Abstract
A biotinylated heparosan hexasaccharide was synthesized using one-pot multi-enzyme strategy, in situ activation and transfer of N-trifluoroacetylglucosamine (GlcNTFA) to heparin backbone significantly improved the synthetic efficiency. The biotinylated hexasaccharide could serve as a flexible core to diversify its conversion into heparan sulfate isoforms with potential biological applications and therapeutics
Heparosan is a linear polysaccharide copolymer consisting of α-1,4-linked D-glucosamine (GlcN) and β-1,4-D-glucuronioc acid (GlcA) disaccharide repeating unit (GlcN-α-1,4-GlcA-β-1,4). It belongs to the glycosaminoglycans (GAGs) family and is found in the capsule polysaccharide of certain pathogentic bacteria. Heparosan also serves as the biosynthetic precursor of heparan sulfates (HSs), which are formed by extensive modifications of the heparosan backbone, namely, sulfations (O-sufations and N-sufations) and epimerization (C5 epimerization of GlcA to L-iduronic acid).1 Heparin and HS bind to a variety of protein ligands and regulate a wide variety of important biological activities, including blood coagulation, bacterial and viral infection, inflammation, growth factor regulation, cell adhesion, cell growth, tumor metastasis, lipid metabolism and diseases of the nervous system.2
Heparin and HS have been widely used as anticoagulant drugs for a century due to their capacity of strong binding to antithrombin.3 Synthetic homogenous heparins can eliminate detrimental effects caused by contamination of heterogeneous heparins purified from natural sources heparins.4 FDA-approved synthetic pentasaccharide (Fondaparinux or Arixtra®) exhibits antithrombin III mediated anti-factor Xa activity for prevention and treatment of venous thrombolytic disorders5 (Figure 1). To date, there is still no efficient antidote available to neutralize the anticoagulant effect of fondaparinux in the case of hemorrhage due to overdose.6 Extensive efforts have been focused on the development of neutralizable strategy for heparin-based antithrombotic treatment.7 It was proposed that stable protein-small molecule complexes, which are formed by strong binding of non-toxic avidin with biotin moiety of biotinylated HS drug molecules, facilitate the drug’s fast clearance from circulation.4a,8 Therefore, the biotinylated-fondaparinux conjugate can be neutralized by avidin to reverse the anticoagulant effect of fondaparinux.
Figure 1.
Fondaparinux (Arixtra®) and its analogues
Although much progress has been made in the chemical synthesis of heparin and HS, there are still significant challenges to synthetic chemists.9 These challenges include: 1) formation of the challenging 1,2-cis linkage from the GlcN donor; 2) tedious and non-productive protection/deprotection manipulations resulting in lengthy steps and low overall yields; 3) difficult access to L-iduronic acid and L-idose; 4) the necessity of choosing a set of orthogonal protecting groups for selective sulfations at desired positions, which often lead to unexpected difficulties.10 During the last decade, numerous methodologies, including the different protection strategies utilized for GlcN donors and acceptors (e.g., the late stage oxidations of glucose into GlcA after glycosylation), have been developed to address such limitations.11 However, large scale chemical synthesis of heparin and HSs in a pure and fully deprotected form is still not a routine practice. Alternatively, recent advances in identification and understanding of heparin biosynthetic enzymes’ substrate specificity in assembly of heparin and HS have made in vitro enzymatic heparin reconstitution practical and realistic.12 In this communication, we present a highly efficient one-pot multi-enzyme system to chemo-enzymatically construct a biotinylated heparosan hexasaccharide in a straightforward means, which can be used for multiple later-stage enzymatic modifications to furnish a library of HS-like isoforms bearing a highly valuable biotin moiety that could allow wide varieties of biological applications. For example, biotin could serve as an anchoring tip to immobilize such a oligosaccharide library onto the streptavidin-coated microarray surface for identification of previously unknown glycan-binding proteins (GBPs) at the molecular-level, providing insight into the potential relationship between sulfation/epimerization patterns and protein binding specificities.13 Another important factor of designing such biotin conjugate arises from the fact that a fondaparinux-like biotin conjugate could be constructed by subsequently selective enzymatic epimerization, N-, 2-O-, 6-O-, 3-O-sulfations (Figure 1).12g This will provide an opportunity for efficient regulation and management of its anticoagulation treatment to prevent hemorrhage by avidin neutralization.14
To date, several enzyme systems for heparosan synthesis have been identified. In Escherichia coli (E. coli), a pair of glycosyltransferases KfiA and KfiC are used to transfer hexosamine and glucuronic acid UDP precursors respectively to the non-reducing end of the growing chain.15 While in Pasteurella multocida (P. multocida), bifunctional glycosytransferase PmHS1 or PmHS2 is used to execute consecutive incorporation of hexosamine and glucuronic acid.16 Interestingly, the heparosan synthases from P. multocida (PmHS1 and PmHS2) possess both UDP-GlcN and UDP-GlcA transfering sites in the same peptide chain, and employ both inverting and retaining polymerization mechanisms. Recently, KfiA from E. coli K5 and PmHS2 from P. multocida have been used in the synthesis of heparosan with a disaccharide as chain initiating scaffold, which is derived from degradation of heparosan by fermentation.12g, 17 However, the use of chemically modified biotinylated GlcA as a chain initiating scaffold, and in situ activation and transfer of N-trifluoroacetylglucosamine (GlcNTFA) to heparosan backbone in one-pot multi-enzyme system have not been reported.
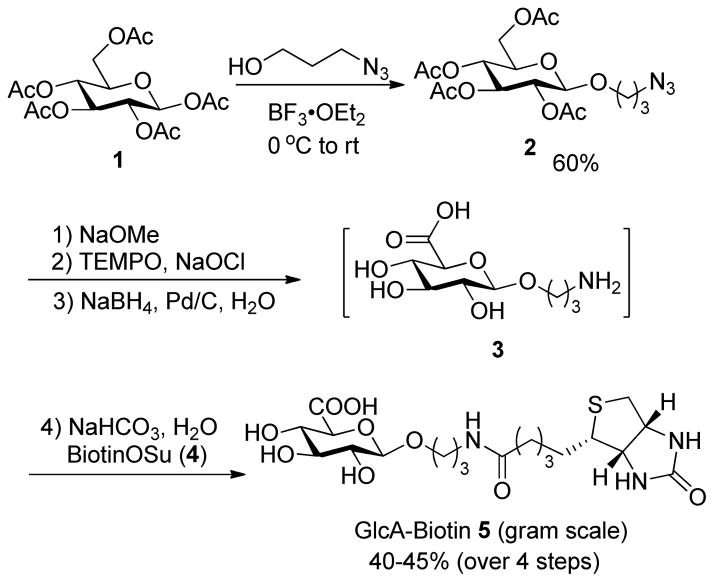
Prior to initiating heparin synthesis, the first issue needed to be addressed is whether the GlcA-biotin conjugate 5 could be tolerated and recognized by either KfiA or PmHS2 to form GlcNTFA-α-1-4-GlcA-biotin disaccharide using the corresponding sugar nucleotide donors. To this end, GlcA-biotin conjugate 5 was chemically synthesized.18 As outlined in Scheme 1, the synthesis started from glucose pentaacetate 1. Lewis acid-mediated glycosylation of 1 with 3-azidopropanol gave azido-glucoside 2 in 60% yield, which then went through Zemplén deacetylation, selective TEMPO-mediated oxidation of primary hydroxyl group of glucose, azide reduction by NaBH4 in the presence of palladium catalyst, and final conjugation with activated biotin succinimide ester (Biotin-OSu) 4. After purification by silica gel column chromatography, GlcA-biotin 5 was provided with 40–45% overall yield (over 4 steps) from compound 2. Remarkably, the synthesis of GlcA-biotin 5 starting from compound 1 required only two silica gel column chromatographic purification steps, dramatically facilitating its synthetic efficacy. In fact, conjugate 5 was easily synthesized in grams scale in one batch.
Scheme 1.
Synthesis of GlcA-biotin conjugate 5
With the substrate 5 in hand, KfiA from E. coli K5 and PmHS2 from P. multocida P-1059 were overexpressed by E. coli BL21 (DE3) expression system and purified, respectively. Specifically, the KfiA gene from E. coli K5 was cloned and expressed as a fusion enzyme with maltose-binding protein (MBP) at the N-terminal and histidine6X-tag at the C-terminal. PmHS2 was cloned and expressed as an N-terminal histidine6X-tagged recombinant protein. In this work, the bi-functional PmHS2 was chosen as the sole chain elongation enzyme owing to its easier over-expression compared to KfiA in E. coli BL21 (DE3) expression system. Initially, one-pot three-enzyme system was used to convert GlcA-biotin conjugate 5 to disaccharide 6. The reaction mixture (pH 7.0) containing GlcA-β-biotin conjugate 5 (1.0 eq.), GlcNTFA (1.2 eq.), ATP (1.2 eq.), UTP (1.2 eq.), MgCl2, N-acetyl-hexosamine 1-kinase (NahK)19, N-acetylglucosamine-1-phosphate uridyltransferase (PmGlmU) 20 and PmHS2 were incubated in one-pot fashion at 37 °C for 2 days to afford disaccharide 6 in 90% yield (Schem 2, step 1). It’s worth mentioning that UDP-GlcNTFA donor is in situ generated using NahK and PmGlmU in the presence of GlcNTFA, ATP and UTP. The use of UDP-GlcNTFA rather than UDP-GlcNAc as donor substrate is based on the consideration of later stage N-sulfation for future preparation of heparin-like oligosaccharide, since trifluoroacetyl (TFA) group could be removed under mild basic conditions. Subsequently, enzymatic glycosylation reaction of 6 (1.0 eq.) with UDP-GlcA (1.0 eq.) using bi-functional PmHS2 in the presence of MnCl2 and Tris-HCl buffer (pH 7.0) at 37°C overnight to afford trisaccharide 7 in 88% yield (Scheme 2, step 2). Encouraged by the results of synthesis of disaccharide 6, we attempted to integrate UDP-GlcNTFA preparation with chain elongation in a one-pot three-enzyme system using simple GlcNTFA as starting substrate to synthesize tetrasaccharide 8 (repeat step 1). As anticipated, the overall reaction rate was dramatically accelerated and the reaction completed within 24 h, providing far more straightforward stoichiometric control of the reaction than the stepwise version. As depicted in Scheme 2, the whole chain elongation was performed by repeating step 1 and step 2 alternatively. The progress of each chain elongation step was conveniently monitored by thin layer chromatography (TLC), in which the reactants and products could well be resolved using the combination of ethyl acetate, methanol, water and acetic acid as developing solvent system (typical TLC in Supporting Information S5). The corresponding products were purified by Biogel@ P2 size-exclusion chromatographic column. Interestingly, the enzymatic reaction normally proceeded at a faster rate as the chain became longer, indicating the preference and dependence of PmHS2’s activity on the acceptor length. In addition, the oligosaccharide acceptors in each elongation step could be cleanly and completely converted by PmHS2 to form the desired products. In last step of hexasaccharide preparation, non-reducing end terminal was capped with GlcNAc residue instead of GlcNTFA to provide a heparosan backbone potentially compatible for future selective epimerization of internal GlcA residue flanked by two GlcNTFA units into corresponding iduronic acid (IdoA) isoform after N-sulfation.12g Remarkably, by capitalizing on our improved one-pot three-enzyme approach and convenient monitoring of the reaction progress by a quick TLC analysis, one run of the whole synthetic scheme starting from 60 mg of GlcA-biotin conjugate 5 yielded 65 mg of the final biotinylated hexasaccharide 10 with 33% overall yield (averaging 80% yield of each step) in two weeks.
Scheme 2.
Sequential one-pot multi-enzyme synthesis of biotinylated heparosan hexasaccharide 10
In summary, we have developed a streamlined, preparative scale and HPLC-free chemoenzymatic synthesis of biotinylated heparosan hexasaccharide. The synthesis features integration of in situ UDP-GlcNTFA generation with heparin backbone elongation in one-pot fashion. As a result of such technical reformation, the overall synthetic efficacy is significantly increased. The flexible core hexasaccharide backbone allows multiple later-stage enzymatic modifications to furnish a library of HS as highly useful molecular probes to dissect the heparin/heparin binding proteins’ interactions in biological settings, owing to that biotin moiety could be fished out by streptavidin. Efforts towards the final construction of fondaparinux-like biotin conjugate, which is formed via consecutive modifications of 10 by N-sulfation, selective epimerization of internal GlcA into iduronic acid (IdoA) and 2-O-, 6-O-, 3-O-sulfations, are currently underway.
Supplementary Material
Acknowledgments
We thank Dr. Zhenming Du and Prof. Markus W. Germann for assistance in NMR characterization of the synthesized compounds. This work was supported by NIH grant R01 GM085267.
Footnotes
Electronic Supplementary Information (ESI) is available, including detailed experimental procedure and characterization of compounds. See DOI: 10.1039/c000000x/
Contributor Information
Peng George Wang, Email: pwang11@gsu.edu.
Tiehai Li, Email: tli8@gsu.edu.
Notes and references
- 1.(a) Esko JD, Kimata K, Lindahl U. Essentials of Glycobiology. (2) 2009;229 [Google Scholar]; (b) Esko JD, Lindahl U. J Clin Invest. 2001;108:169. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Annu Rev Biochem. 1999;68:729. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]; (b) Tumova S, Woods A, Couchman JR. Int J Biochem Cell Biol. 2000;32:269. doi: 10.1016/s1357-2725(99)00116-8. [DOI] [PubMed] [Google Scholar]; (c) Esko JD, Selleck SB. Annu Rev Biochem. 2002;71:435. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]; (d) Lindahl U, Kusche-Gullberg M, Kjellen L. J Biol Chem. 1998;273:24979. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 3.Nutescu EA, Shapiro NL, Chevalier A, Amin AN. Clev Clin J Med. 2005;72(Suppl 1):S2. doi: 10.3949/ccjm.72.suppl_1.s2. [DOI] [PubMed] [Google Scholar]
- 4.(a) Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, Capila I, Lansing JC, Guglieri S, Fraser B, Al-Hakim A, Gunay NS, Zhang Z, Robinson L, Buhse L, Nasr M, Woodcock J, Langer R, Venkataraman G, Linhardt RJ, Casu B, Torri G, Sasisekharan R. Nat Biotechnol. 2008;26:669. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Driguez PA, Potier P, Trouilleux P. Nat Prod Rep. 2014;31:980. doi: 10.1039/c4np00012a. [DOI] [PubMed] [Google Scholar]
- 5.(a) Walenga JM, Jeske WP, Samama MM, Frapaise FX, Bick RL, Fareed J. Expert Opin Inv Drug. 2002;11:397. doi: 10.1517/13543784.11.3.397. [DOI] [PubMed] [Google Scholar]; (b) Turpie AGG. Expert Opin Pharmaco. 2004;5:1373. doi: 10.1517/14656566.5.6.1373. [DOI] [PubMed] [Google Scholar]
- 6.(a) Buller HR, Cohen AT, Lensing AWA, Prins MH, Schulman S, Lassen MR, van Amsterdam RGM. J Throm Haemost. 2004;2:47. [Google Scholar]; (b) Bianchini EP, Fazavana J, Picard V, Borgel D. Blood. 2011;117:2054. doi: 10.1182/blood-2010-06-288522. [DOI] [PubMed] [Google Scholar]; (c) Flight MH. Nat Rev Drug Discov. 2009;8:934. doi: 10.1038/nrd3057. [DOI] [PubMed] [Google Scholar]
- 7.(a) Savi P, Herault JP, Duchaussoy P, Millet L, Schaeffer P, Petitou M, Bono F, Herbert JM. J Throm Haemost. 2008;6:1697. doi: 10.1111/j.1538-7836.2008.03089.x. [DOI] [PubMed] [Google Scholar]; (b) Petitou M, Nancy-Portebois V, Dubreucq G, Motte V, Meuleman D, de Kort M, van Boeckel CA, Vogel GM, Wisse JA. Thromb Haemost. 2009;102:804. doi: 10.1160/TH09-01-0063. [DOI] [PubMed] [Google Scholar]
- 8.Kang YS, Saito Y, Pardridge WM. J Drug target. 1995;3:159. doi: 10.3109/10611869509059215. [DOI] [PubMed] [Google Scholar]
- 9.(a) Sinaÿ P, Jacquinet JC, Petitou M, Duchaussoy P, Lederman I, Choay J, Torri G. Carbohydr Res. 1984;132:C5. doi: 10.1016/s0008-6215(00)90633-5. [DOI] [PubMed] [Google Scholar]; (b) Petitou M, Duchaussoy P, Lederman I, Choay J, Jacquinet JC, Sinaÿ P, Torri G. Carbohydr Res. 1987;167:67. doi: 10.1016/0008-6215(87)80268-9. [DOI] [PubMed] [Google Scholar]; (c) Grootenhuis PDJ, Van Boeckel CAA. J Am Chem Soc. 1991;113:2743. [Google Scholar]; (d) Vanboeckel CAA, Petitou M. Angew Chem Int Edit. 1993;32:1671. [Google Scholar]; (e) Petitou M, Duchaussoy P, Driguez PA, Jaurand G, Herault JP, Lormeau JC, van Boeckel CAA, Herbert JM. Angew Chem Int Edit. 1998;37:3009. doi: 10.1002/(SICI)1521-3773(19981116)37:21<3009::AID-ANIE3009>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]; (f) Petitou M, van Boeckel CAA. Angew Chem Int Edit. 2004;43:3118. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]; (g) Zhou F, Lin J, Chen F, Yu B. Carbohydr Res. 2006;341:1619. doi: 10.1016/j.carres.2006.02.020. [DOI] [PubMed] [Google Scholar]; (h) Tatai J, Fugedi P. Tetrahedron. 2008;64:9865. [Google Scholar]; (i) Lee JC, Lu XA, Kulkarni SS, Wen YS, Hung SC. J Am Chem Soc. 2004;126:476. doi: 10.1021/ja038244h. [DOI] [PubMed] [Google Scholar]
- 10.(a) Karst NA, Linhardt RJ. Curr Med Chem. 2003;10:1993. doi: 10.2174/0929867033456891. [DOI] [PubMed] [Google Scholar]; (b) Dulaney SB, Huang X. Adv Carbohydr Chem Biochem. 2012;67:95. doi: 10.1016/B978-0-12-396527-1.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Orgueira HA, Bartolozzi A, Schell P, Seeberger PH. Angew Chem Int Edit. 2002;41:2128. [PubMed] [Google Scholar]; (b) Codee JDC, Stubba B, Schiattarella M, Overkleeft HS, van Boeckel CAA, van Boom JH, van der Marel GA. J Am Chem Soc. 2005;127:3767. doi: 10.1021/ja045613g. [DOI] [PubMed] [Google Scholar]; (c) Polat T, Wong CH. J Am Chem Soc. 2007;129:12795. doi: 10.1021/ja073098r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Geng YQ, Zhang LH, Ye XS. Chem Commun. 2008:597. doi: 10.1039/b712591g. [DOI] [PubMed] [Google Scholar]; (e) Manabe S, Ishii K, Ito Y. J Am Chem Soc. 2006;128:10666. doi: 10.1021/ja062531e. [DOI] [PubMed] [Google Scholar]; (f) Park J, Kawatkar S, Kim JH, Boons GJ. Org Lett. 2007;9:1959. doi: 10.1021/ol070513b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Arungundram S, Al-Mafraji K, Asong J, Leach FE, Amster IJ, Venot A, Turnbull JE, Boons GJ. J Am Chem Soc. 2009;131:17394. doi: 10.1021/ja907358k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Wang Z, Xu YM, Yang B, Tiruchinapally G, Sun B, Liu RP, Dulaney S, Liu JA, Huang XF. Chem-Eur J. 2010;16:8365. doi: 10.1002/chem.201000987. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Lin F, Lian GY, Zhou Y. Carbohydr Res. 2013;371:32. doi: 10.1016/j.carres.2013.01.003. [DOI] [PubMed] [Google Scholar]; (j) Zulueta MML, Lin SY, Lin YT, Huang CJ, Wang CC, Ku CC, Shi ZH, Chyan CL, Irene D, Lim LH, Tsai TI, Hu YP, Arco SD, Wong CH, Hung SC. J Am Chem Soc. 2012;134:8988. doi: 10.1021/ja302640p. [DOI] [PubMed] [Google Scholar]; (k) Chang CH, Lico LS, Huang TY, Lin SY, Chang CL, Arco SD, Hung SC. Angew Chem Int Edit. 2014;53:9876. doi: 10.1002/anie.201404154. [DOI] [PubMed] [Google Scholar]; (l) Li TH, Ye H, Cao XF, Wang JJ, Liu YH, Zhou LF, Liu Q, Wang WJ, Shen J, Zhao W, Wang P. ChemMedChem. 2014;9:1071. doi: 10.1002/cmdc.201400019. [DOI] [PubMed] [Google Scholar]
- 12.(a) Kuberan B, Beeler DL, Lech M, Wu ZL, Rosenberg RD. J Biol Chem. 2003;278:52613. doi: 10.1074/jbc.M305029200. [DOI] [PubMed] [Google Scholar]; (b) Kuberan B, Lech MZ, Beeler DL, Wu ZL, Rosenberg RD. Nat Biotechnol. 2003;21:1343. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]; (c) Kuberan B, Beeler DL, Lawrence R, Lech M, Rosenberg RD. J Am Chem Soc. 2003;125:12424. doi: 10.1021/ja036737g. [DOI] [PubMed] [Google Scholar]; (d) Xu YM, Cai C, Chandarajoti K, Hsieh PH, Li LY, Pham TQ, Sparkenbaugh EM, Sheng JZ, Key NS, Pawlinski R, Harris EN, Linhardt RJ, Liu J. Nat Chem Biol. 2014;10:248. doi: 10.1038/nchembio.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Liu J, Linhardt RJ. Nat Prod Rep. 2014;31:1676. doi: 10.1039/c4np00076e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Zhou XX, O’Leary TR, Xu YM, Sheng JZ, Liu J. Biocatal Biotransfor. 2012;30:296. [Google Scholar]; (g) Xu Y, Masuko S, Takieddin M, Xu H, Liu R, Jing J, Mousa SA, Linhardt RJ, Liu J. Science. 2011;334:498. doi: 10.1126/science.1207478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rillahan CD, Paulson JC. Ann Rev Biochem. 2011;80:797. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Samama MM, Guinet C. Clin Chem Lab Med. 2011;49:761. [Google Scholar]; (b) Mannucci PM, Franchini M. Ann Med (London, U K) 2011;43:116. doi: 10.3109/07853890.2010.539250. [DOI] [PubMed] [Google Scholar]; (c) Levi M, Eerenberg E, Kamphuisen PW. J Throm Haemost. 2011;9:1705. doi: 10.1111/j.1538-7836.2011.04432.x. [DOI] [PubMed] [Google Scholar]; (d) Kazmi RS, Lwaleed BA. Br J Clin Pharmacol. 2011;72:593. doi: 10.1111/j.1365-2125.2011.04060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Brenner B, Hoffman R. Blood Rev. 2011;25:215. doi: 10.1016/j.blre.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Hodson N, Griffiths G, Cook N, Pourhossein M, Gottfridson E, Lind T, Lidholt K, Roberts IS. J Biol Chem. 2000;275:27311. doi: 10.1074/jbc.M004426200. [DOI] [PubMed] [Google Scholar]
- 16.(a) DeAngelis PL, White CL. J Biol Chem. 2002;277:7209. doi: 10.1074/jbc.M112130200. [DOI] [PubMed] [Google Scholar]; (b) J Bacteriol. 2004;186:8529. doi: 10.1128/JB.186.24.8529-8532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chavaroche AAE, Springer J, Kooy F, Boeriu C, Eggink G. Appl Microbiol Biotechnol. 2010;85:1881. doi: 10.1007/s00253-009-2214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefeber DJ, Kamerling JP, Vliegenthart JFG. Chem-Eur J. 2001;7:4411. doi: 10.1002/1521-3765(20011015)7:20<4411::aid-chem4411>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 19.Cai L, Guan W, Kitaoka M, Shen J, Xia C, Chen W, Wang PG. Chem Commun. 2009:2944. doi: 10.1039/b904853g. [DOI] [PubMed] [Google Scholar]
- 20.(a) Guan W, Cai L, Fang J, Wu B, George Wang P. Chem Commun. 2009:6976. doi: 10.1039/b917573c. [DOI] [PubMed] [Google Scholar]; (b) Zhai Y, Liang M, Fang J, Wang X, Guan W, Liu XW, Wang P, Wang F. Biotechnol Lett. 2012;34:1321. doi: 10.1007/s10529-012-0910-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.