Summary
MicroRNAs are critical post-transcriptional regulators of hematopoietic cell-fate decisions, though little remains known about their role in aging hematopoietic stem cells (HSCs). We found that the microRNA-212/132 cluster (Mirc19) is enriched in HSCs and is up-regulated during aging. Both over-expression and deletion of microRNAs in this cluster leads to inappropriate hematopoiesis with age. Enforced expression of miR-132 in the bone marrow of mice led to rapid HSC cycling and depletion. A genetic deletion of Mirc19 in mice resulted in HSCs that had altered cycling, function, and survival in response to growth factor starvation. We found that miR-132 exerted its effect on aging HSCs by targeting the transcription factor FOXO3, a known aging associated gene. Our data demonstrates that Mirc19 plays a role in maintaining balanced hematopoietic output by buffering FOXO3 expression. We have thus identified it as a potential target that may play a role in age-related hematopoietic defects.
Introduction
Hematopoietic stem cells (HSCs) are the source of most all the immune cells in our body (Orkin and Zon, 2008). A complex gene regulatory network tightly regulates the function and survival of HSCs to ensure balanced and appropriate hematopoietic output (Novershtern et al., 2011). Alteration of the HSC niche and deregulation in cell-intrinsic properties such as HSC self-renewal and cycling, metabolism, and survival can have drastic consequences on hematopoietic output (Passegue et al., 2005; Suda et al., 2011). As an organism ages, the balance between HSC self-renewal, function and survival is drastically altered (Geiger et al., 2013), and this may lead to deleterious consequences such as the inability to effectively combat infection, and the onset of autoimmune disease or hematologic cancers (Frasca and Blomberg, 2011; Henry et al., 2011).
Aged HSCs are characterized by increased self-renewal potential, loss of long-term reconstitution capability, myeloid-biased differentiation and a change in niche localization. As a consequence, aged mice demonstrate an accumulation of phenotypically defined HSCs with a poor ability to home to the bone marrow niche (Geiger et al., 2013). These aged HSCs also develop a requirement for basal autophagy for survival, because replication stress and the accumulation of reactive oxygen species have harmful consequences on HSC function with age (Flach et al., 2014; Tothova et al., 2007). The loss of critical autophagic factors is often associated with altered cell cycling of HSCs, and leads to apoptosis and a rapid loss of HSC numbers in aged mice (Miyamoto et al., 2007; Rubinsztein et al., 2011; Warr et al., 2013). A critical balance between cell cycling and differentiation, and survival of aged HSCs must therefore be established to maintain normal hematopoietic output.
Several genetic and epigenetic factors have been identified as important regulators of hematopoietic stem cell aging (Geiger et al., 2013; Rossi et al., 2012; Sun et al., 2014). To date, however, little is known about the role of noncoding RNAs in the regulation of hematopoietic stem cells with age. MicroRNAs, a class of small-noncoding RNAs, are important post-transcriptional regulators of hematopoietic cell-fate decisions (Baltimore et al., 2008; Chen et al., 2004; Gangaraju and Lin, 2009). They alter cell fate by negatively regulating gene expression through direct binding to the 3′untranslated regions of target mRNAs (Filipowicz et al., 2008). Importantly, as post-transcriptional regulators they function to buffer the protein expression of their targets and confer robustness to biological processes such as lineage commitment (Ebert and Sharp, 2012; Mukherji et al., 2011; Strovas et al., 2014).
Several microRNAs have been found to regulate normal function of HSCs, including cell cycling and engraftment potential (Guo et al., 2010; Lechman et al., 2012; Song et al., 2013; Zhao et al., 2013). However the role of microRNAs in regulating ageing HSC function remains unclear. In this work, we studied a previously unappreciated microRNA cluster, Mirc19, that is enriched in HSCs and up-regulated with age. These two microRNAs share a seed sequence and therefore target many of the same genes. Several groups have demonstrated that Mirc19 is an important regulator of immune function (Lagos et al., 2010; Nakahama et al., 2013; Ni et al., 2014; Shaked et al., 2009). We now show that Mirc19 plays a critical role in maintaining the balance between function and survival of aged HSCs. It does this by buffering the expression of its target FOXO3, one of only a few known genes associated with human longevity (Willcox et al., 2008).
Results
Enforced expression of miR-132 leads to depletion of HSCs and extramedullary hematopoiesis
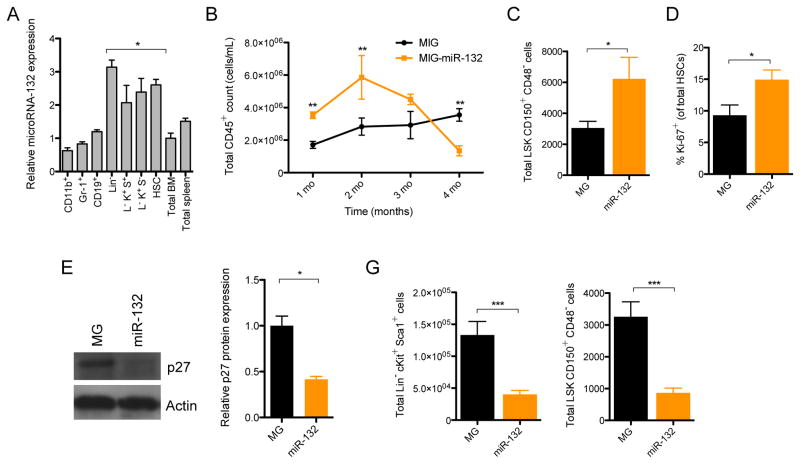
To understand the role of the microRNA-212/132 cluster (Mirc19) in hematopoiesis, we first examined the expression of both microRNAs during hematopoietic differentiation. We determined that both miR-132 and miR-212 were enriched in early hematopoietic progenitors (Lineage−Sca1+cKit+; LSK cells), and in particular, in long-term hematopoietic stem cells (HSCs: LSK CD150+CD48−; Figure 1A and S1A). We initially focused on miR-132 since it was the more enriched of the two microRNAs. To investigate the function of miR-132 in these progenitors, we used a retroviral vector to ectopically express miR-132 in hematopoietic stem and progenitor cells (HSPCs) and transferred these miR-132 over-expressing cells into lethally irradiated wild-type (WT) C57BL/6 recipient mice (Figure S1B–D). We then monitored mature cell output in the peripheral blood of these mice using flow-cytometry to detect the cell-surface markers that identify each cell type. Mice over-expressing miR-132 in the bone marrow compartment (WTmiR-132), when compared to empty vector controls (WTMG), demonstrated a rapid accumulation of CD45+ peripheral blood leukocytes at 2 months post-reconstitution, followed by a progressive decline in the number of these cells by 4 months (Figure 1B). A closer inspection of the bone marrow compartment at 2 months post-reconstitution revealed that WTmiR-132 mice displayed an expansion in the total number of LSK cells and HSCs (Figure 1C and S1D–F). These cells were additionally more proliferative, as measured by the proportion of cells expressing the proliferation marker Ki67, compared to LSK cells and HSCs from age-matched WTMG controls (Figure 1D and S1G). WTmiR-132 HSPCs further demonstrated a down-regulation in protein and RNA expression of several negative cell cycle regulators, including p27 and p57, although no change in p21 transcript expression was observed (Figure 1E and S1H). The mRNA expression of p27 remained down-regulated in WTmiR-132 HSPCs compared to WTMG HSPCs at 4-months post-reconstitution (Figure S1I).
Figure 1.
miR-132 is expressed in hematopoietic stem cells (HSCs) and over-expression alters hematopoiesis. (A) miR-132 expression in mature and progenitor hematopoietic cells. Cell populations were sorted by flow cytometry directly into RNA lysis buffer and miR-132 expression was detected using TaqMan RT-qPCR (n=3). (B) – (F) WT C57BL/6 mice were lethally irradiated and reconstituted with donor bone marrow cells expressing either a control (MG) or a miR-132 over-expressing (miR-132) retroviral vector (n=8–12 mice per group). (B) Total numbers of mature leukocytes (CD45+) in the peripheral blood of MG and miR-132 mice at the indicated time points post-reconstitution. (C) Total number of HSCs (LSK CD150+ CD48−) in the bone marrow of MG and miR-132 mice at 8-weeks post-reconstitution. (D) Percentage of Ki-67+ bone marrow HSCs in MG and miR-132 mice at 8-weeks post-reconstitution. (E) Protein and RNA expression of p27 in the bone marrow of MG and miR-132 mice at 8-weeks post-reconstitution (n=3). (F) Total number of LSK cells and HSCs in the bone marrow of MG and miR-132 mice at 16-weeks post-reconstitution. Data represents at least three independent experiments and is represented as mean ± SEM. See also Figure S1–S3. * denotes p < 0.05, ** denotes p < 0.01 and *** denotes p < 0.001 using a Student’s t test.
We next sought to characterize the cellular basis by which WTmiR-132 mice undergo depletion in peripheral blood leukocytes at 4 months post-reconstitution. Almost two-thirds (29/44) of the WTmiR-132 mice presented with gross pathology characteristic of extramedullary hematopoiesis, including enlarged spleens, and a significant elevation of splenic erythtroid cells (Ter119+) (Figure S2J,K). None of the age-matched WTMG mice presented such a phenotype and no elevation of myeloid cells was found in the peripheral blood of WTmiR-132 mice (Figure S1L). Examination of the bone marrow, however, revealed that WTmiR-132 mice had a severe depletion in the frequency and total number of LSK cells and HSCs compared to WTMG controls (Figure 1F and S1M,N). A similar, more dramatic phenotype was observed at 9 months post-reconstitution in WTmiR-132 mice (Figure S1O). This phenotype of rapid proliferation followed by depletion of HSCs in the bone marrow compartment is an example of HSC exhaustion.
The depletion of HSCs in WTmiR-132 mice had the expected dramatic effect on the numbers of more mature progenitor cells, including multi-potent progenitors (MPPs; LSK CD150−CD48+), lymphoid-primed MPPs (LMPPs; LSK Flt3+), and megakaryocyte and erythroid progenitors (MEPs; Lineage−Sca1−cKit+CD34−FcRγ−). However, no depletion in common myeloid progenitors (CMPs; Lineage−Sca1−cKit+CD34+FcRγ−) or granulocyte-myeloid progenitors (GMPs; Lineage−Sca1−cKit+CD34+FcRγ+) was observed (Figure S1P–T). Importantly, the observed alteration in WTmiR-132 HSCs was intrinsic to the expression of the miR-132 over-expression vector, because no depletion in the proportion of HSCs was evident among the GFP− cells of WTmiR-132 and WTMG mice (Figure S1U). Furthermore, we found that the observed phenotype was specific to the expression of authentic miR-132 because over-expression of a miR-132 mutant lacking the correct miR-132 seed sequence resulted in no observable phenotype at 9 months post-reconstitution when compared to WTMG controls (Figure S1V).
We next sought to investigate the role of miR-212 in HSC maintenance. We found that enforced expression of miR-212 in the bone marrow compartment of mice did not result in a significant change in the total number of bone marrow CD45+ cells or LSK cells compared to controls (Figure S1W,X). However, we found that there was a significant depletion of HSCs at 4-months post-reconstitution in these mice (Figure S1Y), thus suggesting a less severe phenotype than enforced miR-132 expression, which was consistent with the lower amounts of enrichment of miR-212 in HSCs.
Mirc19 has a physiological role of protecting the aging hematopoietic system
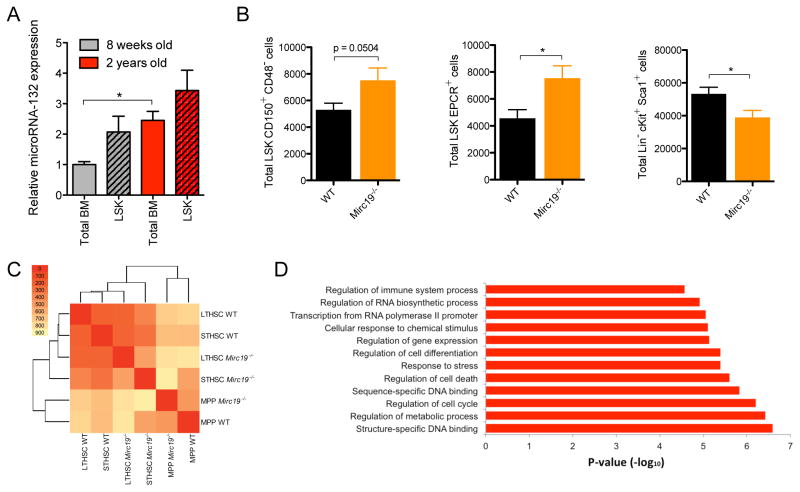
To determine if miR-132 has a physiological role in regulating hematopoietic stem cell function, we obtained mice that have a genetic deletion in both of the microRNAs in the Mirc19 cluster (Mirc19−/−) (Ucar et al., 2012). We observed no apparent defect in the output of mature hematopoietic cells in the peripheral blood, spleen and bone marrow of 12-week old Mirc19−/− mice when compared to age-matched wild-type (WT) controls (Figure S2A–D). We noticed, however, an up-regulation of miR-132 expression in the bone marrow and LSK compartment of aged (2-year old) WT mice compared to young (12-week old) WT mice (Figure 2A), and posited a more important role of miR-132 in maintaining the fidelity of aging HSCs. Consistent with this, we found that unlike in 12-week old mice (Figure S2D), aged (60-week old) Mirc19−/− mice had an elevation in the total number of HSCs (LSK CD150+CD48− and LSK EPCR+) in the bone marrow compartment compared to WT controls (Figure 2B). Surprisingly, this was accompanied by a decrease in the total number of bone marrow LSK cells, which are mostly downstream products of HSCs (Figure 2B). Aged Mirc19−/− mice further presented with enlarged spleens (Figure S2E) and a global depletion of all major mature cell types in the bone marrow compartment (Figure S2F), indicative of a failure of HSCs to maintain normal hematopoietic output and the onset of extramedullary hematopoiesis.
Figure 2.
Genetic deletion of Mirc19 in mice alters hematopoietic output with age. (A) miR-132 expression in total bone marrow and LSK cells from 8-week old and 2-year old C57BL/6 WT mice. miR-132 expression was quantified by TaqMan RT-qPCR (n=2). (B) – (D) Mice with a genetic deletion of Mirc19 (Mirc19−/−) along with WT mice in the C57BL/6 background were analyzed. (B) Total number of HSCs (LSK CD150+ CD48− and LSK EPCR+) and LSK cells in the bone marrow compartment of 60–64 week old WT and Mirc19−/− mice (n=7–12 mice per group). (C) Global expression profiling of WT and Mirc19−/− HSCs, as well as short-term HSCs and MPPs, from 16-week old mice using RNA-seq. The heat map represents the number of differentially expressed genes between the different WT and Mirc19−/− populations. (D) Enriched gene-ontology terms for differently expressed genes between WT and Mirc19−/− progenitors. Data represents at least two independent experiments and is represented as mean ± SEM. See also Figure S4–S6. * denotes p < 0.05, ** denotes p < 0.01 and *** denotes p < 0.001 using a Student’s t test.
To investigate the molecular basis for the role of Mirc19 in HSCs, we performed gene expression analysis by bulk population RNA-sequencing on WT and Mirc19−/− long-term HSCs (LSK CD150+CD48−), short term HSCs (LSK CD150−CD48−) and multipotent progenitors (LSK CD150−CD48+). Approximately 14,000 genes were expressed in each sample (Figure S2N), and clustering based on the number differentially expressed genes revealed close similarity between WT short-term and long-term HSC subsets and Mirc19−/− short-term and long-term HSC subsets, with both these groups differing significantly from the MPP populations (Figure 2C). Differentially expressed genes between the WT and Mirc19−/− HSC populations were enriched for several functional annotations relevant to HSC biology, including regulation of cell-cycle, cell differentiation, response to stress and cell death (Figure 2D).
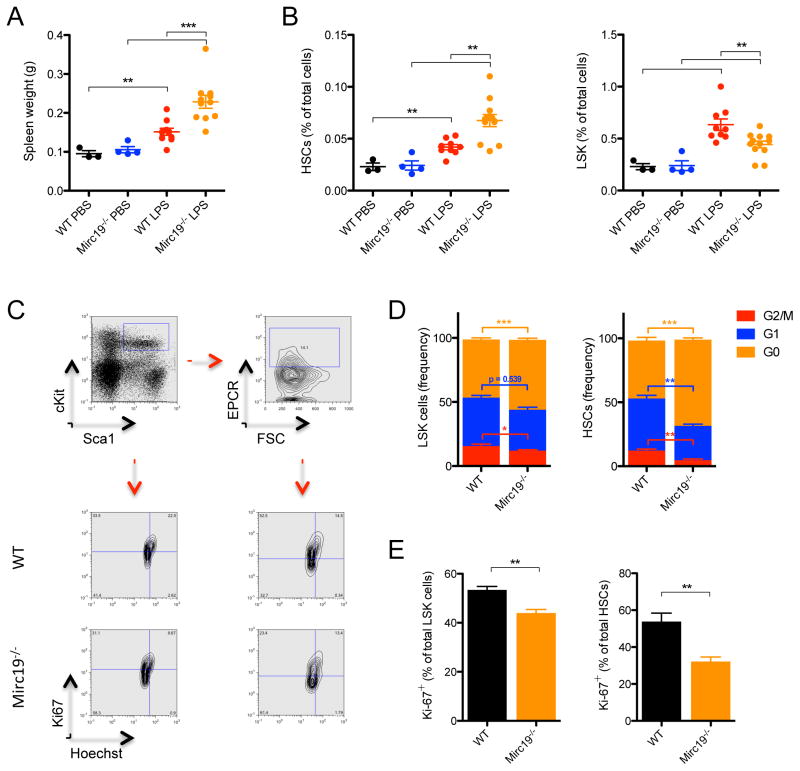
To investigate if the observed phenotype in Mirc19−/− mice is intrinsic to the hematopoietic system, we transferred bone marrow cells from 12-week old WT or Mirc19−/− mice into irradiated WT recipients. After one year, the phenotypes in transplanted mice closely resembled that of aged WT and Mirc19−/− mice, respectively, consistent with a defect intrinsic to the hematopoietic system (Figure S2G). We employed an inflammatory model for hematopoietic aging (Esplin et al., 2011) to determine if this was sufficient to recreate the observed alteration in hematopoiesis. We delivered LPS nine times over one month to 16-week old Mirc19−/− and WT mice. We found that, consistent with the altered hematopoietic output we observed in aged mice, Mirc19−/− mice presented with severely enlarged spleens containing an enrichment of splenic HSCs compared to WT mice also injected with LPS (Figure 3A and S2H). Similar to aged Mirc19−/− mice, LPS treated Mirc19−/− mice also demonstrated an accumulation of HSCs and a decrease in the frequency of LSK cells in the bone marrow compartment compared to LPS treated WT controls (Figure 3B). This skewing of hematopoietic progenitor output in Mirc19−/− mice may be characterized by an increase in the total number of long-term HSCs and a reduction in total number of short-term HSCs and MPPs in the bone marrow compartment (Figure S2I). We can therefore mimic the aging-related hematopoietic defect observed in Mirc19−/− by exposing younger mice to chronic inflammatory stimuli via repetitive LPS injections.
Figure 3.
Genetic deletion of Mirc19 in mice alters hematopoietic output and cycling in response to LPS stimulation. (A) – (E) 6-month old WT and Mirc19−/− mice were treated with 9 evenly-spaced low-dose (1mg/kg of body weight) LPS or PBS injections over one month. (A) Spleen weights of WT and Mirc19−/− mice treated with PBS or LPS. (B) Proportion of HSCs and LSK cells within the bone marrow compartment of WT and Mirc19−/− mice treated with LPS or PBS. (C) Representative flow cytometry plots for cell cycle analysis of HSCs and LSK cells from WT and Mirc19−/− mice following LPS injection. (D) Proportion of bone marrow LSK cells and HSCs in each stage of the cell cycle (G0, G1, G2/M) from WT and Mirc19−/− mice following LPS injection. (E) Proportion of bone marrow LSK cells and HSCs expressing Ki67 from WT and Mirc19−/− mice following LPS injection. Data represents at least two independent experiments and is represented as mean ± SEM. See also Figure S4–S5. * denotes p < 0.05, ** denotes p < 0.01 and *** denotes p < 0.001 using a Student’s t test.
Loss of Mirc19 reduces HSC cycling and improves engraftment potential
The cycling characteristics of HSCs are closely related to their ability to self-renew and differentiate into committed progenitors (Pietras et al., 2011). Furthermore, increased HSC quiescence and an increase in HSC number is characteristic of the aging hematopoietic system (Geiger et al., 2013). We thus sought to determine if the accumulation of HSCs and decrease in output of more committed progenitors in Mirc19−/− mice might be a result of altered cell cycling. We performed cell-cycling analysis using flow cytometry by staining for the proliferation marker Ki67 and utilizing the dsDNA dye Hoescht33342. Under steady-state conditions, we observed no major defect in cell cycling in 16-week old Mirc19−/− mice compared to age-matched WT mice (Figure S2J). However, under conditions of inflammatory stress such as low-grade LPS stimulation, we found that Mirc19−/− HSCs were far less proliferative, with an almost 50% increase in the number of cells in the G0 phase of the cell cycle compared to WT HSCs (Figure 3C–E). Importantly, we observed a substantial decrease in the number of HSCs in G1 and only a small proportion of cells in S/M phases of the cell cycle in Mirc19−/− mice (Figure 3C–E). No change in p27 mRNA expression was observed at steady state between Mirc19−/− and WT HSCs. However, in mice treated with either LPS or 5-fluorouracil, which induce HSC proliferation, the expression of p27 in the bone marrow compartment was up-regulated in Mirc19−/− mice compared to WT mice (Figure S2K,L). It therefore follows that the loss of Mirc19 leads to increased HSC quiescence and an accumulation of HSCs, with a concomitant decrease in the number of more committed progenitors.
The majority of HSCs in the hematopoietic system remain in a dormant state, and disruption of this quiescence can have serious consequences for HSC function (Rossi et al., 2012). To investigate whether the alteration in cell cycling of aged Mirc19−/− HSCs might be related to altered HSC function, we performed competitive transplant assays. Aged (60-weeks) CD45.2 Mirc19−/− or WT HSCs were transplanted with equal numbers of CD45.1 WT HSCs into lethally irradiated CD45.2 recipient mice. The peripheral blood of these mice was analyzed 4 months post-reconstitution for repopulation of major mature cell types. The cells from aged Mirc19−/− mice were more effective at reconstituting most all immune cells than those from aged WT mice, evidenced in total blood leukocytes (CD45+), B-cells (CD19+), myeloid cells (CD11b+) and granulocytes (Gr-1+) in the peripheral blood (Figure 4A). An insignificant difference in the relative proportion of T cells (CD3ε+) was observed (Figure 4A). Competitive transplant of young (12-weeks) Mirc19−/− HSCs yielded no observable functional difference compared to young WT HSCs except for defective repopulation of T cells (Figure 4B); however, secondary transplantation of young Mirc19−/− HSCs yielded a similar phenotype to that observed with primary transplantation of aged Mirc19−/− HSCs (Figure S5I). Consistent with the increased quiescence of aged Mirc19−/− HSCs compared to WT HSCs, Mirc19−/− cells performed better at long-term repopulation. Additionally, HSCs obtained from WTmiR-132 mice, which ectopically over-expressed miR-132, were severely defective in long-term reconstitution in competitive transplant assays compared to control HSCs obtained from WTMG mice (Figure 4C). It therefore appears that Mirc19 is important for tuning the interplay between quiescence and functional output of the aging hematopoietic system.
Figure 4.

Mirc19 regulates long-term reconstitution potential of HSCs with age. (A) and (B) WT or Mirc19−/− CD45.2+ bone marrow cells, calibrated for the total number of phenotypically defined HSCs, were injected in a 1:1 ratio with CD45.1+ WT bone marrow cells into irradiated C57BL/6 CD45.2+ recipients. (A) Ratio of total CD45.2+ cells to CD45.1+ cells at 16-weeks post-reconstitution for various mature immune cell types in the peripheral blood of mice injected with aged WT and Mirc19−/− CD45.2+ cells. Data was normalized to proportion of WT CD45.2+ cells. (B) Ratio of total CD45.2+ cells to CD45.1+ cells at 16-weeks post-reconstitution for various mature immune cell types in the peripheral blood of mice injected with young WT and Mirc19−/− CD45.2+ cells. (C) Control (MG) or miR-132 over-expressing (miR-132) bone marrow HSCs, both expressing a retroviral vector containing eGFP, were injected in a 1:1 ratio with un-infected WT bone marrow HSCs into irradiated C57BL/6 mice. Graphs show the ratio of eGFP+ cells to eGFP− cells for various immune cell types in the peripheral blood of recipients at 16-weeks post-reconstitution. Data represents at two independent experiments and is represented as mean ± SEM. * denotes p < 0.05 using a Student’s t test.
FOXO3 is a target of miR-132 in bone marrow cells
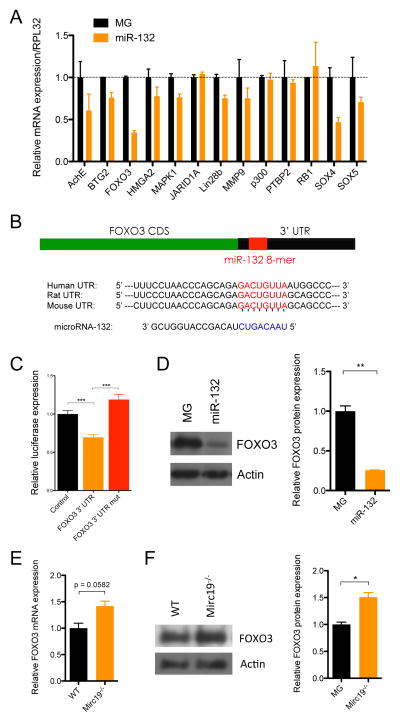
To understand the molecular mechanism of miR-132 action, we characterized the expression of the best computationally predicted targets of miR-132 from TargetScan under conditions of miR-132 over-expression (Friedman et al., 2009). RNA was extracted from lineage-depleted bone marrow cells of WTmiR-132 and WTMG mice and was subjected to quantitative polymerase chain reaction (qPCR) for target genes relevant to HSC function. The most significantly down-regulated targets under conditions of ectopic expression of miR-132 in WTmiR-132 bone marrow, relative to control WTMG bone marrow, were pursued for further analysis. Messenger RNA expression of several genes relevant to hematopoiesis was down regulated in WTmiR-132 bone marrow samples including AchE, FOXO3, Lin28B, MMP9 and SOX4 (Figure 5A). We sought to further investigate the role of FOXO3 in mediating the effect of miR-132 on HSCs as it contains a perfect 8-mer binding site for miR-132 (Figure 5B) and is the most significantly down regulated of these genes. Importantly, we also found a global upregulation of miR-132 targets in our RNA-sequencing analysis of Mirc19−/− and WT HSCs, and this included an upregulation of FOXO3 transcript expression as well as some of its downstream targets (Figure S2O–Q). We validated that miR-132 binds directly to the FOXO3 3′-untranslated region (3′UTR) using a luciferase reporter assay in which the FOXO3 3′UTR was expressed immediately downstream of luciferase. We found that in the presence of miR-132 the expression of this reporter was significantly lower than from a vector lacking the FOXO3 3′UTR (Figure 5C). This binding was specific to miR-132 because mutating the miR-132 binding site on the FOXO3 3′UTR normalized luciferase expression (Figure 5C).
Figure 5.
FOXO3 is a direct target of miR-132 in bone marrow cells. (A) mRNA expression by RT-qPCR of computationally predicted miR-132 targets in control (MG) or miR-132 over-expressing (miR-132) bone marrow cells. (B) Schematic of the predicted miR-132 binding site in the FOXO3 3′UTR. (C) Relative luciferase expression in 293T cells transfected with a miR-132 over-expression vector and either a vector containing luciferase only (control), a vector containing luciferase and the FOXO3 3′UTR immediately downstream (FOXO3 3′UTR), or a vector containing luciferase and a FOXO3 3′UTR with a mutated miR-132 binding site (FOXO3 3′UTR mut). (D) FOXO3 protein expression in bone marrow cells from MG and miR-132 mice obtained by Western Blot. (E) FOXO3 transcript expression in lineage-depleted bone marrow cells obtained from WT or miR-212/132−/− mice. (F) FOXO3 protein expression in lineage-depleted bone marrow cells obtained from WT or Mirc19−/− mice. See also Figure S7A–C. Data represents at least two independent experiments (n=2) and is represented as mean ± SEM. * denotes p < 0.05, ** denotes p < 0.01 and *** denotes p < 0.001 using a Student-T test.
We next quantified FOXO3 protein expression in bone marrow cells from WTmiR-132 and WTMG mice. Consistent with FOXO3 being a target of miR-132, we found protein expression was significantly down regulated in WTmiR-132 mice compared to WTMG mice (Figure 5D). Expression of FOXO1 and FOXO4, closely related family members of FOXO3, remained unchanged in WTmiR-132 bone marrow cells (Figure S3A). Importantly, we also found that FOXO3 mRNA and protein expression was elevated in lineage depleted bone marrow cells from Mirc19−/− mice compared to WT controls (Figure 5E,F). Expression of FOXO4 was also slightly elevated in Mirc19−/− cells and expression of FOXO1 was unchanged (Figure S3B). We additionally performed intracellular staining of FOXO3 and phospho-FOXO3 (p-FOXO3) protein. As expected, we found elevated protein expression of FOXO3 in Mirc19−/− HSCs compared to WT controls. However, we saw only a marginal elevation in p-FOXO3 in Mirc19−/− HSCs, indicating that the majority of extra FOXO3 in these cells is likely in the nucleus in its active, un-phosphorylated state (Figure S3C,D). Together, the data indicate that miR-132 is an important regulator of FOXO3 expression in bone marrow cells. As was observed with miR-132 expression, FOXO3 mRNA expression was increased in bone marrow cells from aged mice compared to those of young mice (Figure 6A), thus suggesting that miR-132 might serve to maintain FOXO3 protein expression within a balanced range for normal hematopoietic function.
Figure 6.
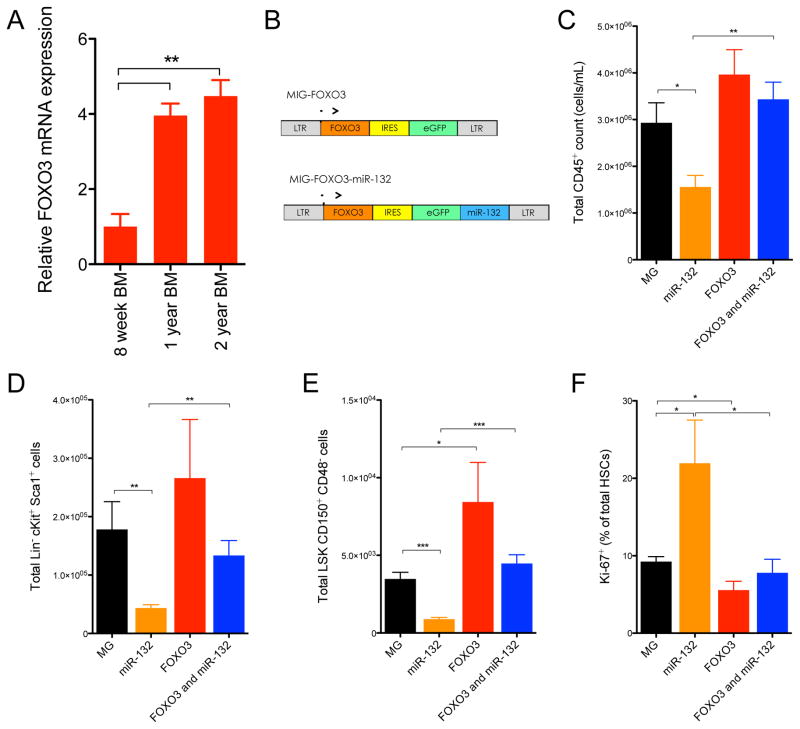
Co-expression of FOXO3 with miR-132 rescues the hematopoietic defects observed with expression of miR-132 alone. (A) FOXO3 mRNA expression by RT-qPCR in total bone marrow cells from 8-week old, 1-year old and 2-year old C57BL/6 WT mice (n=2). (B) Schematic of retroviral vectors constructed for expression of FOXO3 only or for co-expression of FOXO3 along with miR-132. (C) – (F) WT C57BL/6 mice were lethally irradiated and reconstituted with donor bone marrow cells expressing either a control (MG), a miR-132 over-expressing (miR-132), a FOXO3 over-expressing (FOXO3), or a FOXO3 and miR-132 over-expressing (FOXO3 and miR-132) retroviral vector (n=10–12 mice per group). (C) Total peripheral blood CD45+ leukocytes in the respective animals at 16-weeks post-reconstitution. (D) Total LSK cells in the bone marrow compartment of the respective animals at 16-weeks post-reconstitution. (E) Total HSCs in the bone marrow compartment of the respective animals at 16-weeks post-reconstitution. (F) Proportion of bone marrow HSCs expressing Ki-67 in the respective animals at 16-weeks post-reconstitution. Data represents at least two independent experiments and is represented as mean ± SEM. See also Figure S7E–H. * denotes p < 0.05, ** denotes p < 0.01 and *** denotes p < 0.001 using a Student’s t test.
miR-132 regulates HSC cycling and function through FOXO3
To determine if FOXO3 is a key mediator of miR-132 function, we co-expressed FOXO3 with miR-132 in the bone marrow compartment of WT mice to see if it would rescue the phenotype observed with miR-132 expression alone. FOXO3 cDNA lacking a miR-132 target site was cloned into the MSCV-IRES-eGFP (MIG) vector, immediately downstream of the MSCV promoter (Figure 6B). As previously described, miR-132 was cloned downstream of eGFP. Lethally irradiated mice were reconstituted with bone marrow cells transduced with a control vector (WTMIG), or a vector expressing both miR-132 and FOXO3 (WTFOXO3+miR-132), miR-132 only (WTmiR-132) or FOXO3 only (WTFOXO3). The expression of miR-132 and FOXO3 was validated by qPCR and Immunoblot, respectively (Figure S3E–F). We observed the mean fluorescence intensity of eGFP in bone marrow cells expressing only FOXO3 to be lower than that of the other vectors, suggesting that over-expression of FOXO3 above endogenous concentrations may be toxic (Figure S3G). We additionally observed that a larger fraction bone marrow HSCs from WTFOXO3 mice expressed AnnexinV compared to WTMIG controls (Figure S3H). This effect was not observed when FOXO3 was co-expressed with miR-132, presumably because baseline expression of FOXO3 was already down regulated due to miR-132 over-expression.
As expected, a reduction in peripheral blood leukocytes was observed in WTmiR-132 mice compared to WTMIG mice at 4-months post-reconstitution (Figure 6C). Co-expression of FOXO3 with miR-132, however, rescued this defect, as no significant change in peripheral blood leukocytes was observed in WTFOXO3+miR-132 mice compared to WTMIG controls (Figure 6C). Examination of the bone marrow compartment of WTFOXO3 + miR-132 mice revealed total numbers of HSCs and LSK cells comparable to WTMIG controls, demonstrating a rescue of HSC depletion observed with the expression of miR-132 alone (Figure 6D,E). In addition, expression of FOXO3 alone resulted in a moderate elevation in the total number of bone marrow LSK cells compared to control mice (Figure 6E). HSCs from WTFOXO3+miR-132 mice also showed comparable proportions of Ki67 staining to HSCs from WTMIG mice, indicating that these cells were not prone to cycling like WTmiR-132 cells (Figure 5F). WTFOXO3 HSCs demonstrated a significant albeit moderate decrease in the proportion of cycling HSCs compared to controls (Figure 6F). These experiments suggest that co-expression of FOXO3 can rescue the phenotype observed with expression of miR-132 alone. It seems likely that miR-132 regulates hematopoiesis primarily by directly modulating FOXO3 protein expression, although we cannot rule out that FOXO3 overexpression is able to override the miR-132 effect while the true targets of miR-132 in the bone marrow are other genes.
Loss of Mirc19 affects HSC survival through protective autophagy
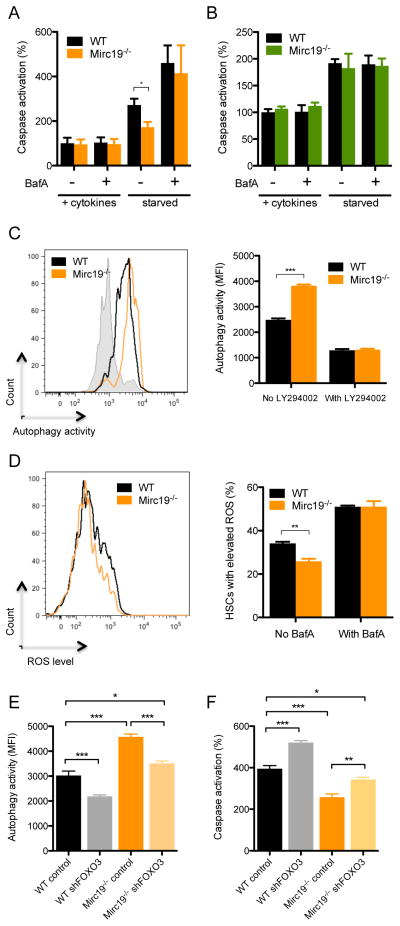
FOXO3 is critical for maintaining the hematopoietic stem cell pool by regulating HSC cell-cycling and resistance to oxidative stress (Miyamoto et al., 2007; Tothova et al., 2007). It is also implicated in maintaining the survival of aging HSCs by directing protective autophagy (Warr et al., 2013). To this end, we found that several autophagy-related genes were up-regulated in Mirc19−/− HSCs compared to WT controls upon inspection of our RNA-sequencing dataset (Figure S2R). Thus, to determine if miR-132 might play a role in altering survival of HSCs, we sorted HSCs from WT and Mirc19−/− mice and cultured them in the presence or absence of survival growth factors and cytokines including mouse stem cell factor (mSCF), mouse interleukin-6 (mIL-6), mIL-3, TPO and Flt3L. We used a luciferase-based assay to monitor caspase activity after 12 hours in culture. In the presence of survival factors, minimal activity was observed in both WT and Mirc19−/− HSCs. However, under starvation conditions, which induces protective autophagy in aged HSCs (Warr et al., 2013), Mirc19−/− HSCs demonstrated a significant reduction in induction of apoptosis compared to WT HSCs (Figure 7A). This is consistent with the more rapid induction of a protective autophagy program due to higher expression of FOXO3 in Mirc19−/− HSCs. Importantly, when autophagy was inhibited by Bafilomycin A (BafA), a known inhibitor of autophagosome fusion to lysosomes, Mirc19−/− and WT HSCs underwent comparable, higher amounts of apoptosis (Figure 7A). As previously reported, FOXO3 expression had no major effect on autophagy and apoptosis of myeloid progenitors (Figure 7B).
Figure 7.
Genetic deletion of Mirc19 results in a FOXO3 dependent alteration in autophagy and survival of HSCs. (A) and (B) Cells obtained from WT and Mirc19−/− mice were cultured with or without growth cytokines (mIL3, mIL6, mSCF, TPO, Flt3L, and G-CSF) or BafA. Caspase activation was measured using a luciferase based assay (n=6). (A) Caspase activation observed in HSCs from WT and Mirc19−/− mice with or without growth factor starvation and BafA treatment. (B) Caspase activation observed in myeloid progenitors from WT and Mirc19−/− mice with or without growth factor starvation and BafA treatment. (C) Autophagy activity in WT and Mirc19−/− HSCs with or without growth factor starvation and LY294002 treatment. Autophagy was measured using a dye that fluorescently labels autophagosomes (cyto-ID autophagy assay) (n=5). (D) ROS accumulation in WT and Mirc19−/− HSCs with or without growth factor starvation and BafA treatment measured using a fluorescence-based ROS detection system (CellROX assay) (n=5). (E) and (F) WT C57BL/6 mice were lethally irradiated and reconstituted with WT or Mirc19−/− donor bone marrow cells expressing either a control or FOXO3 shRNA silencing vector. At 8-weeks post-reconstitution HSCs were sorted from bone marrow and assays were performed. (E) Autophagy activity measured in HSCs from the respective mice in response to growth factor starvation (n=6). (F) Caspase activation measured in HSCs from the respective mice in response to growth factor starvation (n=6). Data represents two independent experiments and is represented as mean ± SEM. See also Figure S7I. * denotes p < 0.05, ** denotes p < 0.01 and *** denotes p < 0.001 using a Student’s t test.
To determine if Mirc19−/− HSCs indeed induce the autophagy machinery more potently than WT HSCs, we utilized a fluorescent reporter for autophagosome formation that was detectable by flow cytometry. The efficacy of this assay in detecting autophagosome formation was validated by comparing signal intensity in WT cells to autophagy deficient cells (Figure S3I). WT and Mirc19−/− HSCs were cultured under growth factor rich or starvation conditions as described above and were stained for the presence of autophagosomes. Under starvation conditions, Mirc19−/− HSCs demonstrated higher amounts of autophagosome formation compared WT HSCs (Figure 7C). In the presence of LY2940002, a PI3-kinase inhibitor and early inhibitor of autophagy, autophagosome formation was decreased to comparable amounts in Mirc19−/− and WT HSCs (Figure 7C). We additionally sought to investigate whether the potent induction of autophagy in Mirc19−/− HSCs may improve survival by altering reactive-oxygen species (ROS) accumulation. We utilized a fluorescent detection system for ROS and found that Mirc19−/− HSCs had lower amounts of ROS accumulation compared to WT HSCs under conditions of starvation (Figure 7D). The accumulation of ROS was elevated to comparable amounts in WT and Mirc19−/− HSCs when autophagy was inhibited with BafA (Figure 7D).
We employed an shRNA silencing strategy for FOXO3 to determine if it was the key mediator of autophagy in Mirc19−/− HSCs. WT and Mirc19−/− HSPCs were transduced with either a control vector (MB) or a FOXO3 shRNA construct (shFOXO3), and were subsequently used to reconstitute lethally irradiated WT mice. At 2 months post-reconstitution, we sorted HSCs from these mice and subjected them to the aforementioned assays for autophagy induction and caspase activation under conditions of growth factor starvation. WT HSCs expressing shFOXO3 demonstrated lower autophagy activity and higher amounts of apoptosis compared to WT HSCs expressing MB (Figure 7E,F). Importantly, silencing of FOXO3 in Mirc19−/− HSCs resulted in a significant reduction in autophagy induction and an increase in caspase activity compared to Mirc19−/− HSCs expressing MB. However, this reduction did not reduce autophagy activity completely to that of WT HSCs expressing MB. This may be due to incomplete silencing of FOXO3 in Mirc19−/− cells or may suggest factors other than FOXO3 might be involved in mediating autophagy induction (Figure 7E,F).
Discussion
MicroRNAs are key regulators of lineage commitment and function in immune cells (Baltimore et al., 2008; Gangaraju and Lin, 2009). Several microRNAs have been implicated in regulating diverse facets of normal HSC maintenance and function, such as cell-cycling (Lechman et al., 2012; Song et al., 2013), apoptosis (Guo et al., 2010), engraftment potential (O’Connell et al., 2010) and resistance to inflammatory stress (Zhao et al., 2013). While much has been done to characterize the functional differences between aged and young HSCs, little is known about how microRNAs might contribute to maintaining balanced hematopoietic output as an organism ages. Our findings suggest that Mirc19, particularly miR-132, is critical in regulating the balance between HSC survival, and proliferation and differentiation. We have demonstrated that it does this primarily by buffering the expression of FOXO3 in the aging hematopoietic system. Deregulation of this cluster, and in turn FOXO3, can have negative consequences on the function of HSCs and the output of mature hematopoietic cells, leading to extramedullary hematopoiesis.
Because the expression of miR-132 was higher in HSCs compared to total bone marrow cells, we utilized both gain-of-function and loss-of-function approaches to investigate its role in HSC function and survival. Ectopic expression of miR-132 resulted in hyper-proliferation and depletion of HSCs within the bone marrow compartment. Enforced expression of miR-212 produced a similar but less dramatic phenotype. This depletion of HSCs with miR-132 over-expression coincided with the onset of extramedullary hematopoiesis, including enlarged spleens and fibrotic bone marrow. We observed a drastic decrease in protein expression of the miR-132 target FOXO3 within the bone marrow compartment of miR-132 over-expressing mice. Consistent with our findings, a genetic deletion of Foxo3 in hematopoietic cells leads to increased HSC proliferation and an age-dependent depletion of the HSC pool with loss of HSC long-term reconstitution potential (Miyamoto et al., 2007). This phenotype is exacerbated by the concomitant deletion of the FOXO family members Foxo1 and Foxo4 (Tothova et al., 2007). We further found several FOXO3 target genes, particularly the negative cell-cycle regulators p21, p57, and p27, to be down regulated in miR-132 over-expressing bone marrow. Importantly, replenishing expression of FOXO3 during miR-132 over-expression rescued the phenotype we observed.
A genetic deletion of Mirc19 led to higher basal expression of FOXO3 in bone marrow cells. Over time, this led to a dramatic increase in the number of HSCs, a decrease in production of more committed progenitors, and a defect in HSC cycling in response to environmental stress, such as lipopolysaccharide treatment. Consistent with the more quiescent state of Mirc19−/− HSCs, we found they were marginally better at long-term reconstitution of the hematopoietic system than WT counterparts. FOXO3 is a known regulator of apoptosis, and we further demonstrated that ectopic expression of FOXO3 from a retroviral vector resulted in a selection for those cells expressing the lowest amount of the vector, presumably because higher amounts of FOXO3 expression were toxic. We have therefore shown that Mirc19 is important in regulating expression of FOXO3, and that when this target is either up-regulated or down regulated, there is a severe alteration in HSC function over time.
The expression of both miR-132 and FOXO3 transcripts is up-regulated with age in murine bone marrow cells and early progenitors. The role of FOXO3 as a longevity-associated gene remains unknown in the hematopoietic system. FOXO3 may be up-regulated in this context due to its vital role in survival through autophagy and in cell cycling. MicroRNAs play an important role in buffering perturbations in the expression of their targets in response to environmental stress (Ebert and Sharp, 2012). Such stress might include inflammation from repetitive exposure to environmental pathogens and hematopoietic aging. Importantly, the abundance of microRNAs in any given cell plays an important role in establishing a threshold for target expression (Mukherji et al., 2011); as such up-regulation of the microRNA may require higher target expression to maintain important physiological functions. Given that both over-expression and deletion of miR-132 in bone marrow cells led to inappropriate hematopoiesis, we believe that miR-132 plays an important role in buffering FOXO3 protein expression within a defined range to maintain normal HSC function as an organism ages. The concomitant increase in the expression of miR-132 alongside FOXO3 in the aging hematopoietic system may be critical for maintaining an important balance between known FOXO3-regulated processes, including cell-cycling and differentiation, and apoptosis of HSCs.
The aging hematopoietic system is characterized by an alteration of the balance between self-renewal and differentiation, which leads to the accumulation of less-functional HSCs, myeloid-biased differentiation and a requirement for basal autophagy for survival (Geiger et al., 2013; Warr et al., 2013). Of note, the loss of critical autophagy factors in the hematopoietic system leads to hyper-proliferation and poor survival of HSCs (Mortensen et al., 2011). Recently, it has also been demonstrated that FOXO3 plays a critical role in inducing protective autophagy of aging HSCs (Warr et al., 2013), which is critical for their survival in response to oxidative stress (Eijkelenboom and Burgering, 2013). The proposed mechanism of FOXO3 regulation of autophagy is through the transcription of glutamine synthase (van der Vos et al., 2012). Consistent with this role of FOXO3 in HSC survival, we found that Mirc19−/− HSCs, when compared to WT HSCs, demonstrated increased resistance to growth-factor starvation as evidenced by the decrease in presence of reactive oxygen species, lower amounts of apoptosis induction, and an increase in accumulation of autophagosomes. We observed an abrogation of this effect when we silenced FOXO3 in Mirc19−/− HSCs, thus demonstrating that this phenotype is mostly due to the up-regulation of FOXO3 in these cells. Importantly, this improved survival of Mirc19−/− cells in response to environmental stress may contribute to the age-dependent accumulation of HSCs in Mirc19−/− mice compared to WT mice.
Our observations demonstrate that the Mirc19−/− cluster is an important regulator of HSC homeostasis by altering cell cycling, function and survival. This is an example of a microRNA playing a physiological role in maintaining the balance of HSC functions during aging. The capacity of this microRNA to buffer expression of FOXO3 is critical given the multiple roles FOXO3 plays in regulating HSC biology. Our findings open the possibility of utilizing miR-132 mimics or antagonists to alter defects in HSC function that might lead to hematopoietic diseases late in life.
Experimental Procedures
DNA constructs
For in-vivo miR-132 over-expression and FOXO3 shRNA experiments, the mature miR-132 or FOXO3 shRNA sequence was placed in the microRNA-155 loop-and-arms format (O’Connell et al., 2010) and cloned into the MSCV-eGFP (MG) and MSCV-TagBFP (MG) vectors, respectively. For FOXO3 rescue experiments, FOXO3 cDNA was cloned into the MSCV-IRES-eGFP (MIG) vector. See supplemental procedures for more details about these vectors. FOXO3 shRNA target sequences are given in Table S1.
For luciferase assays, the microRNA-132 expression cassette was sub-cloned into the pCDNA3 vector. The 3′untranslated regions of relevant gene targets containing the miR-132 binding region were cloned immediately downstream of luciferase in the pMiReport vector as previously described (Chaudhuri et al., 2012).
Cell culture
Cells were cultured in a sterile incubator that was maintained at 37°C and 5% CO2. 293T cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 U/mL streptomycin. Primary cells were cultured in complete RPMI supplemented with 10% FBS, 100 U/mL penicillin, 100 U/mL streptomycin, 50uM β-mercaptoethanol and appropriate growth cytokines as needed for the experiment.
Cell sorting for RNA extraction
For miR-132 expression profiling, bone marrow cells from C57BL/6 mice were depleted of RBCs and sorted for the respective cell populations at the Caltech Flow Cytometry Core Facility. Detailed procedures are provided in the supplemental procedures. RNA was harvested using the miRNAeasy RNA prep kit (Qiagen). For bone marrow samples from MG and miR-132 mice, bone marrow was harvested from the respective mice, lysed of red blood cells, and spun down. RNA was harvested as described above.
Expression profiling and qPCR
We performed real time qPCR (RT-qPCR) with a 7300 Real-Time PCR machine (Applied Biosystems) as previously described (Chaudhuri et al., 2012). TaqMan qPCR was performed for miR-132, miR-212 and snoRNA-202 (control) detection as per manufacturers instructions using TaqMan MicroRNA Assays (Life Technologies). SYBR Green-based RT-qPCR was performed for mRNA of mouse FOXO3, FOXO1, p27, p21, p57, and relevant miR-132 targets following cDNA synthesis using qScript cDNA SuperMix (Quanta) and detection with PerfeCTa qPCR Fastmix with ROX (Quanta) as per manufacturers instructions. Gene-specific primers used for qPCR are listed in Table S2. RNA-seq library construction and analysis are described in the Supplemental Information.
Target prediction and Luciferase reporter assays
Relevant targets for miR-132 were investigated using predictions from TargetScan Mouse 6.2 software (Friedman et al., 2009) and following sorting by probability of conserved targeting (PCT). Luciferase assays for miR-132 targets were performed as previously described (Chaudhuri et al., 2012). Briefly, 4 × 105 cells were plated in 12-well plates for 24 hours and subsequently transfected with either pCDNA or pCDNA-miR-132, a pMiReport vector, and a β-gal expression vector. 48 hours later, cells were lysed using Reporter Lysis Buffer (Promega) and luciferase and β-gal expression was analyzed, respectively, using a Dual Luciferase Kit (Promega) and a chemiluminescent β-gal reporter kit (Roche).
Immunoblotting
Bone marrow samples were prepared as described for RNA preparation. Cell extracts were collected using RIPA lysis buffer (Sigma), and were subjected to gel-electrophoresis and transfer onto a PVDF membrane. Antibody staining was performed using antibodies for FOXO3, p27 and actin. Detailed procedures are given in the supplemental information.
Animals
The California Institute of Technology Institutional Animal Care and Use Committee approved all experiments. C57BL/6 WT and miR-212/132−/− mice were bred and housed in the Caltech Office of Laboratory Animal Resources (OLAR) facility. Bone marrow reconstitution experiments were performed as previously described (Chaudhuri et al., 2012) with the aforementioned vectors and are explained in more detail in the Supplemental Procedures. Recipient mice were monitored for health and peripheral blood was analyzed for mature blood cell types each month up till the experimental end-point at either 16 or 36 weeks post-reconstitution. At each end-point, immune organs were harvested for further analysis as described. The number of mice for each experimental cohort is described in the figure legends. Each experiment was repeated at least twice, and in many cases three or four times.
Competitive transplant experiments
Bone marrow cells from age and gender-matched WT CD45.2+ C57BL/6 mice, Mirc19−/− CD45.2+ C57BL/6 mice and WT CD45.1+ C57BL/6 mice were harvested and depleted of RBCs as described above. A 1:1 ratio of WT CD45.1+ HSCs with either WT or Mirc19−/− CD45.2+ HSCs were subsequently injected into lethally irradiated (1000 rads) WT CD45.2+ CD57BL/6 mice. Mice were monitored for up to 20 weeks post-reconstitution and relevant tissues were harvested for further analysis by flow cytometry.
Flow cytometry
Relevant tissues were harvested and cells were homogenized and subsequently depleted of red blood cells as described above. Flurophore-conjugated antibodies were used for the indicated markers, and detected using a MACSQuant10 Flow Cytometry machine (Miltenyi). Detailed procedures are given in the supplemental information.
Autophagy and reactive-oxygen species assays
HSCs were sorted as described above from either WT or Mirc19−/− C57BL/6 mice, or from reconstituted mice with donor WT or Mirc19−/− bone marrow infected with either MB or shFOXO3 retroviral constructs. Cells were then cultured with the appropriate growth factors and cytokines, or authophagy inhibitors, and processed for caspase activity (Promega), the presence of ROS (Life Technologies), or autophagy activity (Enzo Life Sciences). Detailed procedures are given in the supplemental information.
Statistical tests
All statistical analysis was done in Graphpad Prism software using an unpaired Student’s t test. Data was reported as mean ± SEM. Significance measurements were marked as follows: * p < 0.05, ** p < 0.01, ** p < 0.001, or ns for not significant.
Data access
The RNA-seq data used in this study can be accessed from the Gene Expression Omnibus under the accession project ID GSE66352.
Supplementary Material
Acknowledgments
We thank Diana Perez at the Caltech Flow Cytometry core facility, and Igor Antoshechkin and Vijaya Kumar at the Caltech Genetics and Genomics Laboratory for their assistance. We also thank Michael T. Bethune for helpful discussions throughout the preparation of this manuscript. This work was supported by an NIH RO1AI079243 (D.B.), National Research Service Award CA183220 (A.M.) and HL110691 (J. L. Z), the UCLA/Caltech Medical Scientist Training Program (A.M and J. L. Z.), the Human Frontiers Science Foundation (M.M), and the Broad Institute (M.S.K and A.R).
Footnotes
Author contributions
A.M, J.L.Z. and D.B. designed the study. A.M. conducted all the experimental work with assistance from N.S., M.M., M.S.K., R.P.G, and X.D. and with guidance from J.L.Z. G.K.M. and A.M. performed bioinformatics analysis. E.E. and K.C. contributed the miR-212/132−/− mice. A.M. and D.B. wrote the manuscript with contributions from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nature immunology. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- Chaudhuri AA, So AY, Mehta A, Minisandram A, Sinha N, Jonsson VD, Rao DS, O’Connell RM, Baltimore D. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4233–4238. doi: 10.1073/pnas.1200677109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science (New York, NY) 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nature reviews Molecular cell biology. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- Esplin BL, Shimazu T, Welner RS, Garrett KP, Nie L, Zhang Q, Humphrey MB, Yang Q, Borghesi LA, Kincade PW. Chronic exposure to a TLR ligand injures hematopoietic stem cells. Journal of immunology (Baltimore, Md : 1950) 2011;186:5367–5375. doi: 10.4049/jimmunol.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews. Genetics. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Flach J, Bakker ST, Mohrin M, Conroy PC, Pietras EM, Reynaud D, Alvarez S, Diolaiti ME, Ugarte F, Forsberg EC, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512:198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca D, Blomberg BB. Aging affects human B cell responses. Journal of clinical immunology. 2011;31:430–435. doi: 10.1007/s10875-010-9501-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nature reviews Molecular cell biology. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nature reviews Immunology. 2013;13:376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, Purton LE, Fleming HH, Cobb B, Merkenschlager M, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CJ, Marusyk A, DeGregori J. Aging-associated changes in hematopoiesis and leukemogenesis: what’s the connection? Aging. 2011;3:643–656. doi: 10.18632/aging.100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nature cell biology. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, Hiramatsu H, Restuccia U, Bachi A, Voisin V, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell stem cell. 2012;11:799–811. doi: 10.1016/j.stem.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell stem cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, Stranks AJ, Glanville J, Knight S, Jacobsen SE, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. The Journal of experimental medicine. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nature genetics. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahama T, Hanieh H, Nguyen NT, Chinen I, Ripley B, Millrine D, Lee S, Nyati KK, Dubey PK, Chowdhury K, et al. Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11964–11969. doi: 10.1073/pnas.1311087110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B, Rajaram MV, Lafuse WP, Landes MB, Schlesinger LS. Mycobacterium tuberculosis decreases human macrophage IFN-gamma responsiveness through miR-132 and miR-26a. Journal of immunology (Baltimore, Md : 1950) 2014;193:4537–4547. doi: 10.4049/jimmunol.1400124. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. The Journal of experimental medicine. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Warr MR, Passegue E. Cell cycle regulation in hematopoietic stem cells. The Journal of cell biology. 2011;195:709–720. doi: 10.1083/jcb.201102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L, Lin KK, Boles NC, Yang L, King KY, Jeong M, Mayle A, Goodell MA. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell stem cell. 2012;11:302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa-Geffen A, Soreq H. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J, Avigan DE, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell stem cell. 2013;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strovas TJ, Rosenberg AB, Kuypers BE, Muscat RA, Seelig G. MicroRNA-based single-gene circuits buffer protein synthesis rates against perturbations. ACS synthetic biology. 2014;3:324–331. doi: 10.1021/sb4001867. [DOI] [PubMed] [Google Scholar]
- Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell stem cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, Wang H, Le T, Faull KF, Chen R, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell stem cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nature communications. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vos KE, Eliasson P, Proikas-Cezanne T, Vervoort SJ, van Boxtel R, Putker M, van Zutphen IJ, Mauthe M, Zellmer S, Pals C, et al. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nature cell biology. 2012;14:829–837. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J, Passegue E. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JL, Rao DS, O’Connell RM, Garcia-Flores Y, Baltimore D. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. eLife. 2013;2:e00537. doi: 10.7554/eLife.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.