Abstract
Although conventional high-throughput screens performed in vitro with purified protein kinases are powerful tools to discover new kinase inhibitors, they are far from ideal for determining efficacy in vivo. As a complementary approach, cell-based, target-driven secondary screens may help predict in vivo compound potency and specificity as well as evaluate bioavailability and toxicity. Here the authors report a simple protocol for treating K562 Bcr-Abl-expressing cells with small-molecule kinase inhibitors in 96-well filter-bottom plates followed by in-plate cell lysis. The lysates were assayed via a solid-phase kinase assay, allowing determination of apparent IC50 for known Bcr-Abl inhibitors as well as facilitating the screening of a small kinase inhibitor library. This approach may have further applications in generating lysates for analyzing kinase activity and inhibition in other nonadherent suspension cell lines.
Keywords: cell lysis, secondary assay, Bcr-Abl, imatinib, lead identification
INTRODUCTION
Protein tyrosine kinases (PTKS) are key players in signal transduction, and their deregulation has been implicated in various human diseases, including autoimmune disorders and malignancies. As such, vast compound libraries have been tested in primary screens to assay the inhibition of deregulated PTKs. Primary screens for kinase inhibitors typically use an in vitro assay with a purified recombinant tyrosine kinase to assess relative inhibition. Although this allows many compounds to be screened at reduced cost, many promising lead compounds from in vitro experiments are eliminated at later, more costly stages of drug development due to lack of efficacy, nonspecific targeting, and/or unfavorable ADMET.1 Cell-based, target-driven secondary assays may help eliminate compounds with poor ADMET and/or in vivo efficacy and also facilitate the evaluation of compounds not directly targeting the kinase active site.
The Food and Drug Administration (FDA)–approved agents imatinib, nilotinib, and dasatinib have revolutionized the treatment of chronic myelogenous leukemia (CML)2,3 by inhibiting Bcr-Abl, the fusion gene product of the Ph1 reciprocal 9:22 translocation with constitutive tyrosine kinase activity.4,5 However, even more compounds are in clinical trials because mutations in the Bcr-Abl kinase cause resistance to these drugs.6,7 With new compounds constantly being developed to overcome resistance in CML as well as to target oncogenic tyrosine kinases involved in other diseases, the demand for new PTK inhibitors will continue for the foreseeable future.
We have previously described a solid-phase kinase assay using peptides immobilized on 96-well hydrogel plates to detect the activity of recombinant c-Abl or Bcr-Abl present in lysates from the CML-derived cell line K562.8 Here, aiming to measure targeted kinase activities in a more physiological context, we developed a protocol for treating K562 cells in 96-well filter plates with kinase inhibitors to obtain consistent amounts of lysate and detect Bcr-Abl activity with the solid-phase kinase assay. This simple, cell-based secondary assay may offer a rapid and inexpensive tool to select lead compounds from primary high-throughput screens while examining key drug properties such as cell permeability and toxicity, expediting the drug development process.
MATERIALS AND METHODS
Materials
Imatinib mesylate was provided by Dr. Wendy Stock (University of Chicago, Chicago, IL). The 80-compound Screen-Well™ Kinase Inhibitor Library was purchased from BioMol (Enzo Life Sciences, Plymouth Meeting, PA). A collection of pyrido[2,3-d]pyrimidine-7-one class multitargeted kinase inhibitors was synthesized by the methods of Klutchko et al9 and Boschelli et al.10 All compounds were dissolved and further diluted in molecular biology grade DMSO (Sigma-Aldrich, St. Louis, MO).
Cell culture
K562 cells11 (American Type Culture Collection, Manassas, VA) were grown at 37°C and 5% CO2 in RPMI-1640 media (Sigma-Aldrich) with 4 mM L-glutamine (Sigma-Aldrich) and 10% (v/v) fetal bovine serum (Gemini Bio-Products, West Sacramento, CA).
Bulk cell lysis and concentration analysis
K562 cells were lysed using PhosphoSafe Extraction Reagent (PER, Novagen, San Diego, CA) with 1× Protease Inhibitor Cocktail (PIC, Sigma-Aldrich). In brief, cells grown in T-175 flasks (Greiner Bio-One, Monroe, NC) were washed twice with 1 mL of cold phosphate-buffered saline (PBS; Fisher Scientific, Pittsburgh, PA) per 106 cells and incubated on ice for 20 min in 50 μL PER with 1× PIC per 106 cells. After brief agitation by vortex mixing, lysate was aliquoted into precooled 1.5-mL microcentrifuge tubes and centrifuged for 5 min at 16,000 g at 4°C. The protein concentration of the supernatant was analyzed with Coomassie Protein Assay Reagent (Pierce, Rockford, IL) by comparing absorbance at 595 nm to a bovine serum albumin standard.
Cell lysis in 96-well filter plate
The bottoms of 1-mL AcroPrep™ 96-well Filter Plates (Pall, East Hills, NY) with 3.0-μm glass fiber prefilters and 0.2-μm Bio-Inert filters were sealed with 3 layers of Parafilm (Pechiney Plastic Packaging, Menasha, WI). The plates were then filled with 106 cells per well suspended in 810 μL media. The cells were treated with 90 μL per well of either 10% DMSO or compound solution and shaken gently at 37°C for 1 or 2 h. Following drug treatment, the cells were lysed in the wells. First, the Parafilm was removed and the media were removed by centrifugation at 100 g for 5 min into a receiver plate. Then the cells were washed twice with 250 μL cold PBS per well by repeated centrifugation at 800 g at 4°C for 5 min. After a third 5-min centrifugation to remove residual PBS, 50 μL PER with 1× PIC was added per well, and plates were shaken on a Microplate Genie (Scientific Industries, Bohemia, NY) for 30 s and then incubated on ice for 20 min. The plates were shaken for another 30 s, and then cell lysates were collected in a chilled, sterile U-shaped 96-well plate (Greiner Bio-One) through centrifugation at 800 g at 4°C for 10 min. Another 25 μL PER with 1× PIC was added per well, the plate was briefly shaken, and flowthrough was collected by a 10-min centrifugation. The lysates were reserved on ice, and unused portions of lysate were flash frozen in liquid nitrogen and stored at −80°C. Protein concentrations were measured as above.
Peptide synthesis
Cys-Abltide (CEAIYAAPFAKKK)12 and Cys-Abl ligand (CGGAPTYSPPPPPLL)13 were synthesized on a Prelude Parallel Peptide Synthesizer (Protein Technologies, Tucson, AZ) using solid-phase Fmoc chemistry. The peptides were purified with an Agilent 1200 Series LC/MS (Santa Clara, CA) on an RP-C18 column and analyzed with an Applied Biosystems 4700 MALDI TOF/TOF MS (Foster City, CA). The peptides were dissolved in H2O, and concentrations were determined using Beer’s law at 280 nm.
Kinase reaction
The kinase assay was performed as previously described.8 Briefly, Cys-Abltide and Cys-Abl ligand were attached to ezrays™ plates (Matrix Technologies, Hudson, NH) and reacted with either 25 μg bulk lysate or 24 μL (~30 μg) lysate from the filter plate. Phosphorylated Cys-Abltide was probed with mouse antiphosphotyrosine recombinant clone 4G10 (Upstate, Charlottesville, VA) and horseradish peroxidase–conjugated antimouse IgG secondary antibody (GE Healthcare, Piscataway, NJ) and visualized using either SuperSignal West chemiluminescent substrate (Pierce, Waltham, MA) exposed to autoradiography film or Amplex Red substrate (Invitrogen, Carlsbad, CA) scanned by a Bio-Rad FX Pro Plus using the Alexa 532 filter (532 nm/588 nm). Quantitation of each well was performed with Quantity One software v4.6.2, with the diameter of the circular volume equivalent to the diameter of a well.
Assay evaluation
For assay evaluation, the cells were treated with 0 or 100 μM imatinib as positive and negative controls, respectively. The quantitated results were used to calculate the signal-to-background (S/B) and signal-to-noise (S/N) ratios, as well as the Z factor.14 Z factor ranges from negative values to 1, and a value greater than 0.5 is generally accepted as indicating that an assay is sufficiently robust for high-throughput screening (HTS). The S/B and S/N ratios and Z′ factor were evaluated as follows14:
where μpos and σpos are the mean and standard deviation of the positive controls, and μneg and σneg are the mean and the standard deviation of the negative controls, respectively.
IC50 calculation
Prism v5.01 (GraphPad, La Jolla, CA) was used to calculate the IC50 values. Briefly, chemiluminescence and chemifluorescence were quantitated in QuantityOne, and the values were normalized to untreated samples. The drug concentrations were log transformed, and a sigmoidal dose-response (variable-slope) curve was fitted to the medians.
RESULTS
96-well filter plate lysis validation
Previously, we developed a solid-phase kinase assay to measure the activity and inhibition of Bcr-Abl from K562 cell extracts using peptide substrates covalently attached to a 96-well hydrogel plate.8 It provides a simple and robust platform to screen small molecules for inhibition of Bcr-Abl in vitro. To accelerate the examination of Bcr-Abl inhibition in cells, we developed a protocol to treat K562 cells in 96-well filter-bottom plates with various small-molecule inhibitors. The treated cells are then lysed in the filter-bottom plates and collected in a 96-well plate to yield cell extracts for the solid-phase kinase assay (Fig. 1A).
FIG. 1.
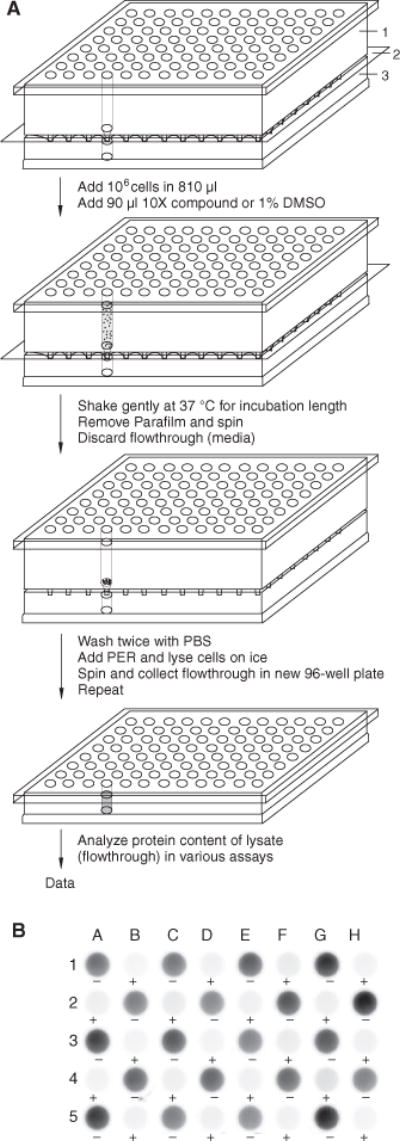
96-well filter plate lysis and lysate analysis. (A) Scheme for lysing cells in 96-well filter plates. (1) 96-well filter plate, (2) Parafilm, and (3) U-bottom 96-well plate. After sealing the bottom of the 96-well filter plate with Parafilm, cells are added and incubated with a compound or DMSO. When the incubation is complete, the media are centrifuged into a receiver plate, and the cells are washed twice with cold phosphate-buffered saline (PBS). PhosphoSafe Extraction Reagent (PER) with 1× protease inhibitor cocktail (PIC) is added to the cell pellets for lysis. Cell lysates are collected by centrifugation at 4°C, and an additional aliquot of PER with 1× PIC is added to each well and spun through to collect any remaining lysate. (B) Solid-phase kinase assay for Bcr-Abl activity. The lysates (24 μL/sample) were added to immobilized Cys-Abltide and Cys-Abl ligand on hydrogel plates to determine Bcr-Abl activity. The wells were probed with antiphosphotyrosine and visualized with Amplex Red fluorescence for Abltide phosphorylation.
To measure whether lysis occurred reproducibly throughout the filter plate between positive and negative controls, cells in 48 wells were treated for 2 h with either 1% (v/v) DMSO (positive controls) or 100 μM imatinib (negative controls) and lysed. The protein concentrations were 1.25 ± 0.15 μg/μL and 1.24 ± 0.10 μg/μL for the positive and negative control groups, respectively. Lysate concentrations were consistent between treatment group and nontreatment group.
The lysates were then analyzed for Bcr-Abl activities in the solid-phase kinase assay.8 In this kinase assay, a peptide substrate specific for Abl kinase (Cys-Abltide) and an Abl SH3 domain affinity peptide (Cys-Abl ligand) are tethered to a hydrogel surface through Michael addition chemistry. After performing the kinase reaction with the cell lysates, peptides are probed with an antiphosphotyrosine antibody and a horseradish peroxidase–conjugated secondary antibody and then visualized with either chemiluminescence or chemifluorescence. In the kinase assay, the lysates phosphorylated Cys-Abltide in each of 24 positive control wells and showed minimal background in all negative controls (Fig. 1B).
Several statistics were calculated for the kinase assay: the S/N ratio was 10.3, and the S/B ratio was 32.9. The Z factor considers both dynamic range and variability across multiple samples, with a value of 1 representing a theoretical perfect assay and values over 0.5 being suitable for high-throughput screens. In prior work using either purified c-Abl or Bcr-Abl in bulk K562 lysates,8 we attained Z factors between 0.5 and 0.9 for the solid-phase kinase assay. In our present study, we were able to obtain a Z factor of 0.4, most likely due to well-to-well variations in the protein concentrations. Although unlikely to provide a robust tool for high-throughput screens in its present form, the assay appears to have sufficient sensitivity and specificity to compare compounds in a medium-throughput assay.
Apparent imatinib IC50
To evaluate and compare the in vitro and in vivo inhibition of Bcr-Abl, we performed 2 kinase assays in parallel to determine the apparent IC50 values of imatinib. Here, in vitro inhibition is defined as adding compounds to K562 lysates during the kinase assay. In vivo inhibition is defined as treating K562 cells with the same compound prior to lysis, then analyzing the lysates in the kinase assay. To compare Bcr-Abl inhibition in these contexts, a 2-fold dilution series of imatinib from 256 μM to 0.0156 μM was examined for kinase inhibition in vitro (Fig. 2A, left) and in vivo (Fig. 2A, right). Kinase activity detected with chemifluorescence showed similar patterns of dose-dependent inhibition. The wells were quantitated and the medians were used to calculate the apparent IC50 values of in vitro versus in vivo inhibition, yielding 0.35 μM and 0.17 μM respectively, in close agreement with previously published values (Fig. 2B).15,16
FIG. 2.
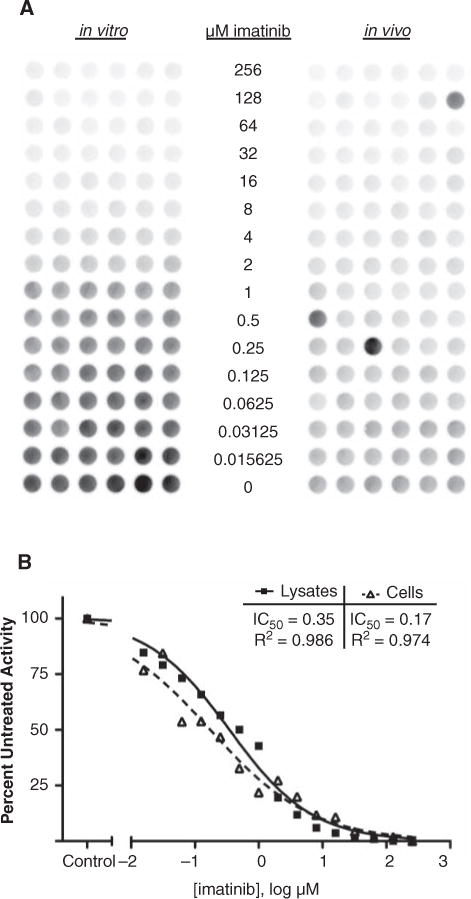
Imatinib dose responses in vitro and in vivo. (A) In vitro kinase assays were performed with a 2-fold dilution series of imatinib (256-0.015625 μM) added to K562 lysates during the kinase reaction. In vivo kinase assays were performed with lysates from cells that had been treated for 1 h with an identical dilution series of imatinib prior to lysis. The resulting phosphorylated Abltide was probed with antiphosphotyrosine and visualized with Amplex Red fluorescence. The image was obtained and analyzed using Bio-Rad QuantityOne software. (B) The IC50 curve was calculated for the in vitro and in vivo treatments in GraphPad Prism (v5.01) using the median from each dilution as quantified in QuantityOne. R2 represents the correlation of the data.
Cell-based model screens
Pyridopyrimidine series
After validating the lysis protocol and studying the sensitivity and reproducibility of the kinase assay, we screened 16 pyridopyrimidine dual-specificity kinase inhibitors alongside imatinib, dasatinib, and 2 dasatinib-related compounds, using chemiluminescence for visualization. Compounds in this library were previously observed to display superior inhibition of Bcr-Abl compared to imatinib when applied to K562 lysates.8 When K562 cells were treated with 0.04, 0.4, 4, or 40 nM of the compounds for 1 h, lysed, and then assayed for Bcr-Abl kinase activity, several significantly inhibited Bcr-Abl even at sub-nanomolar concentrations (Fig. 3), and all of them displayed apparent IC50 values lower than imatinib, which appeared to induce no inhibition at 40 nM.
FIG. 3.

Model screen of potent kinase inhibitors. Compounds that strongly inhibited purified c-Abl in previous screens8 were further characterized by in vivo assays. Briefly, cells were treated for 1 h prior to lysis. The collected lysates were added to the hydrogel plate with immobilized Cys-Abltide and Cys-Abl ligand. The resulting phosphorylated Cys-Abltide was probed with antiphosphotyrosine and visualized with chemiluminescence. Quantitation was performed using QuantityOne, and the IC50 curve was calculated using GraphPad Prism (v5.01).
BioMol ScreenWell™ Kinase Library
The BioMol ScreenWell™ Kinase Library is a collection of 80 compounds that show activity against diverse serine/threonine and tyrosine kinases.8 To interrogate this library for indirect and/or synergistic effects on Bcr-Abl activity, we sensitized the in vivo and in vitro kinase assays by combining 5 μM imatinib along with 200 μM of each compound. Substrate phosphorylation was detected with chemiflurorescence (Fig. 4A,B). Patterns of relative inhibition were distinct between the 2 screens, with several unique hits in vivo and in vitro. When combined with 5 μM imatinib, PP1 and PP2 demonstrated significant inhibition, but many other compounds also caused a measurable decrease in Bcr-Abl activity. Surprisingly, some compounds appeared to increase Bcr-Abl activity over that of 5 μM imatinib alone. It is possible that these compounds that appeared to increase Bcr-Abl activity may prevent the protein from folding into its inactive conformation, thereby hindering binding to imatinib. Alternatively, they may target antagonistic kinases in vivo.17 For quantitation, the activity of each compound was normalized to the mean of the control wells treated with 5 μM imatinib, which we defined as 100%. The normalized activities of the BioMol compounds from an in vivo screen were compared to normalized activities from an in vitro screen (Fig. 4C). Hits for each assay were separately determined to be 1 standard deviation below the mean of the screen. Five molecules—staurosporine, damnacanthal, PP1, PP2, and quercetin dihydrate—are active in both assays.
FIG. 4.
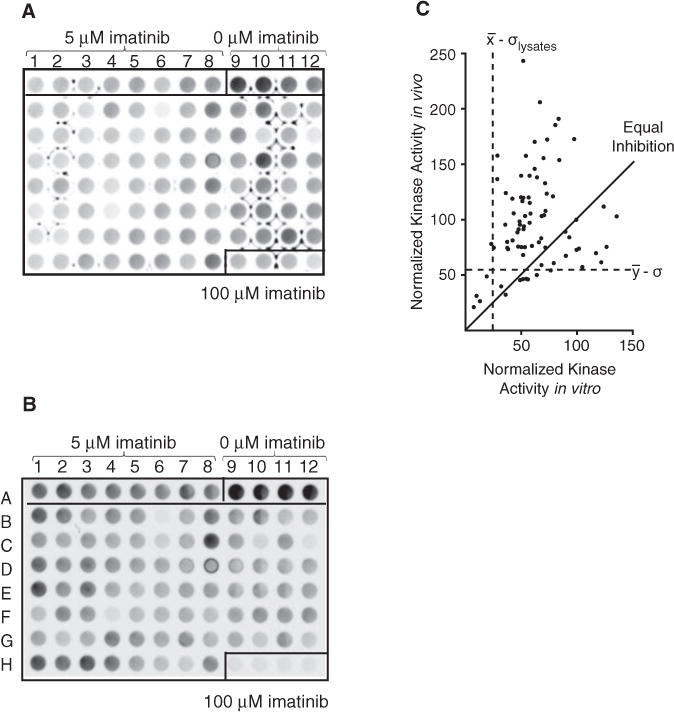
Model screen of BioMol kinase inhibitor library. (A) In vivo assay. K562 cells (106) were treated for 1 h with 0 (A9–A12), 5 (A1–A8), or 100 (H9-H12) μM imatinib alone or 5 μM imatinib in combination with 200 μM kinase inhibitors from the BioMol library (B1–H8) before lysing. Cys-Abltide and Cys-Abl ligand immobilized on hydrogel plates were reacted with the collected lysates, and the resulting phosphorylation was probed with antiphosphotyrosine and visualized with Amplex Red chemifluorescence. The stray marks are artifacts sometimes observed in low-intensity samples scanned with a higher photomultiplier voltage setting. These artifacts have no effect on quantitation, as the volumes are measured using circles that correspond to the diameter of the wells. (B) In vitro assay. K562 cell lysates were mixed with 0 (A9–A12), 5 (A1–A8), or 100 (H9–H12) μM imatinib alone or 5 μM imatinib in combination with 200 μM kinase inhibitors from the BioMol library (B1–H8) and added onto immobilized substrates on the hydrogel plates. The phosphorylation was with antiphosphotyrosine and visualized with Amplex Red chemifluorescence. (C) Comparison across screens. In vitro and in vivo screens, as defined in Figure 3, were performed and independently normalized to the respective 5-μM treated sample. Hits for each screen were determined separately as outliers less than 1 standard deviation below the mean (dashed lines). The solid line shows hypothetical equal inhibition in both screens.
In summary, we report here a medium-throughput screening protocol in which cells are treated with compounds in 96-well filter-bottom plates followed by in-plate lysis and analysis in a solid-phase kinase assay. Compared to the previous in vitro compound treatment, this protocol provides more information about the permeability and cellular efficacy of potential kinase inhibitors. Although this assay was developed to measure Bcr-Abl activity in K562 lysates, it is likely to be useful for other receptor and nonreceptor protein tyrosine kinases,18 given the availability of specific kinase substrates, and could also be employed for additional nonadherent cells, such as the widely used Ba/F3 cells.19 This protocol provides a medium-throughput scheme for generating lysates to accelerate sample preparation for cell-based assays requiring high-quality lysates and should be adaptable to robotic handling for increased throughput and consistency in the future.
Acknowledgments
We thank Won Jun Rhee and Stacey L. Kigar for synthesis and purification of the peptides, as well as Drs. Wendy Stock and Bayard Clarkson for generously providing reagents. Funding for this work was provided by National Institutes of Health grants R33 CA10323 and R01 HG003864. SJK was a Fletcher Scholar of the Cancer Research Foundation and a Leukemia & Lymphoma Society Scholar.
References
- 1.Bleicher KH, Bohm HJ, Muller K, Alanine AI. Hit and lead generation: beyond high-throughput screening. Nat Rev Drug Discov. 2003;2:369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 2.Das J, Chen P, Norris D, Padmanabha R, Lin J, Moquin RV, et al. 2-Aminothiazole as a novel kinase inhibitor template: structure-activity relationship studies toward the discovery of N-(2-chloro-6-methylphenyl)-2-[[6-[4-(2-hydroxyethyl)-1-piperazinyl)]-2-methyl-4-pyrimidinyl]amino)]-1,3-thiazole-5-carboxamide (dasatinib, BMS-354825) as a potent pan-Src kinase inhibitor. J Med Chem. 2006;49:6819–6832. doi: 10.1021/jm060727j. [DOI] [PubMed] [Google Scholar]
- 3.Zimmermann J, Buchdunger E, Mett H, Meyer T, Lydon NB. Potent and selective inhibitors of the Abl-kinase: phenylamino-pyrimidine (PAP) derivatives. Bioorgan Med Chem Lett. 1997;7:187–192. [Google Scholar]
- 4.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining [letter] Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 5.Kantarjian HM, Talpaz M, Giles F, O’Brien S, Cortes J. New insights into the pathophysiology of chronic myeloid leukemia and imatinib resistance. Ann Intern Med. 2006;145:913–923. doi: 10.7326/0003-4819-145-12-200612190-00008. [DOI] [PubMed] [Google Scholar]
- 6.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 7.Nardi V, Azam M, Daley GQ. Mechanisms and implications of imatinib resistance mutations in BCR-ABL. Curr Opin Hematol. 2004;11:35–43. doi: 10.1097/00062752-200401000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Wu D, Mand MR, Veach DR, Parker LL, Clarkson B, Kron SJ. A solid-phase Bcr-Abl kinase assay in 96-well hydrogel plates. Anal Biochem. 2008;375:18–26. doi: 10.1016/j.ab.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klutchko SR, Hamby JM, Boschelli DH, Wu Z, Kraker AJ, Amar AM, et al. 2-Substituted aminopyrido[2,3-d]pyrimidin-7(8H)-ones: structure-activity relationships against selected tyrosine kinases and in vitro and in vivo anticancer activity. J Med Chem. 1998;41:3276–3292. doi: 10.1021/jm9802259. [DOI] [PubMed] [Google Scholar]
- 10.Boschelli DH, Wu Z, Klutchko SR, Showalter HD, Hamby JM, Lu GH, et al. Synthesis and tyrosine kinase inhibitory activity of a series of 2-amino-8H-pyrido[2,3-d]pyrimidines: identification of potent, selective platelet-derived growth factor receptor tyrosine kinase inhibitors. J Med Chem. 1998;41:4365–4377. doi: 10.1021/jm980398y. [DOI] [PubMed] [Google Scholar]
- 11.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 12.Songyang Z, Carraway KL, III, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 13.Pisabarro MT, Serrano L. Rational design of specific high-affinity peptide ligands for the Abl-SH3 domain. Biochemistry. 1996;35:10634–10640. doi: 10.1021/bi960203t. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 15.Deininger MW, Druker BJ. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol Rev. 2003;55:401–423. doi: 10.1124/pr.55.3.4. [DOI] [PubMed] [Google Scholar]
- 16.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 17.Wang JY. Eph tumour suppression: the dark side of Gleevec. Nat Cell Biol. 2006;8:785–786. doi: 10.1038/ncb0806-785. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh G, Lee AG, Palecek SP. Hydrogel-based protein array for quantifying epidermal growth factor receptor activity in cell lysates. Anal Biochem. 2009;393:201–214. doi: 10.1016/j.ab.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol. 2007;19:55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- 20.Azam M, Nardi V, Shakespeare WC, Metcalf CA, Bohacek RS, Wang Y, et al. Activity of dual SRC-ABL inhibitors highlights the role of BCR/ABL kinase dynamics in drug resistance. Proc Natl Acad Sci U S A. 2006;103:9244–9249. doi: 10.1073/pnas.0600001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 22.Dorsey JF, Cunnick JM, Lanehart R, Huang M, Kraker AJ, Bhalla KN, et al. Interleukin-3 protects Bcr-Abl-transformed hematopoietic progenitor cells from apoptosis induced by Bcr-Abl tyrosine kinase inhibitors. Leukemia. 2002;16:1589–1595. doi: 10.1038/sj.leu.2402678. [DOI] [PubMed] [Google Scholar]
- 23.Dorsey JF, Jove R, Kraker AJ, Wu J. The pyrido[2,3-d]pyrimidine derivative PD180970 inhibits p210Bcr-Abl tyrosine kinase and induces apoptosis of K562 leukemic cells. Cancer Res. 2000;60:3127–3131. [PubMed] [Google Scholar]
- 24.von Bubnoff N, Veach DR, Miller WT, Li W, Sanger J, Peschel C, et al. Inhibition of wild-type and mutant Bcr-Abl by pyridopyrimidine-type small molecule kinase inhibitors. Cancer Res. 2003;63:6395–6404. [PubMed] [Google Scholar]
- 25.Tipping AJ, Baluch S, Barnes DJ, Veach DR, Clarkson BM, Bornmann WG, et al. Efficacy of dual-specific Bcr-Abl and Src-family kinase inhibitors in cells sensitive and resistant to imatinib mesylate. Leukemia. 2004;18:1352–1356. doi: 10.1038/sj.leu.2403416. [DOI] [PubMed] [Google Scholar]
- 26.Wisniewski D, Lambek CL, Liu C, Strife A, Veach DR, Nagar B, et al. Characterization of potent inhibitors of the Bcr-Abl and the c-kit receptor tyrosine kinases. Cancer Res. 2002;62:4244–4255. [PubMed] [Google Scholar]
- 27.O’Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, Mestan J, et al. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res. 2005;65:4500–4505. doi: 10.1158/0008-5472.CAN-05-0259. [DOI] [PubMed] [Google Scholar]
- 28.Veach DR, Namavari M, Pillarsetty N, Santos EB, Beresten-Kochetkov T, Lambek C, et al. Synthesis and biological evaluation of a fluorine-18 derivative of dasatinib. J Med Chem. 2007;50:5853–5857. doi: 10.1021/jm070342g. [DOI] [PubMed] [Google Scholar]


