelt T., editors. Handbook of Chemical Neuroanatomy. Elsevier; 1996. pp. 371–468. [Google Scholar]
- Grealish S., Diguet E., Kirkeby A., Mattsson B., Heuer A., Bramoulle Y., Van Camp N., Perrier A.L., Hantraye P., Björklund A., Parmar M. Human ESC-derived dopamine neurons show similar preclinical efficacy and potency to fetal neurons when grafted in a rat model of Parkinson’s disease. Cell Stem Cell. 2014;15:653–665. doi: 10.1016/j.stem.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkeby A., Grealish S., Wolf D.A., Nelander J., Wood J., Lundblad M., Lindvall O., Parmar M. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012;1:703–714. doi: 10.1016/j.celrep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Kriks S., Shim J.W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel J.H., Mori T., Nielsen K.J., Callaway E.M. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K., Amat F., Moussavi F., Wang C., Wickersham I., Wall N.R., Taniguchi H., Tasic B., Huang Z.J., He Z. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F., Callaway E.M. Design and generation of recombinant rabies virus vectors. Nat. Protoc. 2013;8:1583–1601. doi: 10.1038/nprot.2013.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen A.T., Thompson L., Kirik D., Björklund A., Lindvall O., Kokaia M. Functional properties and synaptic integration of genetically labelled dopaminergic neurons in intrastriatal grafts. Eur. J. Neurosci. 2005;21:2793–2799. doi: 10.1111/j.1460-9568.2005.04116.x. [DOI] [PubMed] [Google Scholar]
- Thompson L., Barraud P., Andersson E., Kirik D., Bjorklund A. Identification of dopaminergic neurons of nigral and ventral tegmental area subtypes in grafts of fetal ventral mesencephalon based on cell morphology, protein expression, and efferent projections. J. Neurosci. 2005;25:6467–6477. doi: 10.1523/JNEUROSCI.1676-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønnesen J., Kokaia M. Electrophysiological investigations of synaptic connectivity between host and graft neurons. Prog. Brain Res. 2012;200:97–112. doi: 10.1016/B978-0-444-59575-1.00005-3. [DOI] [PubMed] [Google Scholar]
- Tønnesen J., Parish C.L., Sørensen A.T., Andersson A., Lundberg C., Deisseroth K., Arenas E., Lindvall O., Kokaia M. Functional integration of grafted neural stem cell-derived dopaminergic neurons monitored by optogenetics in an in vitro Parkinson model. PLoS ONE. 2011;6:e17560. doi: 10.1371/journal.pone.0017560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini G. Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J. Comp. Neurol. 1995;356:457–480. doi: 10.1002/cne.903560312. [DOI] [PubMed] [Google Scholar]
- Van der Werf Y.D., Witter M.P., Groenewegen H.J. The intralaminar and midline nuclei of the thalamus. Anatomical and functional evidence for participation in processes of arousal and awareness. Brain Res. Brain Res. Rev. 2002;39:107–140. doi: 10.1016/s0165-0173(02)00181-9. [DOI] [PubMed] [Google Scholar]
- Vivar C., Potter M.C., Choi J., Lee J.Y., Stringer T.P., Callaway E.M., Gage F.H., Suh H., van Praag H. Monosynaptic inputs to new neurons in the dentate gyrus. Nat. Commun. 2012;3:1107. doi: 10.1038/ncomms2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall N.R., De La Parra M., Callaway E.M., Kreitzer A.C. Differential innervation of direct- and indirect-pathway striatal projection neurons. Neuron. 2013;79:347–360. doi: 10.1016/j.neuron.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M., Zhu L., Ogawa S.K., Vamanrao A., Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Wickersham I.R., Lyon D.C., Barnard R.J., Mori T., Finke S., Conzelmann K.K., Young J.A., Callaway E.M. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


