Abstract
OBJECTIVES:
Risk stratification in Barrett's esophagus (BE) is challenging. We evaluated the ability of a panel of genetic markers to predict progression to high-grade dysplasia (HGD) or esophageal adenocarcinoma (EAC).
METHODS:
In this case–control study, we assessed a measure of genetic instability, the mutational load (ML), in predicting progression to HGD or EAC. Cases had nondysplastic BE or low-grade dysplasia (LGD) at baseline and developed HGD/EAC ≥1 year later. Controls were matched 2:1, had nondysplastic BE or LGD, and no progression at follow-up. Formalin-fixed, paraffin-embedded tissue was microdissected for the epithelium. Loss of heterozygosity (LOH) and microsatellite instability (MSI) were assessed. ML was calculated from derangements in 10 genomic loci. High-clonality LOH mutations were assigned a value of 1, low-clonality mutations were assigned a value of 0.5, and MSI 0.75 at the first loci, and 0.5 for additional loci. These values were summed to the ML. Receiver operator characteristic (ROC) curves were created.
RESULTS:
There were 69 patients (46 controls and 23 cases). Groups were similar in age, follow-up time, baseline histology, and the number of microdissected targets. Mean ML in pre-progression biopsies was higher in cases (2.21) than in controls (0.42; P<0.0001). Sensitivity was 100% at ML ≥0.5 and specificity was 96% at ML ≥1.5. Accuracy was highest at 89.9% for ML ≥1. ROC curves for ML ≥1 demonstrated an area under the curve (AUC) of 0.95.
CONCLUSIONS:
ML in pre-progression BE tissue predicts progression to HGD or EAC. Although further validation is necessary, ML may have utility as a biomarker in endoscopic surveillance of BE.
INTRODUCTION
Barrett's esophagus (BE) is a precursor to esophageal adenocarcinoma (EAC), with an estimated cancer incidence of 3–6/1,000 person-years (1, 2, 3). EAC has a high mortality and a poor 5-year survival rate if it is not diagnosed and treated at an early stage (4). It is thought that the progression of BE to EAC occurs through a dysplasia to carcinoma sequence. Consequently, incidence of EAC is increased in BE with high-grade dysplasia (HGD) compared with nondysplastic BE (NDBE) (5, 6). Currently, dysplasia is the only clinical biomarker that is commonly used to stratify risk in BE (7, 8, 9). Patients with BE undergo periodic endoscopic surveillance to detect dysplasia and adenocarcinoma, but it is unclear whether this practice is beneficial (10, 11, 12). In fact, recent evidence suggests that current surveillance practices may not prevent cancer death in BE patients (13).
Endoscopic screening and surveillance relies on histology from random esophageal biopsies, but this process is imperfect owing to sampling error and inconsistencies in histological grading. Sampling error may in part explain why two-thirds of all adenocarcinomas can occur during the first year of follow-up (14). Once samples are collected, another potential source of error arises from the interpretation of histology. Agreement between pathologists regarding the degree of dysplasia is relatively poor, particularly for low-grade dysplasia (LGD) (15, 16, 17, 18). As a result, current surveillance in BE patients is inadequate to accurately predict who will progress to EAC. The need for other modalities of risk stratification, in light of the increasing incidence of EAC, has led to the development of advanced endoscopic methods and biomarkers (19, 20, 21, 22, 23). Biomarkers could be especially helpful in identifying high-risk individuals, so that surveillance can be performed in an effective and cost-conscious manner (24) to facilitate earlier therapeutic interventions.
Molecular biomarkers have promise in identifying early neoplastic transformation, as the development of EAC in BE patients is associated with increasing genetic instability. In this study, we assessed the utility of a measure of genetic aberration, the mutational load (ML), to predict subsequent progression in BE. Previous work demonstrated that ML correlated with increasingly severe histology (25). Therefore, we performed a case–control study to determine whether the degree of ML present in pre-progression BE tissue predicted the risk of progression to HGD or EAC. We hypothesized that high ML in pre-progression tissue would be associated with an increased risk of developing HGD or EAC.
METHODS
Design
We performed a case–control study comparing ML in pre-progression tissue of BE patients who progressed to either HGD or EAC with those who did not progress. Cases (i.e., progressors) were BE subjects with no dysplasia or LGD at baseline who subsequently developed HGD or EAC, with a minimum of at least 1 year between the baseline biopsy and the follow-up biopsy demonstrating progression. Similar to the cases, controls (i.e., nonprogressors) had either NDBE or LGD at baseline, but had no progression of BE at the follow-up biopsy. All dysplasia readings were confirmed by a second expert pathologist. Controls were frequency-matched 2:1 to cases by age, sex, index biopsy histology, and length of follow-up (time from index to outcome biopsy). Study subjects were recruited from three sources: Massachusetts General Hospital, Boston, MA; Allegheny General Hospital, Pittsburgh, PA; and PathGroup, Brentwood, TN. Institutional review board approval was obtained to collect specimens and patient data from each of these sites, and associated institutional review board numbers were 2011-P-002116 (27 October 2011), RC-5743 (25 July 2013), and 28827/1 (22 November 2013), respectively.
A limited, deidentified data set, including age, sex, and endoscopic and histological evaluation, was abstracted from the electronic medical records. In addition, archival FFPE (formalin-fixed, paraffin-embedded) tissue from the pre-progression endoscopy was retrieved for analysis by the central lab.
Tissue processing
FFPE tissue from biopsies taken from cases and controls at the index time point was assessed for ML. Hematoxylin and eosin (H&E)-stained FFPE slides were examined microscopically to identify representative patient histology, either intestinal metaplasia or LGD, for microdissection of histological targets to determine ML. In patients with NDBE, microscopic sites of epithelial cells with no features of dysplasia were selected as targets, and in patients with LGD cells demonstrating dysplasia were selected. H&E-stained slides were used to guide microdissection of recut, unstained, 4-micron-thick, FFPE slides. Slides were microdissected for the maximum number of histological targets available upon microscopic inspection. Microdissection was performed manually, targeting areas in which epithelial cells constituted 90% or more of the total cells removed. By microscopic estimation, no more than 10% of microdissected cells were stromal or inflammatory cells. Accuracy of all microdissections was carefully reviewed. DNA from microdissected targets was prepared either through the crude lysate method or QIAamp DNA micro kit (QIAGEN Sciences, Germantown, MD).
ML assessments were made with a previously reported panel of 10 genomic loci using 24 DNA markers (26, 27, 28, 29, 30, 31). The following genomic loci (with associated tumor suppressor genes) were included in the panel: 1p (CMM1, L-myc), 3p (VHL, HoGG1), 5q (MCC, APC), 9p (CDKN2A), 10q (PTEN, MXI1), 17p (TP53), 17q (NME1), 18q (DCC), 21q (TFF1, PSEN2), and 22q (NF2). The presence of loss of heterozygosity (LOH) and new alleles consistent with microsatellite instability (MSI) was investigated using PCR and quantitative capillary electrophoresis of DNA extracted from each microdissected target (26, 28, 29, 32). PCR-amplifiable DNA was used to identify targets of adequate quality for use in assessing ML. If the extracted DNA was of low quality, then the DNA specimen was considered as nonassessable for that target and was not included in the ML calculation. Laboratory personnel and pathologists performing the microdissection and scoring the genetic loci were blinded to the progressor status of the tissue.
ML analysis
The ML is a summary construct to quantify the degree of cumulative genetic derangement present at all 10 genomic loci assessed. ML was determined for each tissue target by considering the presence and clonality of LOH mutations and the presence of MSI at each genomic locus. Development of the ML algorithm has been previously described (26, 32). In brief, all LOH mutations at a genomic locus were assigned a numerical value based on the extent (clonality) of LOH. Clonality was determined using the ratio of allele peak heights in DNA from microdissected targets. LOH mutations were high clonality when greater than 75% of the DNA had LOH mutation, and low clonality when 50–75% of the DNA had LOH mutation. A value of 0.5 was assigned for low-clonality mutations and 1 for high-clonality mutations. The value of the first MSI at a genomic locus was 0.75, and for each additional MSI thereafter was 0.5. The values for low- and high-clonality LOH mutations and MSI were used to calculate the highest weighted value at each locus, which were summed together for all loci in a tissue target. The resulting cumulative value was defined as the overall ML for that tissue target, ranging from 0 to 10 in value (a high-clonality LOH mutation, weighted 1, was the highest possible weighted value at each of the 10 genomic loci). The maximum ML present in all tissue targets for a patient's baseline index biopsy was defined as the ML for the pre-progression tissue.
Statistical analysis
ML in pre-progression tissues of cases and controls was compared to assess its ability to predict progression to HGD or EAC. Chi-square and Fisher's exact tests were used to compare categorical variables, and Wilcoxon rank-sum test was used for continuous variables. A box-and-whisker plot of ML in cases and controls was constructed. Receiver operator characteristic (ROC) curves were created to assess the diagnostic utility of various cutoffs of ML for progression, and the utility of subgroups of loci was assessed for predicting progression. The difference between ROC curves using different loci was tested using the nonparametric method established by Venkatraman and Begg (33). Logistic regression was performed including age, gender, and baseline index histology (i.e., NDBE and LGD) as covariates. Effects of these covariates were tested using the likelihood ratio test. Confidence intervals for AUROC were calculated using the DeLong method (34). Linear discriminant analysis was performed to assess the effect of differential weighting of results from individual loci on assay accuracy. Given the variability of assessable targets, Monte Carlo simulations were used to determine the optimal number of microdissected histological targets needed to achieve maximum accuracy. Simulations were performed by randomly selecting one, two, three, or more than three targets. The average performance of more than 100 simulations for each target number, per patient, at index time points was calculated. Two-tailed tests with the significance level of P<0.05 were used. All analyses were performed with the R statistical programming language (r-project.org).
RESULTS
A total of 69 patients were included in the analysis, including 46 nonprogressors (controls) and 23 progressors (cases). There was no difference in average age between the two groups (Table 1). Typical of Barrett's patients, the majority was male. Both groups had on average two assessable microdissected histological targets (range 1–8) per patient from the baseline (pre-progression) exam. There was no significant difference between the proportion of cases and controls harboring LGD at index biopsy (30.4 vs. 17.4%, P=0.23). The mean follow-up time between index and outcome biopsies was 4 years for both cases and controls.
Table 1. Description of Barrett's esophagus nonprogressor and progressor patients included in the study.
| Nonprogressors (n=46) | Progressors (n=23) | |
|---|---|---|
| Mean age (years) | 62.5 | 63.9 |
| Male | 35 (76%) | 22 (96%) |
| Mean targets per patient | 1.70 | 1.87 |
| Range follow-up time (minimum–maximum years) between baseline and outcome biopsies | 1.1–9.0 | 1.2–11.5 |
| Mean follow-up time (years) between baseline and outcome biopsies | 4.3 | 3.9 |
| Median follow-up time (years) between baseline and outcome biopsies | 4.1 | 3.1 |
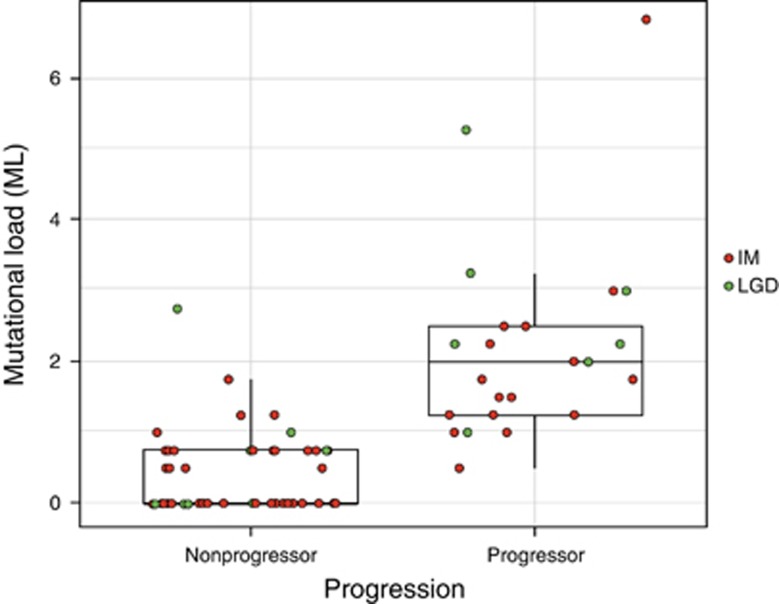
The mean per-patient ML at index biopsy was significantly higher in cases (mean ML=2.21) compared with controls (mean ML=0.42, P<0.0001) (Figure 1). The ML of progressors ranged from 0.5 to 6.75 and the ML of nonprogressors ranged from 0.00 to 2.75. No case had an ML of 0 in their pre-progression tissue, compared with 25/46 (54%) of controls.
Figure 1.
Mutational load (ML) of each nonprogressor and progressor patient at baseline index time point. ML per patient was the maximum ML in all microdissected histological targets at baseline time point for the patient. The most severe histology found in each patient's clinical pathology report at that same index time point is indicated as intestinal metaplasia (IM) or low-grade dysplasia (LGD) (IM=orange circle; LGD=green circle).
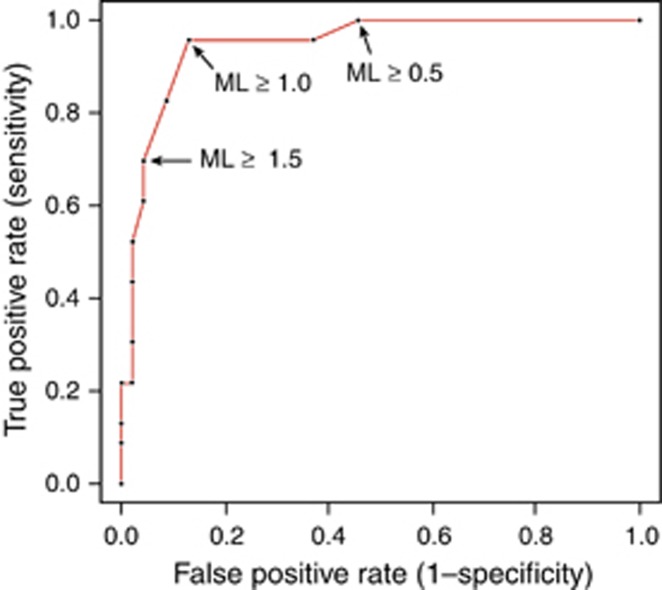
To understand the operating characteristics, the test was evaluated with different per-patient ML cutoffs ranging from 0.5 to 1.5 (Table 2). As expected, sensitivity decreased and specificity increased with increasing cutoffs for ML. There was 100% sensitivity for ML cutoff ≥0.5 and 96% specificity for ML cutoff ≥1.5. After adjusting for covariates, including age, gender, and histology of patient at index time point (NDBE or LGD), the adjusted odds ratio (OR) for identifying cases at ML ≥1 was 165.8 (P<0.0001), compared with an unadjusted odds ratio of 146.7 (P<0.0001). All covariates were statistically insignificant in both univariate and multivariate odds ratio analysis. Accuracy of the test was highest at 89.9% (95% confidence interval (CI) 80.2–95.8) at ML ≥1. ROC curves were constructed with varying ML thresholds (Figure 2), and the corresponding area under the curve (AUC) at ML ≥1 was 0.95 (95% CI 0.89–1.0).
Table 2. Mutational load (ML) performance characteristics based on various per-patient ML thresholds derived from ROC curve.
| Per-patient ML threshold | |||
|---|---|---|---|
| Performance characteristic | ≥0.5 | ≥1 | ≥1.5 |
| Sensitivity (%) | 100.0 | 95.7 | 69.6 |
| Specificity (%) | 54.3 | 87.0 | 95.7 |
| Accuracy (%) | 69.6 | 89.9 | 87.0 |
| LR+ | 2.2 | 7.3 | 16.0 |
| LR− | 0.0 | 0.1 | 0.3 |
LR+, positive likelihood ratio; LR−, negative likelihood ratio; ROC, receiver operator characteristic.
Figure 2.
Receiver operator characteristic (ROC) for the performance of mutational load (ML) in predicting progression at baseline index time points for each patient. Area under the curve (AUC)=0.95 (95% confidence interval (CI) 0.89–0.99).
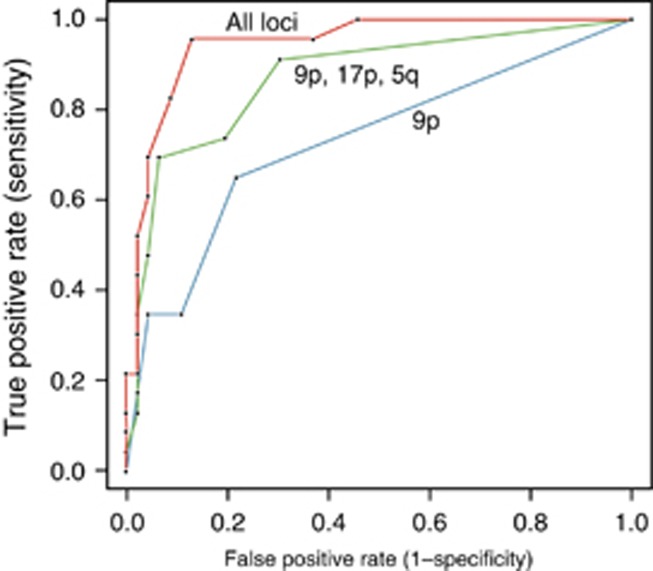
Given that some genomic loci may be more predictive of progression than others, we assessed the frequency of mutations across the panel. There was a fairly equal distribution of mutations across all genomic loci in both groups (Supplementary Table 1 online). All 10 loci showed a markedly higher rate of mutation in cases compared with controls. The most frequent locus in the control group was 9p. In the cases, the most frequent loci were 9p, 17p, and 5q.
We then assessed the utility of simplified models by using only the most predictive loci. Three ROC curves were compared: the first, using all loci; the second, using data only from the three most predictive loci (9p, 17p, or 5q); and the third, using 9p alone (Figure 3). The best ROC curve was achieved when all 10 loci were used with AUC 0.95 (95% CI 0.89–1.0). ROC curves for the subset of 9p, 17p, or 5q had an AUC of 0.87 (95% CI 0.78–0.96). The worst curve was with 9p only, with an AUC of 0.73 (95% CI 0.61–0.85). Comparing the three ROC curves statistically demonstrated a P value of 0.002 when comparing the 10 loci with 9p alone, a P value of 0.07 when comparing 9p with 9p/17p/5q, and a P value of 0.17 when comparing 9p/17p/5q with all loci.
Figure 3.
Receiver operator characteristic (ROC) for performance of mutational load (ML) in predicting progression at baseline time points for each patient based on subsets of genomic loci included in ML calculations.
The optimal number of microdissected histologic targets needed for assessing ML performance was investigated by Monte Carlo simulation. ROC curves showed that the AUC for one target was 0.88 (95% CI 0.83–0.94), compared with 0.94 (95% CI 0.89–0.99) for two targets, and 0.95 (95% CI 0.90–1.0) for three or more targets. Therefore, classification performance markedly improved when two or more microdissected histological targets were used for ML calculation, but it only modestly changed with additional targets.
Finally, when a modified ML calculation was tested by using variable weightings for each genomic locus, there was improved performance of ML with an AUC of 0.97, compared with the current method of ML calculation that had an AUC of 0.95. However, the difference between the two AUCs was not statistically significant (P=0.42), suggesting that the simple predefined scoring algorithm for ML, as described above, was about as accurate as the modified loci-level weighted approach.
DISCUSSION
We performed a case–control study comparing ML in pre-progression tissue of BE subjects destined to develop HGD or EAC with controls who did not progress, to assess the diagnostic value of the ML in predicting progression. The AUC of the ROC curve for the assay at a cutoff of ≥1 was 0.95. The test had the highest sensitivity at a threshold of ML ≥0.5. All loci that were tested for LOH and MSI provided value to predict progression to HGD or EAC. Test performance was highest when two or more microdissected histological targets were used. All 10 loci appeared to contribute to the assay's accuracy and modified calculations with optimized weightings of genomic loci did not markedly improve accuracy, suggesting that the simple summative model is appropriate to carry forward for clinical work. Therefore, our preliminary study demonstrates the potential utility of using ML as a biomarker that predicts progression to dysplasia in patients with BE.
The genetic instability associated with the progression of BE to EAC provides the conceptual underpinnings for the use of ML as a biomarker (35, 36, 37, 38, 39). Previous studies showed that increasing ML, as used in our study, correlated with worsening BE histological classification (32). This work demonstrated that ML distinguished patients with EAC from those without EAC, but did not determine when the biomarker appeared before occurrence of HGD or EAC. Given these findings, it was important to understand the temporal relationship between changes in ML and progression to advanced neoplasia. For a biomarker to have clinical utility in BE, it needs to distinguish between high-risk and low-risk patients well before progression to carcinoma, to allow for early intervention. As the mean time between the baseline and follow-up biopsies showing progression was 4 years in our study, our data suggest that the changes in ML predate the development of histological progression by a sufficient period, to allow this assay to be a clinically useful biomarker for predicting risk of progression.
Previous investigators have assessed genetic changes associated with progression in BE. Multiple loci differentiating between benign and malignant BE tissue have been identified in cross-sectional studies (30, 31, 40). Longitudinal studies in BE have often focused on a single genomic locus such as 17p (41, 42), and as a result may have limited clinical utility. A prospective 10-year study (43) found that 9p LOH, 17p LOH, and DNA content abnormalities were best at predicting progression to EAC (RR 38.7; 95% CI 10.8–138.5; P<0.001), but it was limited owing to the small number of cancers occurring in the sample. Another biomarker panel using a combination of LGD and abnormal DNA copy number was able to identify 24% of BE progressors before the development of EAC (21). DNA methylation-based assays have shown a sensitivity of up to 50% in predicting progression to EAC (44, 45, 46). Other genetic studies have focused on microRNA expression (47, 48), genetic instability, and clonal expansion (39), and transcribed ultraconserved non-coding RNAs (49), among other techniques (50, 51), to determine elements of genetic aberration that can be used as predictors of carcinogenesis in BE.
The assay in our study was 100% sensitive at a threshold of ML ≥0.5 and 89.9% accurate at a ML of ≥1 in differentiating between cases and controls. High sensitivity is a key for a molecular marker to detect disease states such as EAC that can have dire clinical consequences if missed. Therefore, this assay at a threshold of ML ≥0.5 demonstrates adequate promise to proceed with the next phase of biomarker development (52), a larger prospective trial, to more completely determine the operating characteristics of the test.
Our study provides strong preliminary evidence for a robust biomarker calculated from genetic aberrations at multiple loci that can stratify risk in pre-progression BE tissue, and shows that these aberrations occur well before the onset of advanced neoplasia. One of the limitations of this study is that ML was derived from microdissected histological targets from archived pathology specimens. There are potential problems with inadequate biopsies or missing dysplastic lesions, as these areas can be endoscopically difficult to detect, and therefore sampling error may occur. However, sampling error leading to misclassification of patients with dysplasia as nondysplastic BE presumably would bias our results toward the null, making it less likely that we would observe the strongly positive results reported here. In addition, there is known variability between pathologists on histologic classification, which defined case and control groups. Histologic misclassification is an unavoidable problem when developing tissue-based biomarkers if histologic readings are to be the “gold standard” against which the biomarker is to be measured. In addition, the size of the data set limited the evaluation of genomic loci-level-based weighting in the modified ML score, and made it impossible to test for DNA marker-level-based ML scores that could potentially outperform the current ML system. Finally, as the rate of progression in BE is low, further examination of ML in larger cohorts will be necessary to more completely understand the clinical utility of the assay in low-risk populations.
Dysplasia surveillance in BE is a challenging and costly problem. Current surveillance methods based on histological classification are inaccurate and inadequate in predicting progression to HGD or EAC in patients with low-risk BE. Biomarkers are desperately needed, which accurately stratify risk in BE patients. Better risk stratification will have implications in the frequency of surveillance endoscopy, as well as treatment decisions for ablative therapy in high-risk individuals. The results of this study provide support for the potential use of ML as a predictive biomarker in low-risk BE patients to assess for the risk of progression to malignancy.
Study Highlights
Guarantor of the article: Nicholas J. Shaheen, MD, MPH.
Specific author contributions: Study design, data interpretation, manuscript drafting, and critical revision: Swathi Eluri; project conception, study design, and critical revision: William R. Brugge, Ebubekir S. Daglilar, Derek C. Welch, and Todd M. Barr; project conception, study design, data interpretation, and critical revision: Sara A. Jackson; study design, data interpretation, and critical revision: Mindi A. Styn, Keith M. Callenberg, Lucas C. Duits, and Jacques J. Bergman; project conception, study design, data interpretation, manuscript drafting, critical revision: Nicholas J. Shaheen; all authors approved the final draft submitted.
Financial support: Interpace Diagnostics in part funded efforts to obtain archived pathology specimens, and performed the ML analysis.
Potential competing interests: NJS receives research funding from Interpace. SAJ, MAS, and KMC are full-time employees of Interpace.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/ajg
Supplementary Material
References
- Thomas T, Abrams KR, De Caestecker JS, et al. Meta analysis: cancer risk in Barrett's oesophagus. Aliment Pharmacol Ther. 2007;26:1465–1477. doi: 10.1111/j.1365-2036.2007.03528.x. [DOI] [PubMed] [Google Scholar]
- Yousef F, Cardwell C, Cantwell MM, et al. The incidence of esophageal cancer and high-grade dysplasia in Barrett's esophagus: a systematic review and meta-analysis. Am J Epidemiol. 2008;168:237–249. doi: 10.1093/aje/kwn121. [DOI] [PubMed] [Google Scholar]
- Haggitt RC, Tryzelaar J, Ellis FH, et al. Adenocarcinoma complicating columnar epithelium-lined (Barrett's) esophagus. Am J Clin Pathol. 1978;70:1–5. doi: 10.1093/ajcp/70.1.1. [DOI] [PubMed] [Google Scholar]
- Lerut T, De Leyn P, Coosemans W, et al. Surgical strategies in esophageal carcinoma with emphasis on radical lymphadenectomy. Ann Surg. 1992;216:583. doi: 10.1097/00000658-199211000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi A, Puli S, El-Serag HB, et al. Incidence of esophageal adenocarcinoma in patients with Barrett's esophagus and high-grade dysplasia: a meta-analysis. Gastrointest Endosc. 2008;67:394–398. doi: 10.1016/j.gie.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Shaheen NJ, Sharma P, Overholt BF, et al. Radiofrequency ablation in Barrett's esophagus with dysplasia. New Engl J Med. 2009;360:2277–2288. doi: 10.1056/NEJMoa0808145. [DOI] [PubMed] [Google Scholar]
- Abdalla M, Dhanekula R, Greenspan M, et al. Dysplasia detection rate of confirmatory EGD in nondysplastic Barrett's esophagus. Dis Esophagus. 2014;27:505–510. doi: 10.1111/j.1442-2050.2012.01431.x. [DOI] [PubMed] [Google Scholar]
- Anaparthy R, Sharma P. Progression of Barrett oesophagus: role of endoscopic and histological predictors. Nat Rev Gastroenterol Hepatol. 2014;11:525–534. doi: 10.1038/nrgastro.2014.69. [DOI] [PubMed] [Google Scholar]
- Weston AP, Sharma P, Topalovski M, et al. Long-term follow-up of Barrett's high-grade dysplasia. Am J Gastroenterol. 2000;95:1888–1893. doi: 10.1111/j.1572-0241.2000.02234.x. [DOI] [PubMed] [Google Scholar]
- Choi SE, Hur C. Screening and surveillance for Barrett's esophagus: current issues and future directions. Curr Opin Gastroenterol. 2012;28:377–381. doi: 10.1097/MOG.0b013e328353d58e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spechler SJ. Barrett esophagus and risk of esophageal cancer: a clinical review. JAMA. 2013;310:627–636. doi: 10.1001/jama.2013.226450. [DOI] [PubMed] [Google Scholar]
- Falk GW, Ours TM, Richter JE. Practice patterns for surveillance of Barrett's esophagus in the United States. Gastrointest Endosc. 2000;52:197–203. doi: 10.1067/mge.2000.107728. [DOI] [PubMed] [Google Scholar]
- Corley DA, Mehtani K, Quesenberry C, et al. Impact of endoscopic surveillance on mortality from Barrett's esophagus-associated esophageal adenocarcinomas. Gastroenterology. 2013;145:312–9. doi: 10.1053/j.gastro.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvid-Jensen F, Pedersen L, Drewes AM, et al. Incidence of adenocarcinoma among patients with Barrett's esophagus. New Engl J Med. 2011;365:1375–1383. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- Shaheen NJ, Richter JE. Barrett's oesophagus. Lancet. 2009;373:850–861. doi: 10.1016/S0140-6736(09)60487-6. [DOI] [PubMed] [Google Scholar]
- Gatenby P, Soon Y. Barrett's oesophagus: evidence from the current meta-analyses. World J Gastrointest Pathophysiol. 2014;5:178–187. doi: 10.4291/wjgp.v5.i3.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs-Kelly E, Mendelin JE, Bennett AE, et al. Poor interobserver agreement in the distinction of high-grade dysplasia and adenocarcinoma in pretreatment Barrett's esophagus biopsies. Am J Gastroenterol. 2008;103:2333–2340. doi: 10.1111/j.1572-0241.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- Ormsby A, Petras R, Henricks W, et al. Observer variation in the diagnosis of superficial oesophageal adenocarcinoma. Gut. 2002;51:671–676. doi: 10.1136/gut.51.5.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto MI, Anandasabapathy S, Brugge W, et al. In vivo endomicroscopy improves detection of Barrett's esophagus-related neoplasia: a multicenter international randomized controlled trial (with video) Gastrointest Endosc. 2014;79:211–221. doi: 10.1016/j.gie.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar KB, Spechler SJ. Controversies in Barrett esophagus. Mayo Clin Proc. 2014;89:973–984. doi: 10.1016/j.mayocp.2014.01.022. [DOI] [PubMed] [Google Scholar]
- Bird-Lieberman EL, Dunn JM, Coleman HG, et al. Population-based study reveals new risk-stratification biomarker panel for Barrett's esophagus. Gastroenterology. 2012;143:927–35. doi: 10.1053/j.gastro.2012.06.041. [DOI] [PubMed] [Google Scholar]
- Fels Elliott DR, Fitzgerald RC. Molecular markers for Barrett's esophagus and its progression to cancer. Curr Opin Gastroenterol. 2013;29:437–445. doi: 10.1097/MOG.0b013e328362282f. [DOI] [PubMed] [Google Scholar]
- Timmer MR, Sun G, Gorospe EC, et al. Predictive biomarkers for Barrett's esophagus: so near and yet so far. Dis Esophagus. 2013;26:574–581. doi: 10.1111/dote.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon LG, Mayne GC, Hirst NG, et al. Cost-effectiveness of endoscopic surveillance of non-dysplastic Barrett's esophagus. Gastrointest Endosc. 2014;79:242–56. doi: 10.1016/j.gie.2013.07.046. [DOI] [PubMed] [Google Scholar]
- Khara HS, Jackson SA, Nair S, et al. Assessment of mutational load in biopsy tissue provides additional information about genomic instability to histological classifications of Barrett's esophagus. J Gastrointest Cancer. 2014;45:137–145. doi: 10.1007/s12029-013-9570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellsworth E, Jackson SA, Thakkar SJ, et al. Correlation of the presence and extent of loss of heterozygosity mutations with histological classifications of Barrett's esophagus. BMC Gastroenterol. 2012;12:181. doi: 10.1186/1471-230X-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan K, Garde J, Walker SJ, et al. LOH at the sites of the DCC, APC, and TP53 tumor suppressor genes occurs in Barrett's metaplasia and dysplasia adjacent to adenocarcinoma of the esophagus. Hum Pathol. 1999;30:1508–1514. doi: 10.1016/s0046-8177(99)90175-2. [DOI] [PubMed] [Google Scholar]
- Raja S, Finkelstein SD, Baksh FK, et al. Correlation between dysplasia and mutations of six tumor suppressor genes in Barrett's esophagus. Ann Thorac Surg. 2001;72:1130–1135. doi: 10.1016/s0003-4975(01)03005-3. [DOI] [PubMed] [Google Scholar]
- Lin X, Finkelstein SD, Zhu B, et al. Loss of heterozygosities in Barrett esophagus, dysplasia, and adenocarcinoma detected by esophageal brushing cytology and gastroesophageal biopsy. Cancer. 2009;117:57–66. doi: 10.1002/cncy.20010. [DOI] [PubMed] [Google Scholar]
- Barrett MT, Galipeau PC, Sanchez CA, et al. Determination of the frequency of loss of heterozygosity in esophageal adenocarcinoma by cell sorting, whole genome amplification and microsatellite polymorphisms. Oncogene. 1996;12:1873–1878. [PubMed] [Google Scholar]
- Sanz-Ortega J, Hernandez S, Saez MC, et al. 3p21, 5q21, 9p21 and 17p13.1 allelic deletions are potential markers of individuals with a high risk of developing adenocarcinoma in Barrett's epithelium without dysplasia. Hepato-gastroenterology. 2003;50:404–407. [PubMed] [Google Scholar]
- Khara HS, Jackson SA, Nair S, et al. Assessment of mutational load in biopsy tissue provides additional information about genomic instability to histological classifications of Barrett's esophagus. J Gastrointest Cancer. 2014;45:137–145. doi: 10.1007/s12029-013-9570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman E, Begg CB. A distribution-free procedure for comparing receiver operating characteristic curves from a paired experiment. Biometrika. 1996;83:835–848. [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- Garewal HS, Sampliner R, Liu Y, et al. Chromosomal rearrangements in Barrett's esophagus. A premalignant lesion of esophageal adenocarcinoma. Cancer Genet Cytogenet. 1989;42:281–286. doi: 10.1016/0165-4608(89)90096-4. [DOI] [PubMed] [Google Scholar]
- Raskind WH, Norwood T, Levine DS, et al. Persistent clonal areas and clonal expansion in Barrett's esophagus. Cancer Res. 1992;52:2946–2950. [PubMed] [Google Scholar]
- Dolan K, Garde J, Gosney J, et al. Allelotype analysis of oesophageal adenocarcinoma: loss of heterozygosity occurs at multiple sites. Br J Cancer. 1998;78:950–957. doi: 10.1038/bjc.1998.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson TG, Maley CC, Li X, et al. Chromosomal instability and copy number alterations in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:3305–3314. doi: 10.1158/1078-0432.CCR-08-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Li X, et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 2004;64:7629–7633. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- Ahmad J, Arthur K, Maxwell P, et al. A cross sectional study of p504s, CD133, and Twist expression in the esophageal metaplasia dysplasia adenocarcinoma sequence. Dis Esophagus. 2014;28:276–282. doi: 10.1111/dote.12181. [DOI] [PubMed] [Google Scholar]
- Reid BJ, Prevo LJ, Galipeau PC, et al. Predictors of progression in Barrett's esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–2848. doi: 10.1111/j.1572-0241.2001.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan K, Morris AI, Gosney JR, et al. Loss of heterozygosity on chromosome 17p predicts neoplastic progression in Barrett's esophagus. J Gastroenterol Hepatol. 2003;18:683–689. doi: 10.1046/j.1440-1746.2003.03048.x. [DOI] [PubMed] [Google Scholar]
- Galipeau PC, Li X, Blount PL, et al. NSAIDs modulate CDKN2A, TP53, and DNA content risk for progression to esophageal adenocarcinoma. PLoS Med. 2007;4:e67. doi: 10.1371/journal.pmed.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Cheng Y, Gu W, et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett's esophagus. Cancer Res. 2009;69:4112–4115. doi: 10.1158/0008-5472.CAN-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Bhagat TD, Yang X, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956–66. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement G, Braunschweig R, Pasquier N, et al. Methylation of APC, TIMP3, and TERT: a new predictive marker to distinguish Barrett's oesophagus patients at risk for malignant transformation. J Pathol. 2006;208:100–107. doi: 10.1002/path.1884. [DOI] [PubMed] [Google Scholar]
- Bansal A, Lee IH, Hong X, et al. Feasibility of mcroRNAs as biomarkers for Barrett's Esophagus progression: a pilot cross-sectional, phase 2 biomarker study. Am J Gastroenterol. 2011;106:1055–1063. doi: 10.1038/ajg.2011.37. [DOI] [PubMed] [Google Scholar]
- Revilla-Nuin B, Parrilla P, Lozano JJ, et al. Predictive value of MicroRNAs in the progression of barrett esophagus to adenocarcinoma in a long-term follow-up study. Ann Surg. 2013;257:886–893. doi: 10.1097/SLA.0b013e31826ddba6. [DOI] [PubMed] [Google Scholar]
- Fassan M, Dall'Olmo L, Galasso M, et al. Transcribed ultraconserved noncoding RNAs (T-UCR) are involved in Barrett's esophagus carcinogenesis. Oncotarget. 2014;5:7162–7171. doi: 10.18632/oncotarget.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brankley SM, Fritcher EG, Smyrk TC, et al. Fluorescence in situ hybridization mapping of esophagectomy specimens from patients with Barrett's esophagus with high-grade dysplasia or adenocarcinoma. Hum Pathol. 2012;43:172–179. doi: 10.1016/j.humpath.2011.04.018. [DOI] [PubMed] [Google Scholar]
- Davelaar AL, Calpe S, Lau L, et al. Aberrant TP53 detected by combining immunohistochemistry and DNA-FISH improves Barrett's esophagus progression prediction: a prospective follow-up study. Genes Chromosomes Cancer. 2014;54:82–90. doi: 10.1002/gcc.22220. [DOI] [PubMed] [Google Scholar]
- Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.