Abstract
Many animal and human studies indicated that dietary ω-3 fatty acids could have beneficial roles on brain development, memory, and learning. However, the exact mechanisms involved are far from being clearly understood, especially for α-linolenic acid (ALA), which is the precursor for the ω-3 elongation and desaturation pathways. This study investigated the alterations induced by different intakes of flaxseed oil (containing 50% ALA), during gestation and lactation, upon the expression of genes involved in neurogenesis, memory-related molecular processes, and DNA methylation, in the brains of mouse offspring at the end of lactation (postnatal day 19, P19). In addition, DNA methylation status for the same genes was investigated. Maternal flaxseed oil supplementation during lactation increased the expression of Mecp2, Ppp1cc, and Reelin, while decreasing the expression of Ppp1cb and Dnmt3a. Dnmt1 expression was decreased by postnatal flaxseed oil supplementation but this effect was offset by ALA deficiency during gestation. Mecp2 DNA methylation was decreased by maternal ALA deficiency during gestation, with a more robust effect in the lactation-deficient group. In addition, linear regression analysis revealed positive correlations between Mecp2, Reelin, and Ppp1cc, between Gadd45b, Bdnf, and Creb1, and between Egr1 and Dnmt1, respectively. However, there were no correlations, in any gene, between DNA methylation and gene expression. In summary, the interplay between ALA availability during gestation and lactation differentially altered the expression of genes involved in neurogenesis and memory, in the whole brain of the offspring at the end of lactation. The Mecp2 epigenetic status was correlated with ALA availability during gestation. However, the epigenetic status of the genes investigated was not associated with transcript levels, suggesting that either the regulation of these genes is not necessarily under epigenetic control, or that the whole brain model is not adequate for the exploration of epigenetic regulation in the context of this study.
Keywords: Flaxseed oil, α-Linolenic acid, Perinatal nutrition, Brain development, DNA methylation, Lactation
1. Introduction
Memory is the process by which new information is being encoded, stored, and then retrieved (Abel and Lattal, 2001). Specificity in gene expression, protein synthesis and structural properties of neurons and synapses, is required for all three stages (Alberini, 2009). It has been acknowledged that dietary ω-3 polyunsaturated fatty acids (ω-3 PUFAs) could improve learning and memory, and also alter gene expression and proteome profiles in the whole brain or certain brain regions, such as cerebrum and hippocampus, therefore altering neurogenesis and synaptic plasticity (Kitajka et al., 2004; Lee et al., 2012).
α-Linolenic acid (ALA; 18:3 n-3) is the precursor for the ω-3 PUFAs elongation and desaturation pathways. This 18-carbon fatty acid possesses three double bonds and is commonly found in plant-derived dietary oils such as flaxseed, canola, and soybean oils. All the other common dietary ω-3 PUFAs have longer chains, and are found mainly in fish and fish oils, including the 20-carbon eicosapentaenoic acid (EPA; 20:5 n-3) and the 22-carbon docosahexaenoic acid DHA (Surette, 2013). Although numerous studies have defined the roles of DHA in brain development, less is known about the roles of ALA.
ALA availability has been shown to be important for brain development and its function during gestation and early postnatal life. During the perinatal period, the neonate's brain is experiencing a substantial acceleration in growth, cellular proliferation, and neuronal and glial differentiation (Hoffman et al., 2009). Also, perinatal ALA deficiency induced gene expression changes in the brain of adult rats (Kitajka et al., 2004). We have previously indicated that, in mice, ALA availability during gestation and lactation altered cell proliferation, early neuronal differentiation, and apoptosis, in the hippocampus of the offspring (Niculescu et al., 2011). These outcomes were associated with postnatal ALA supplementation, but were offset by gestational ALA deficiency. We also reported that the perinatal ALA availability was associated with epigenetic alterations in Fads2 DNA methylation in maternal and offspring livers (Niculescu et al., 2013).
Recent studies indicated the importance of epigenetic mechanisms in the alteration of neuronal gene expression patterns, which are required for synaptic plasticity or memory formation (reviewed in Sultan and Day, 2011). Epigenetic mechanisms include interrelated processes such as DNA methylation, histone modifications and RNA interference (Wood et al., 2005; Feng et al., 2010; Lubin et al., 2011).
In the current study, we explored the impact of maternal perinatal ALA availability on the expression of memory-associated genes in offspring brains, and the associated modifications in DNA methylation. Because memory and learning are complex processes that are not confined to only a brain area (Vousden et al., 2014; van Groen et al., 2014), we sought to first determine, in a whole brain model, whether such alterations could be detected, and whether this approach could warrant additional and more specific explorations.
2. Materials and methods
All reagents were purchased from Sigma–Aldrich (St. Louis, MO, USA) if not otherwise specified.
2.1. Animals, diets, and tissue collection
This study was approved by the University of North Carolina Institutional Animal Care and Use Committee. This work is a continuation of our previously published studies (Niculescu et al., 2011, 2013). Briefly, mouse C57BL/6J females (10 wk old; Jackson Laboratory, Bar Harbor, ME, USA) were randomly assigned into two initial feeding groups (intervention 1) for 30 days prior, and during gestation. One group was fed a defined control diet (AIN-93G, DYETS, Bethlehem, PA) containing soybean oil as fat source (ALA control, C, n = 12), while the other group was given an AIG-93G modified diet (DYETS) with corn oil as the only source of fatty acids (ALA deficient, D, n= 12). After 30 days, the females were bred overnight with males that were maintained at all times on the C diet. One day prior to delivery date, the two groups of females were randomly split into two subgroups (intervention 2). Half from each group remained on the same diet (n = 6), and the other half were switched to a modified AIN-93G diet containing flaxseed oil (ALA supplemented, S, n = 6), until postnatal day 20 (P19, considering the date of delivery as P0). On P19, all mothers and their pups were sacrificed. The pups were decapitated, the brains were extracted, snap frozen in liquid nitrogen, and stored at −80°C. In this study, the brain samples from the male pups were used for further analysis. However, due to the lack of male pups, only n = 5 male brains were included here in the CC, CS and DS respectively, while the DD group had n = 6 samples. For further reference, each group is designated by two letters: first letter reflects the diet received prior and during gestation, and the second letter reflects the maternal diet received during lactation.
The fatty acid composition of each diet has been previously published (Niculescu et al., 2011). Particularly, the diets contained 18:2 n-6 (linoleic acid, LA) and 18:3 n-3 (ALA) with the following concentrations: D diet, 87.2 and 1.5 nmol/mg; C diet, 86.9 and 10.8 nmol/mg; S diet, 23.1 and 75.3 nmol/mg.
2.2. Real-time RT-PCR
RNA was extracted from the male offspring brains (all at P19) with a QIAcube instrument (Qiagen, Valencia, CA, USA) using AllPrep® DNA/RNA mini kit (Qiagen), according to manufacturer's protocol. After the concentration and quality were determined with a NanoPhotometer (Implen GmbH, Munich, Germany), real time reverse transcriptase PCR (RT-PCR) method was used for the assessment of gene expression. First, cDNA synthesis was performed using a QuantiTect reverse transcription kit (Qiagen), on an Eppendorf Mastercycler ProS (Eppendorf, Hamburg, Germany). The amplification was performed on an Eppendorf Mastercycler Realplex2 ep Gradient S real-time PCR cycler, using the QuantiTect SYBR Green PCR kit (Qiagen). QuantiTect primer assays (Qiagen) were purchased for the following genes: Dnmt1, Dnmt3a, Mecp2, Gadd45b, Ppp1cb, Ppp1cc, Ppp3ca, Arc, Bdnf, Creb1, Reelin, Egr1 and 18s (used as internal reference for each sample). The real-time PCR reactions were run in triplicate, and data were retrieved as CT values normalized to 18S, then log2 transformed for subsequent statistical analysis. For each gene final data were expressed as ratios between each sample and the average of the CC group.
2.3. Bisulfite pyrosequencing
DNA from male offspring brains (all at P19) was extracted with a QIAcube instrument (Qiagen, Valencia, CA, USA) using AllPrep® DNA/RNA mini kit (Qiagen), following manufacturer's protocol. Up to 1 μg DNA from each sample was treated with sodium bisulfite using the same instrument and the corresponding protocol (EpiTect kit, Qiagen).
A pyrosequencing DNA methylation assay was designed for Mecp2 promoter using PyroMark Assay Design 2.0 software (Qiagen). The other 8 PyroMark CpG assays were directly purchased from Qiagen. Primers and sequences to be analyzed are indicated in Table 1. The amplification of templates was performed on an Eppendorf Mastercycler ProS using a PyroMark PCR kit and following the recommended protocol (Qiagen). The PCR conditions were: 95 °C for 15 min; 45 cycles of 94 °C for 30s, 56°C for 30 s, 72°C for 30s; final extension at 72°C for 10 min, and 4°C hold. The biotinylated strand of the amplified DNA was subjected to pyrosequencing on a PyroMark MD machine (Qiagen), as described previously (Mehedint et al., 2010). Each sample and sequence were run in triplicate, and only pyrosequencing reactions that passed the quality test were included in the analysis. For each sample and sequence, percentage methylated cytosine for each CpG site was expressed as the average of triplicates, and the values across all sites were averaged to express the average methylation across each sequence.
Table 1.
Assay information for bisulfite pyrosequencing.
| Gene & position | Assay | Sequence to analyze | Number of CpGs |
|---|---|---|---|
| Mecp2 promoter | In-house designed | F:AGTTTGGGTTTTATAATTAATGAAGGG | 7 |
| R[5-Biotin]: ACCTTAACCATCCCACTCACAATCTC | |||
| S: AGGTGTAGTAGTATATAGG | |||
| Sequence to analyze: | |||
| TTGGTCGGGAGGGCGGGGCGCGACGTTTGTCGTGCGGGG | |||
| Mecp2intron1 | Mm_Mecp2_01_PM | CGCGCGCAGCCCCAACTGGCGAAGCCCAGACGA | 5 |
| Reelin promoter | Mm_Reln_01 _PM | GGACCCGACAGGCGAGCTTCGCCGGACTCTGTATTTACGCGT | 6 |
| Egr1 promoter | Mm_Egr1_08_PM | CTCCACCCTGCGACCCGCTCCGGCATCGCGAGCGC | 6 |
| Arc exon1 | Mm_Arc_02_PM | AACTTGGACGGCTACGTGCCCACCGGCGACTCACAGCGC | 5 |
| Ppp1cb promoter | Mm_Ppp1cb_01_PM | CGAAACGCCGCGTGACTCGTAGGTGAGAACGCCG | 7 |
| Ppp1cc intron1 | Mm_Ppp1cc_03_PM | CGGCGCGCGGGTGGGTGGCGGGGCAGGCCCGGCCGGT | 7 |
| Dnmt3a promoter | Mm _Dnmt3a_04_PM | CGCGTCGCCACCCACAGCCAGGTGCCGCGGT | 5 |
| Dnmt1 promoter | Mm_Dnmt1_01_PM | GCGCATGCGCGCAACGGA | 4 |
F: forward primer; R, reverse primer; S, sequencing primer. CpG denotes a cytosine followed by a Guanine (CpG site).
2.4. Statistical analysis
Statistical analysis was carried out using both JMP 10 software (SAS, Cary, NC, USA) and MeV 4.9. (Dana-Farber Cancer Institute, Boston, MA, USA). MeV was used for the assessment of false discovery rates (FDR, Benjamini-Hochberg method) in the case of both gene expression and DNA methylation assessment. For each variable the p value (p < 0.05) was adapted accordingly to reflect a FDR of 5%. MeV was also used for Cluster Affinity Search Technique (CAST) on gene expression. For DNA methylation, because of its beta distribution, multiple comparisons were performed using JMP with the nonparametric Wilcoxon Each Pair test against adjusted p values calculated by the Kruskal–Wallis test (MeV). Gene expression was analyzed in JMP using two-way ANOVA (for the gestation and lactation factors), followed by Tukey HSD test for multiple comparisons, against adjusted p values. In addition, bivariate linear fit analysis was performed (JMP) for the correlation between two variables, with a threshold for significance of p < 0.05.
3. Results
3.1. Perinatal ALA availability alters the expression of genes associated with memory and DNA methylation
The expression of eight genes involved in memory and brain development, and of four genes involved in the regulation of DNA methylation, was determined in the whole brains of pups at the end of lactation (P19). The expression of three genes associated with memory was altered by ALA supplementation during lactation (Reelin, Ppp1cb, and Ppp1cc), regardless the ALA status during pregnancy (Fig. 1A). The expression of Reelin was increased in both CS and DS groups as compared to CC (ratio 5.44 ± 1.65 SE, and ratio 7.96 ± 1.60, respectively). While DS alterations were statistically significant, the CS alterations did not reach statistical significance (p = 0.017 vs. the FDR-corrected threshold p = 0.008). However, the expression values in both ALA supplemented groups were significantly higher than in the DD group. The expression of Ppp1cb was significantly lower in the CS group as compared to CC (ratio 0.23±0.05 SE). Ppp1cc expression was higher in the CS and DS groups as compared to both CC and DD groups (ratio 5.70 ± 1.25 SE vs. CC, and ratio 6.72 ± 1.76 SE vs. CC, respectively).
Fig. 1.
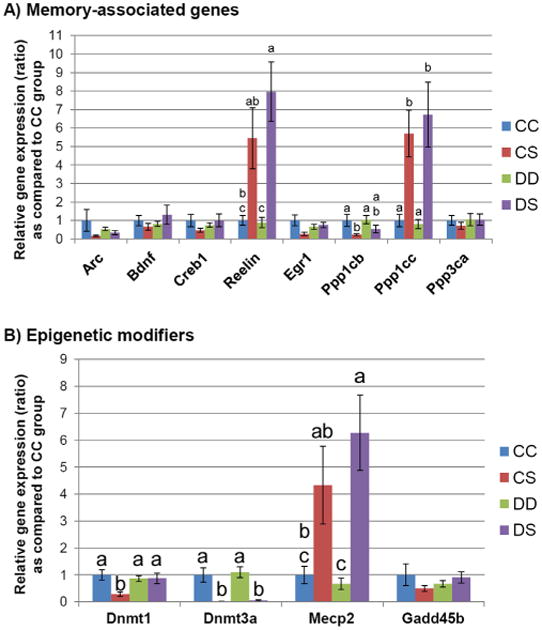
Gene expression assessment. At the end of lactation (P19), gene expression for DNA methylation regulatory genes and memory-associate genes was measured in male offspring brains, as described in Section 2. (A) The expression of eight genes involved in memory and brain development is indicated for each treatment group (whole P19 male brains). Columns marked with different letters denote statistical significance as assessed by Tukey HSD test. The following p values, adjusted against 5% FDR were used: Reelin p < 0.008, Ppp1cb p < 0.03, and Ppp1cc p < 0.008. Error bars indicate standard error (SE). (B) The expression of four genes involved in epigenetic regulation was measured for each treatment group. Columns marked with different letters denote statistical significance as assessed by Tukey HSD test. The following p values, adjusted against 5% FDR were used: Dnmt1 p < 0.03, Dnmt3a p < 0.008, and Mecp2 p < 0.008. Error bars indicate standard error (SE).
Among the genes involved in the epigenetic regulation of gene expression, three genes (Dnmt1, Dnmt3a, and Mecp2) were altered by perinatal ALA availability (Fig. 1B). Dnmt1 expression was decreased only in the CS group when compared to all other three treatment groups (ratio 0.29 ± 0.08 SE vs. CC). The expression of Dnmt3a was markedly lower in both ALA supplemented groups (CS and DS) as compared to either the CC or the DD groups (ratio 0.012 ± 0.003 SE vs. CC, and ratio 0.053 ± 0.03 SE vs. CC). The expression of Mecp2 was higher in ALA-supplemented groups. However, only the DS group reached statistical significance against both CC and DD groups (ratio 6.27 ± 1.40 SE vs. CC), while the increased expression in the CS group (ratio 4.33±1.44 SE vs. CC) was not statistically significant (p = 0.055 vs. the FDR-corrected threshold p = 0.008) when compared to the CC group, but only against the DD group.
3.2. Correlations between the expression of memory-associated genes and epigenetic regulators
We sought to determine whether the expression of genes involved in memory and neuronal development were correlated with the expression of epigenetic regulators. Using CAST analysis, we first identified clusters of genes that were candidates for such correlations. The identified clusters were then subject of linear regression for the assessment of statistical significance and coefficient of determination (R2), against a significance threshold p = 0.05. Several clusters were identified.
Mecp2, Reelin, and Ppp1cc formed one cluster (Fig. 2), as both the expression of Reelin (Fig. 2A, R2 = 0.98 and p < 0.0001) and Ppp1cc (Fig. 2B, R2 = 0.92 and p <0.0001) were highly correlated with Mecp2.
Fig. 2.

Correlations between Mecp2, Reelin, and Ppp1cc gene expression. CAST analysis followed by linear regression analysis (all samples) indicated that Mecp2 expression was highly correlated with the expression of Reelin (panel A) and Ppp1cc (panel B). The inset at the top shows the cluster identified by CAST analysis. R2 indicates the coefficient of determination for linear regression.
A second cluster consisted of Creb1, Gadd45b, and Bdnf (Fig. 3). Linear regression analysis indicated that both Bdnf (Fig. 3A, R2 = 0.65 and p < 0.0001) and Creb1 (Fig. 3B, R2 = 0.87 and p < 0.0001) were associated with Gadd45b.
Fig. 3.
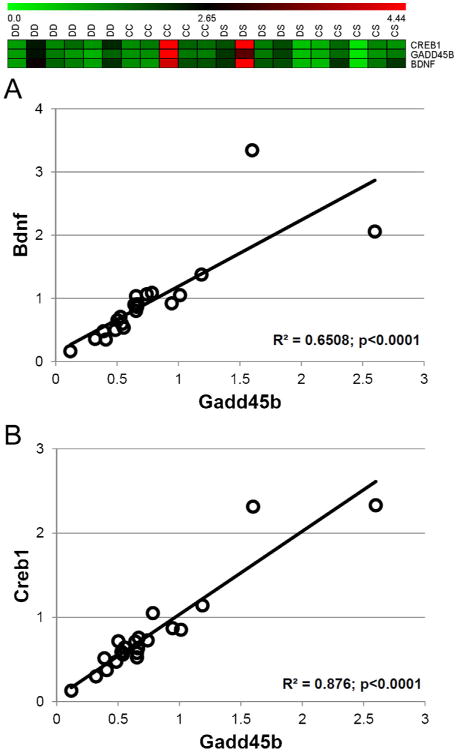
Correlations between Gadd45b, Bdnf, and Creb1 gene expression. CAST analysis followed by linear regression analysis (all samples) indicated that Gadd45b expression was highly correlated with the expression of Bdnf (panel A) and Creb1 (panel B). The inset at the top shows the cluster identified by CAST analysis. R2 indicates the coefficient of determination for linear regression.
In addition, Dnmt1 expression was significantly correlated with Egr1 across all samples (Fig. 4, R2 = 0.86 and p < 0.0001).
Fig. 4.
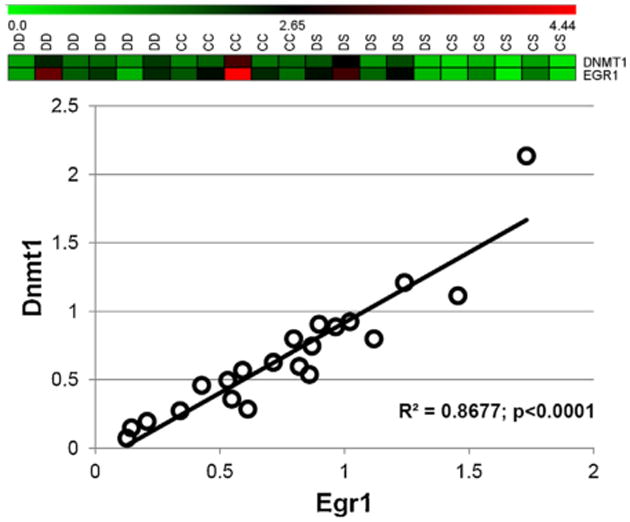
Dnmt1 expression is correlated with Egr1 expression. CAST analysis followed by linear regression analysis (all samples) indicated that Dnmt1 expression was highly correlated with the expression of Egr1. The inset at the top shows the cluster identified by CAST analysis. R2 indicates the coefficient of determination for linear regression.
3.3. Effects of the maternal ALA treatment on DNA methylation in the offspring brains
The assessment of DNA methylation status was performed by bisulfite pyrosequencing for the genes and sequences indicated in Table 1. Mecp2 promoter DNA methylation (Fig. 5) was altered by perinatal ALA availability in the CS and DD groups. Mecp2 promoter methylation in the DD group was significantly lower (% methylation 2.82 ± 0.35 SE) than in both CS and CC groups (% methylation 4.43 ± 0.19 SE, and 5.56 ± 0.44 SE, respectively), when tested against a FDR-adjusted value of p = 0.022. However, no differences between groups were noted for the Mecp2 intron 1 methylation. For all other DNA methylation assays, no significant changes were noted when testing against FDR-adjusted p values (data not shown).
Fig. 5.
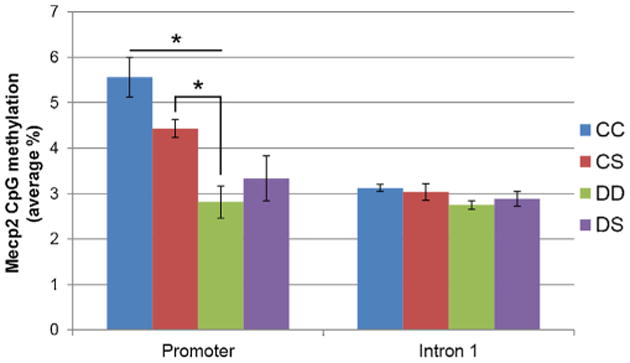
ALA availability alters Mecp2 promoter DNA methylation but not its intron I methylation. DNA was extracted from P19 whole brains, and subject of bisulfite pyrosequencing. Mecp2 DNA methylation was lower in the DD group, when compared to either the CC or the CS groups, indicating the overall ALA deficiency during gestation and lactation could trigger epigenetic alterations. Horizontal bars with asterisk above indicate statistical significance (p<0.05) between the respective two groups, as assessed by the Wilcoxon Each Pair test against adjusted p values calculated by the Kruskal–Wallis test.
3.4. Lack of correlation between gene expression and DNA methylation
Using a linear regression model, we sought to determine whether gene expression correlates with the DNA methylation status for the genes presented in Table 1. Using a threshold for statistical significance of p < 0.05, no correlations were found for any of the genes, including Mecp2 (data not shown).
4. Discussion
The findings of this study support the hypothesis that maternal ALA availability during pregnancy and lactation alters the expression of memory-associated genes and of those involved in the DNA methylation machinery, in the offspring brain at P19 (Fig. 1). In addition, ALA deficiency decreased the DNA methylation within the Mecp2 promoter, but not within intron 1 (Fig. 5). Interestingly, among the six genes that had substantial alterations in their transcript levels, five (Reelin, Ppp1cb, Ppp1cc, Dnmt3a, and Mecp2) were dependent exclusively on postnatal maternal ALA availability (regardless gestational ALA intakes), while Dnmt1 was decreased only in the CS group, indicating that postnatal ALA supplementation triggered its decreased expression, which was nevertheless offset by maternal ALA deficiency during gestation. The increased expression of Ppp1cc, Reelin and Mecp2 in the CS and DS groups support, in part, our previous findings about the enhanced neuronal differentiation due to postnatal ALA supplementation (Niculescu et al., 2011).
Reelin has been associated with the maintenance of polarity in cortical neurons and in the structural development of juvenile prefrontal circuits and memory formation (Forster, 2014; Iafrati et al., 2014). Moreover, the cognitive decline registered in aged rats was also associated with a reduction of Reelin expression (Stranahan et al., 2011).
Ppp1cc encodes one of the catalytic subunit (PP1γ) of nuclear protein-phosphatase 1 (Pp1), which was reported to decrease phosphorylation and ubiquitination of cAMP response element-binding protein (CREB), and thereby promote neurogenesis (Mu et al, 2013, 2011). While the role of the Ppp1cc subunit has not been sufficiently studied in brain, controversial roles have been assigned to the PP1 complex in regard to memory. In CD1 mice, PP1 levels were positively associated with olfactory memory (Winding et al., 2011). In contrast, increased PP1 activity was implicated in deficits in learning and memory induced by lead exposure in postnatal rats (Rahman et al., 2012), and in the epigenetic suppression of fear memory and synaptic plasticity in the amygdala (Koshibu et al., 2011). Moreover, the regulation of PP1 transcription could be epigenetically controlled by its DNA methylation (Miller and Sweatt, 2007). However, in our study, we did not find any DNA methylation differences between groups, which indicated that Ppp1cc expression control might be independent of its DNA methylation status in the context of maternal ALA availability.
In contrast with the increased Ppp1cc expression by postnatal ALA supplementation, Ppp1cb was decreased in same groups (CS and DS). This gene encodes the PP1 catalytic subunit PP1β (Saadat et al., 1994), and its nuclear inhibition was associated with increased memory performance (Graff et al., 2010). Again, our study did not identify any epigenetic modifications between groups.
The alterations induced by ALA availability to DNA methyltransferases (Dnmt1 and Dnmt3a), and to Mecp2 expression were more specific (Fig. 1B). Dnmt1 expression was decreased only in the CS group but not in the DS group, indicating that the influence of postnatal ALA supplementation upon Dnmt1 is offset by gestational ALA deficiency. Dnmt1 is a maintenance DNA methyltransferase required for the propagation of DNA methylation patterns to replicated DNA (Jin and Robertson, 2013). Because Dnmt1 is over expressed in post-mitotic neurons, then down-regulated in adult neurons (Inano et al., 2000), one could speculate that the Dnmt1 reduced expression in the CS group could reflect the more advanced differentiated state of neurons, which we previously reported to be the case for hippocampus (Niculescu et al., 2011). However, in the same study, we also reported that the CS group had increased cellular proliferation. Therefore, in this study, it is difficult to interpret the biological meaning of Dnmt1 decreased expression with ALA supplementation.
Dnmt3a expression was decreased in both postnatal ALA supplemented groups (CS and DS), irrespective of the ALA intakes during gestation. This suggests that, at P19, Dnmt3a expression in postnatal brains is not dependent upon gestational ALA levels. Because Dnmt3a is transiently expressed during neuronal differentiation (Watanabe et al., 2006), ALA-induced under-expression could suggest an increased differentiation state of the offspring brains exposed to postnatal ALA supplementation.
Mecp2 is an important epigenetic factor in the maintenance and development of the neurotransmission in cortical and hippocampal regions of the brain, through the regulation of synaptic gene expression (Na et al., 2013). Strongly involved in the development of central nervous system, Mecp2 deficiency was associated in mouse models with reduced neuronal dendritic formation (Schule et al., 2008). In the present study Mecp2 was over-expressed by postnatal ALA supplementation, and the increase was more robust statistically in the DS group (Fig. 1B). Because the increase in the CS group was not statistically significant due to one outlier, it is difficult to ascertain whether gestational ALA exposure was an independent factor. While Mecp2 promoter DNA methylation was decreased in the DD group as compared with either CC or CS groups, but not when compared with the DS group, one might speculate that its methylation status would be dependent primarily on the gestational ALA availability, rather than its postnatal intake levels. Linear regression indicated no association between Mecp2 expression levels and its DNA methylation status (data not shown).
Although the dietary treatments altered the expression of only six genes out of twelve, we sought to also investigate the relationship between the expression of genes, independent of dietary interventions. Few studies have addressed the issue of correlated expression between genes involved in brain development and memory. Therefore, such correlations could be informative regarding potential gene–gene interactions required during postnatal brain development. Several positive correlations were noted between the expressions of different genes. Mecp2 correlated with both Reelin and Ppp1cc (Fig. 2), suggesting that Mecp2 could be involved in the promoter activation of these two genes. Indeed, in humans MeCP2 binds to the Reelin promoter, and binding is enhanced by the hydroxy-methylation of Reelin promoter (Zhubi et al., 2014). It is not yet known whether Mecp2 has a similar role for the activation of Ppp1cc. Independent of ALA availability, the expression of Gadd45b was correlated with Bdnf and Creb1. Gadd45b, involved in the active DNA-demethylation required for adult neurogenesis (Ma et al., 2009), binds in humans to BDNF promoter and induces its over-expression (Gavin et al., 2012). Creb1 was reported to enhance Gadd45B expression in the mouse brain, but this role was highly contextual (Lemberger et al., 2008). Also independent of ALA availability, Dnmt1 correlated with Egr1 (Fig. 4). Egr1 is a zinc-finger protein and transcription factor involved in differentiation and proliferation. In brain, it regulates neuroplasticity and memory formation (Veyrac et al., 2014), potentially via a succession of events involving DNA methyltransferases Dnmt1, Dnmt3b, histone deacetylase Hdac1, and histone methyltransferase G9a (Cartron et al., 2013).
The present study has important limitations. While the investigation of whole brain can bring insightful information about global trends in brain development, its highly-organized and heterogeneous structure creates the premises for false negative results, if the alterations are localized to specific areas, or if opposite alterations are present in different brain regions. Therefore, the results of this study warrant further explorations that would interrogate potential alterations in specific brain areas associated with memory, such as amygdala, frontal cortex, septum, and hippocampus. Secondly, in the absence of a timed study, it is very difficult to decipher the sequence of events in regard to the correlations observed. Therefore, any inference of potential mechanisms or speculation regarding the role of such correlations should be made with caution.
The mechanisms by which ALA could induce alterations in gene expression are poorly understood. While ALA is a precursor for the synthesis of other ω-3 PUFAs, it can also have a distinct role in cell signaling (Darios and Davletov, 2006; Ren and Chung, 2007). ALA's role in the regulation of gene expression is mediated either through peroxisome proliferator activated receptor (PPAR-dependent) mechanisms, PPAR-independent mechanisms (reviewed in Innis, 2003), or by altering the phosphorylation of mitogen-activated protein kinases (MAPKs) involved in cell proliferation, differentiation and apoptosis (Blondeau et al., 2001; Ren and Chung, 2007). The activation of the ERK-dependent MAPK pathway component plays a central role in cell proliferation and differentiation via the nuclear c-fos activation mechanisms on gene expression (Nishioka et al., 2007). The expression of DNA methyltransferases is, in part, regulated by the activation of one or more MAPK pathways. ERK-MAPK inhibitors down-regulate Dnmt1 (Lu et al., 2007) and Dnmt3a (Deng et al., 2003). However, another MAPK pathway (JNK-dependent phosphorylation) may also be required for Dnmt1 promoter activation via c-jun phosphorylation (within the AP-1 transcription complex) (Slack et al., 2001). However, since we previously reported that ALA availability alters the content of EPA and DPA in the brains of the offspring (Niculescu et al., 2011), it is not clear whether ALA's role in altering the expression of genes is direct (via signaling), or due to alterations in other ω-3 species.
Our study suggested that ALA availability during gestation and lactation differentially altered the expression of genes involved in memory formation and neurogenesis, and in the epigenetic regulation of gene expression. Some of these alterations could be the result of the interplay between ALA availability in the two developmental periods, while others depended exclusively on postnatal ALA supplementation. With the notable exception of Mecp2 promoter methylation, ALA availability did not alter the methylation of the aforementioned genes, as measured in P19 whole brain extracts. Also, Mecp2 methylation was not correlated with its expression. Whether potential correlations between gene expression and DNA methylation could be altered by ALA availability in specific brain areas, it remains to be investigated further.
Acknowledgments
This work was funded, in part, by a grant to MDN from the UNC Center of Excellence in Children's Nutrition sponsored by Mead Johnson Nutrition.
Abbreviations
- ALA
α-linolenic acid
- P19
postnatal day 19
- DHA
docosahexaenoic acid
- DPA
docosapentaenoic acid
- EPA
eicosapentaenoic acid
- LA
linoleic acid
References
- Abel T, Lattal KM. Molecular mechanisms of memory acquisition, consolidation and retrieval. Curr Opin Neurobiol. 2001;11:180–187. doi: 10.1016/s0959-4388(00)00194-x. [DOI] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau N, Widmann C, Lazdunski M, Heurteaux C. Activation of the nuclear factor-κb is a key event in brain tolerance. J Neurosci. 2001;21:4668–4677. doi: 10.1523/JNEUROSCI.21-13-04668.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron PF, Blanquart C, Hervouet E, Gregoire M, Vallette FM. HDAC1-mSin3a-NCOR1, Dnmt3b-HDAC1-Egr1 and Dnmt1-PCNA-UHRF1-G9a regulate the NY-ESO1 gene expression. Mol Oncol. 2013;7:452–463. doi: 10.1016/j.molonc.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darios F, Davletov B. Omega-3 and omega-6 fatty acids stimulate cell membrane expansion by acting on syntaxin 3. Nature. 2006;440:813–817. doi: 10.1038/nature04598. [DOI] [PubMed] [Google Scholar]
- Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster E. Reelin, neuronal polarity and process orientation of cortical neurons. Neuroscience. 2014;269C:102–111. doi: 10.1016/j.neuroscience.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Koshibu K, Jouvenceau A, Dutar P, Mansuy IM. Protein phosphatase 1-dependent transcriptional programs for long-term memory and plasticity. Learn Mem. 2010;17:355–363. doi: 10.1101/lm.1766510. [DOI] [PubMed] [Google Scholar]
- Hoffman DR, Boettcher JA, Diersen-Schade DA. Toward optimizing vision and cognition in term infants by dietary docosahexaenoic and arachidonic acid supplementation: a review of randomized controlled trials. Prostaglandins Leukot Essent Fatty Acids. 2009;81:151–158. doi: 10.1016/j.plefa.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Iafrati J, Orejarena MJ, Lassalle O, Bouamrane L, Chavis P. Reelin, an extracellular matrix protein linked to early onset psychiatric diseases, drives postnatal development of the prefrontal cortex via GluN2B-NMDARs and the mTOR pathway. Mol Psychiatry. 2014;19:527. doi: 10.1038/mp.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inano K, Suetake I, Ueda T, Miyake Y, Nakamura M, Okada M, Tajima S. Maintenance-type DNA methyltransferase is highly expressed in postmitotic neurons and localized in the cytoplasmic compartment. J Biochem. 2000;128:315–321. doi: 10.1093/oxfordjournals.jbchem.a022755. [DOI] [PubMed] [Google Scholar]
- Innis SM. Perinatal biochemistry and physiology of long-chain polyunsaturated fatty acids. J Pediatrics. 2003;143:1–8. doi: 10.1067/s0022-3476(03)00396-2. [DOI] [PubMed] [Google Scholar]
- Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajka K, Sinclair AJ, Weisinger RS, Weisinger HS, Mathai M, Jayasooriya AP, Halver JE, Puskas LG. Effects of dietary omega-3 polyunsaturated fatty acids on brain gene expression. Proc Natl Acad Sci U S A. 2004;101:10931–10936. doi: 10.1073/pnas.0402342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshibu K, Graff J, Mansuy IM. Nuclear protein phosphatase-1: an epigenetic regulator of fear memory and amygdala long-term potentiation. Neuroscience. 2011;173:30–36. doi: 10.1016/j.neuroscience.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Lee J, Park S, Lee JY, Yeo YK, Kim JS, Lim J. Improved spatial learning and memory by perilla diet is correlated with immunoreactivities to neurofilament and alpha-synuclein in hilus of dentate gyrus. Proteome Sci. 2012;10:72. doi: 10.1186/1477-5956-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberger T, Parkitna JR, Chai M, Schutz G, Engblom D. CREB has a context-dependent role in activity-regulated transcription and maintains neuronal cholesterol homeostasis. FASEB J. 2008;22:2872–2879. doi: 10.1096/fj.08-107888. [DOI] [PubMed] [Google Scholar]
- Lu R, Wang X, Chen ZF, Sun DF, Tian XQ, Fang JY. Inhibition of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway decreases DNA methylation in colon cancer cells. J Biol Chem. 2007;282:12249–12259. doi: 10.1074/jbc.M608525200. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Gupta S, Parrish RR, Grissom NM, Davis RL. Epigenetic mechanisms: critical contributors to long-term memory formation. Neuroscientist. 2011;17:616–632. doi: 10.1177/1073858411386967. [DOI] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehedint MG, Niculescu MD, Craciunescu CN, Zeisel SH. Choline deficiency alters global histone methylation and epigenetic marking at the Re1 site of the calbindin 1 gene. FASEB J. 2010;24:184–195. doi: 10.1096/fj.09-140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Mu J, Krafft PR, Zhang JH. Hyperbaric oxygen therapy promotes neurogenesis: where do we stand? Med Gas Res. 2011;1:14. doi: 10.1186/2045-9912-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Ostrowski RP, Soejima Y, Rolland WB, Krafft PR, Tang J, Zhang JH. Delayed hyperbaric oxygen therapy induces cell proliferation through stabilization of cAMP responsive element binding protein in the rat model of MCAo-induced ischemic brain injury. Neurobiol Dis. 2013;51:133–143. doi: 10.1016/j.nbd.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na ES, Nelson ED, Kavalali ET, Monteggia LM. The impact of MeCP2 loss- orgain-of-function on synaptic plasticity. Neuropsychopharmacology. 2013;38:212–219. doi: 10.1038/npp.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu MD, Lupu DS, Craciunescu CN. Maternal alpha-linolenic acid availability during gestation and lactation alters the postnatal hippocampal development in the mouse offspring. Int J Dev Neurosci. 2011;29:795–802. doi: 10.1016/j.ijdevneu.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu MD, Lupu DS, Craciunescu CN. Perinatal manipulation of alphalinolenic acid intake induces epigenetic changes in maternal and offspring livers. FASEB J. 2013;27:350–358. doi: 10.1096/fj.12-210724. [DOI] [PubMed] [Google Scholar]
- Nishioka C, Ikezoe T, Takeshita A, Yang J, Tasaka T, Yang Y, Kuwayama Y, Komatsu N, Togitani K, Koeffler HP, Taguchi H. ZD6474 induces growth arrest and apoptosis of human leukemia cells, which is enhanced by concomitant use of a novel MEK inhibitor, AZD6244. Leukemia. 2007;21:1308–1310. doi: 10.1038/sj.leu.2404647. [DOI] [PubMed] [Google Scholar]
- Rahman A, Khan KM, Al-Khaledi G, Khan I, Al-Shemary T. Over activation of hippocampal serine/threonine protein phosphatases PP1 and PP2A is involved in lead-induced deficits in learning and memory in young rats. Neurotoxicology. 2012;33:370–383. doi: 10.1016/j.neuro.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Ren J, Chung SH. Anti-inflammatory effect of alpha-linolenic acid and its mode of action through the inhibition of nitric oxide production and inducible nitric oxide synthase gene expression via NF-kappaB and mitogen-activated protein kinase pathways. J Agric Food Chem. 2007;55:5073–5080. doi: 10.1021/jf0702693. [DOI] [PubMed] [Google Scholar]
- Saadat M, Kakinoki Y, Mizuno Y, Kikuchi K, Yoshida MC. Chromosomal localization of human, rat, and mouse protein phosphatase type 1 beta catalytic subunit genes (PPP1CB) by fluorescence in situ hybridization. Jpn J Genet. 1994;69:697–700. doi: 10.1266/jjg.69.697. [DOI] [PubMed] [Google Scholar]
- Schule B, Armstrong DD, Vogel H, Oviedo A, Francke U. Severe congenital encephalopathy caused by MECP2 null mutations in males: central hypoxia and reduced neuronal dendritic structure. Clin Genet. 2008;74:116–126. doi: 10.1111/j.1399-0004.2008.01005.x. [DOI] [PubMed] [Google Scholar]
- Slack A, Pinard M, Araujo FD, Szyf M. A novel regulatory element in the dnmt1 gene that responds to co-activation by Rb and c-Jun. Gene. 2001;268:87–96. doi: 10.1016/s0378-1119(01)00427-9. [DOI] [PubMed] [Google Scholar]
- Stranahan AM, Haberman RP, Gallagher M. Cognitive decline is associated with reduced reelin expression in the entorhinal cortex of aged rats. Cereb Cortex. 2011;21:392–400. doi: 10.1093/cercor/bhq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Day JJ. Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011;3:157–181. doi: 10.2217/epi.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette ME. Dietary omega-3 PUFA and health: stearidonic acid-containing seed oils as effective and sustainable alternatives to traditional marine oils. Mol Nutr Food Res. 2013 doi: 10.1002/mnfr.201200706. [DOI] [PubMed] [Google Scholar]
- van Groen T, Miettinen P, Kadish I. Axonal tract tracing for delineating interacting brain regions: implications for Alzheimer's disease-associated memory. Future Neurol. 2014;9:89–98. doi: 10.2217/fnl.13.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrac A, Besnard A, Caboche J, Davis S, Laroche S. The transcription factor Zif268/Egr1, brain plasticity, and memory. Prog Mol Biol Transl Sci. 2014;122:89–129. doi: 10.1016/B978-0-12-420170-5.00004-0. [DOI] [PubMed] [Google Scholar]
- Vousden DA, Epp J, Okuno H, Nieman BJ, Van Eede M, Dazai J, Ragan T, Bito H, Frankland PW, Lerch JP, Henkelman RM. Whole-brain mapping of behaviourally induced neural activation in mice. Brain Struct Funct. 2014 doi: 10.1007/s00429-014-0774-0. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Uchiyama K, Hanaoka K. Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience. 2006;142:727–737. doi: 10.1016/j.neuroscience.2006.07.053. [DOI] [PubMed] [Google Scholar]
- Winding C, Sun Y, Hoger H, Bubna-Littitz H, Pollak A, Schmidt P, Lubec G. Serine/threonine-protein phosphatase 1 alpha levels are paralleling olfactory memory formation in the CD1 mouse. Electrophoresis. 2011;32:1675–1683. doi: 10.1002/elps.201000615. [DOI] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Chen Y, Dong E, Cook EH, Guidotti A, Grayson DR. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]


