Abstract
Data-independent acquisition (DIA) strategies provide a sensitive and reproducible alternative to data-dependent acquisition (DDA) methods for large-scale quantitative proteomic analyses. Unfortunately, DIA methods suffer from incompatibility with common multiplexed quantification methods, specifically stable isotope labeling approaches such as isobaric tags and stable isotope labeling of amino acids in cell culture (SILAC). Here we expand the use of neutron-encoded (NeuCode) SILAC to DIA applications (NeuCoDIA), producing a strategy that enables multiplexing within DIA scans without further convoluting the already complex MS2 spectra. We demonstrate duplex NeuCoDIA analysis of both mixed-ratio (1:1 and 10:1) yeast and mouse embryo myogenesis proteomes. Analysis of the mixed-ratio yeast samples revealed the strong accuracy and precision of our NeuCoDIA method, both of which were comparable to our established MS1-based quantification approach. NeuCoDIA also uncovered the dynamic protein changes that occur during myogenic differentiation, demonstrating the feasibility of this methodology for biological applications. We consequently establish DIA quantification of NeuCode SILAC as a useful and practical alternative to DDA-based approaches.
SWATH mass spectrometry (SWATH-MS), and other data-independent acquisition (DIA) approaches, have become increasingly popular alternatives to data-dependent acquisition (DDA) methods for large-scale proteomic studies. Data-dependent strategies rely on survey scan (MS1) acquisition to select precursors for subsequent MS/MS analysis. Data-independent approaches, however, do not require the procurement of MS1 scans; instead, discrete m/z ranges of peptide precursors are analyzed through the repeated sampling of successive isolation windows, or swaths, over the course of chromatographic elution.1–5 The sequential acquisition of MS2 spectra, typically over an 800 to 1000 Th precursor mass range using isolation windows 10–25 Th wide, is theoretically advantageous as it enables the generation and detection of fragment ions from all precursors present within the experimental m/z boundaries. This both eliminates the requirement of MS1 scan detectability/selection for precursor MS/MS analysis and permits peptide quantification from MS2 scans (which inherently have less chemical noise); in some cases, this affords DIA methods greater reproducibility and sensitivity than typical MS1-based techniques.4–6 Although DIA strategies do not achieve sensitivity comparable to targeted methods, such as selected or parallel reaction monitoring (SRM/PRM),7 they can achieve a similar quantitative dynamic range and sample a larger number of peptides in a single experiment.4,5 For these reasons, existing DIA-MS workflows are well-suited to proteomic applications in which accurate quantitation and a large number of reproducible identifications are required.
One shortcoming of DIA has been its lack of compatibility with common quantification methods, especially stable isotope labeling approaches such as isobaric tagging and stable isotope labeling of amino acids in cell culture (SILAC). Isobaric tagging offers multiplexed analyses; however, abundance information is encoded in the MS/MS scan. For DIA methods that coisolate and cofragment multiple precursors, the generated reporter ions are convoluted and uninformative. Typical SILAC methods produce unique fragment ions upon dissociation of the duplex or triplex precursor clusters; however, the ~4 Da mass spacing of these clusters not only significantly convolutes MS2 spectra but can also spread the clusters across multiple DIA windows, preventing their coisolation and subsequent quantification.
We have recently described a method of neutron-encoded (NeuCode) SILAC in which isotopologues of lysine metabolically incorporate subtle mass differences (≤36 mDa) in two or more complex biological samples.8 Using typical DDA shotgun analyses, we have demonstrated the ability to perform multiplexed quantification (up to 18-plex) with NeuCode-labeled samples through the acquisition of high-resolution (R ≥ 100 000) MS1 scans.9 The characteristic NeuCode signatures, however, are also present in all C-terminal fragment ions produced upon dissociation of always coisolated NeuCo-delabeled precursors. These NeuCode signatures are easily detected by high-resolution MS/MS analysis but, due to their minuscule mass difference, do not clutter the MS/MS spectra with added complexity.10,11 Here we report the use of NeuCode SILAC labels for multiplexed quantification within a DIA strategy (NeuCoDIA), allowing the coupling of isotope-based quantification with the emergent DIA approach.
MATERIALS AND METHODS
Sample Preparation
Mixed-ratio yeast samples were prepared by mixing 13C6 15N2 (+8.0412 Da)-labeled and D8 (+8.0502 Da)-labeled yeast peptides in 1:1 or 10:1 ratios by mass. The mouse myoblast/myotube differentiation samples were prepared by mixing biological replicates of 6C13/2N15 (+8.0412 Da)-labeled myotube peptides and 8D (+8.0502 Da)-labeled myoblast peptides in a 1:1 ratio, by mass.
Liquid Chromatography–Mass Spectrometry
All experiments were performed using an EASY-nLC system (Thermo Fisher Scientific, San Jose, CA) coupled to an Orbitrap Fusion (Q-OT-qIT) mass spectrometer (Thermo Fisher Scientific, San Jose, CA).
MS1 NeuCode Quantification
MS1 Quant instrument methods consisted of a survey scan (450–950Th) analyzed in the Orbitrap at a resolution of 240k (at 200m/z). Tandem MS scans were collected at top speed mode with 5 s cycles. Precursor peptides were isolated, subjected to HCD fragmentation (NCE 30), and analyzed in the ion trap at rapid scan speed.
NeuCoDIA Quantification
For this study, DDA peptide identification and NeuCoDIA quantification were combined into a single instrument method. Each experiment consisted of a survey scan (which covered all mass ranges) analyzed in the Orbitrap at a resolution of 60k (at 200m/z). This survey scan was followed by tandem MS scans collected at top speed mode with 1 s cycles that only selected precursors from the 100 Th mass range of interest. Precursor peptides were isolated, subjected to HCD fragmentation (NCE 30), and analyzed in the ion trap at rapid scan speed. These scans were followed by tandem MS scans which sequentially isolated, fragmented (HCD, NCE 30), and analyzed (Orbitrap, 120k resolution at 200m/z) four identically sized m/z windows within their designated mass range (e.g., the 450–550 Th analysis will conduct MS/MS analyses on the 449–476 Th, 474–501 Th, 499–526 Th, and 524–551 Th ranges).
Data Analysis
Data was processed using the in-house software suite COMPASS.12 MS1 NeuCode quantification was performed as described previously.8 NeuCoDIA quantification was achieved using in-house software (Supporting Information). For the evaluation of our myogenesis study, enzymes belonging to major catabolic pathways were manually curated from Gene Ontology, KEGG, and BioCyc annotations.
RESULTS AND DISCUSSION
NeuCoDIA
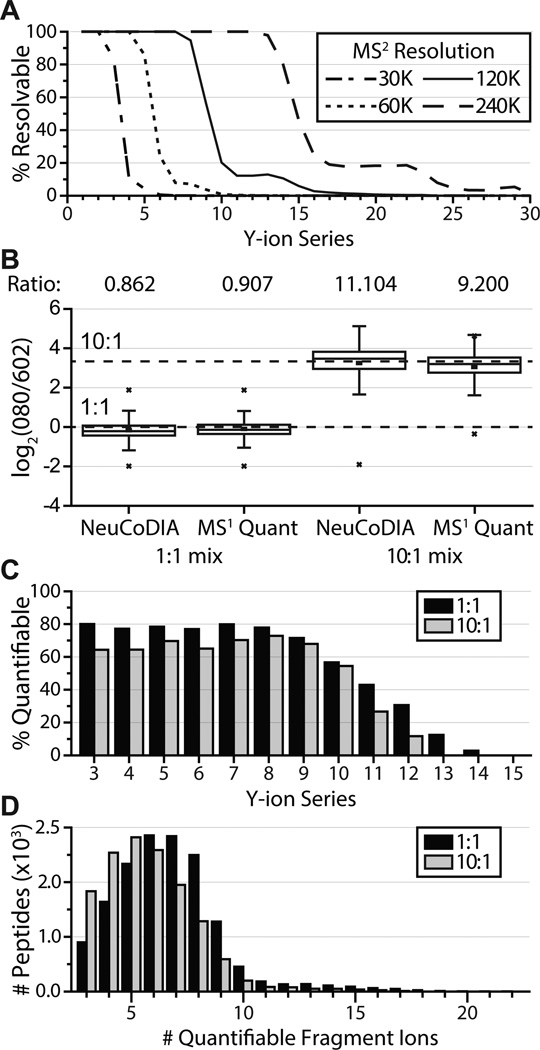
The ultrahigh resolution provided by Fourier transform mass spectrometry (FTMS) instruments, such as the Orbitrap or Fourier transform ion cyclotron resonance (FTICR),13–15 enables the recognition of closely spaced (36 mDa) duplex NeuCode peaks, as has been demonstrated in previous studies.7,11,12 To assess the feasibility of duplex NeuCode quantification using product ions, we first calculated the percentage of C-terminal fragments that could theoretically be resolved using an orbitrap analyzer at resolutions of 30k, 60k, 120k, and 240k at m/z 200 (Figure 1a). This calculation was generated using a list of 29 012 NeuCode-labeled (13C6 15N2-heavy lysine) LysC-digested yeast peptides identified in a separate DDA analysis. At a resolving power of 240k, the NeuCode doublet can theoretically be resolved for all fragment ions containing less than 13 residues; however, the time cost for this long transient (~512 ms)16 is detrimental to a DIA study, where it is desirable to acquire as many data points as possible for quantification across extracted ion chromatograms (XICs). A resolution of 120k (~256 ms transient)16 is a suitable compromise, as it enables the theoretical resolution of many fragment ions (at least all y-type ions that are 8 residues or shorter) in half the analysis time.
Figure 1.
NeuCoDIA utilizes fragment ion doublets to attain high quantitative accuracy and precision. (A) Theoretical calculations depict the percentage of NeuCode doublet (36 mDa) fragment ions that can be resolved at resolutions of 30k, 60k, 120k, and 240k (full width at 10% maximum peak height). Calculations based off of a target list (n = 29 012) generated from DDA analysis of LysC yeast peptides. (B) Boxplots show the measured (box and whiskers) and true (dashed lines) peptide ratios for NeuCoDIA and DDA analyses at mixing ratios of 1:1 and 10:1 (n = 12 986, 13 393, 11 803, 12 112, left to right). Boxplots indicate the median (stripe), 25th–75th percentile (interquartile range, box), 1.5 times the interquartile range (whiskers), the 1st and 99th percentiles (x symbols), and the minimum/maximum values (lines, when applicable). Median ratios are provided above the boxplots. (C) Percentage of y-type ions that were observed and quantified by NeuCoDIA analysis given each y-type ion fragment that could theoretically be produced by the peptides quantified in the 1:1 and 10:1 mixtures (n = 12 986 and 11 803, respectively). (D) Number of fragment ions utilized for quantification given all peptides quantified in the 1:1 and 10:1 NeuCoDIA analyses.
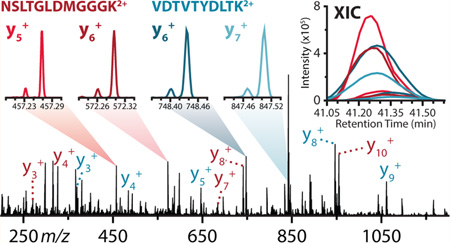
To test this theory, we conducted a NeuCode DIA (NeuCoDIA) analysis comprising a set of individual experiments, each covering a 100 Th precursor m/z window (i.e., 350–450 Th, …, 750–850 Th). Each experiment rotated between the isolation and subsequent fragmentation of four successive 27 Th windows, such that MS2 analysis (120k resolution) was performed across the entire 100 Th precursor m/z range (e.g., 299–326 Th, 324–351 Th, 349–376 Th, 374–401 Th). The 2 Da overlap between windows was implemented to ensure isolation and fragmentation of all precursor species and their isotopes, as was applied in Gillet et al.4 To test the quality of quantification from the data-independent scans, an independent set of confirmed peptides present in the sample was generated by embedding an MS1 scan, along with a series of DDA-triggered tandem MS scans, between each set of NeuCoDIA scans (Supplementary Figure 1 in the Supporting Information).
Quantitative data were extracted from these experiments with in-house software, which utilized in silico fragmentation to calculate and facilitate the generation of XICs for all theoretically quantifiable fragment ion pairs (i.e., all fragments containing a lysine residue) from each detected peptide. For the sake of accuracy and specificity, only fragment ions containing three or more residues were considered for quantification (Supplementary Figure 2 in the Supporting Information). Following the alignment and filtering of all fragment ion XICs attained for a unique peptide target, the total ion current (TIC) was calculated from each product ion XIC; quantification was then performed by summing these TIC values for each NeuCode channel. Peptides were only considered quantifiable if they had three or more quantifiable fragment ions.
Validation Using Mixed-Ratio Yeast
To test the efficacy of NeuCoDIA, we employed defined, mixed-ratio (1:1 and 10:1) samples of NeuCode-labeled yeast. Yeast cells were grown in media supplemented with either 13C6 15N2-heavy lysine (+8.0142 Da) or D8-heavy lysine (+8.0502 Da), such that the isotopologues became completely incorporated into the respective proteins. Subsequent enzymatic digestion with LysC ensured that all peptides contained at least one lysine residue, producing a mass difference of at least 36 mDa between otherwise identical peptides from the two yeast cultures.
NeuCoDIA analysis was able to quantify 12 986 (95.4%) and 11 803 (85.9%) of the peptides identified within the experiments, recording median ratios of 0.862 and 11.104 for the 1:1 and 10:1 mixtures, respectively (Figure 1b, Supplementary Figure 3 in the Supporting Information). Despite the complex MS2 spectra generated upon fragmentation of all precursors within the 27 Th isolation windows, the NeuCoDIA strategy permitted identification and quantification of 60%–80% of all theoretically quantifiable y-type ions containing at least 9 residues (Figure 1c). This enabled quantification from a median of 7 and 6 fragment ions per peptide target for the 1:1 and 10:1 yeast mixtures, respectively (Figure 1d).
We compared the precision and accuracy of MS2-based NeuCoDIA with that achieved by MS1-based DDA. For the DDA analysis of NeuCode samples, MS1 scans were acquired at 240k resolution in the Orbitrap, followed by tandem MS scans analyzed at rapid speed in the ion trap. With DDA quantification, 13 393 (85.6%) and 12 112 (88.4%) of the yeast peptides identified were quantified at median ratios of 0.907 and 9.200 for the 1:1 and 10:1 mixtures, respectively (Figure 1b and Supplementary Figure 3 in the Supporting Information). This is comparable to the ratios recorded for NeuCoDIA and, together with the comparable standard deviations reported for the two methods (0.679 vs 0.629 and 1.39 vs 0.914, log2, for the NeuCoDIA vs NeuCode 1:1 and 10:1 analyses, respectively; see Supplementary Figure 3 in the Supporting Information for more details), these results highlight the consistency between DIA and DDA quantification of NeuCode-labeled samples. Although NeuCoDIA offers no distinct advantage over DDA in terms of quantitative accuracy, the method’s ability to perform peptide quantification independent of precursor sampling and identification allows NeuCoDIA to quantify peptides with greater reproducibility than MS1-based DDA (Supplementary Figure 4 in the Supporting Information).
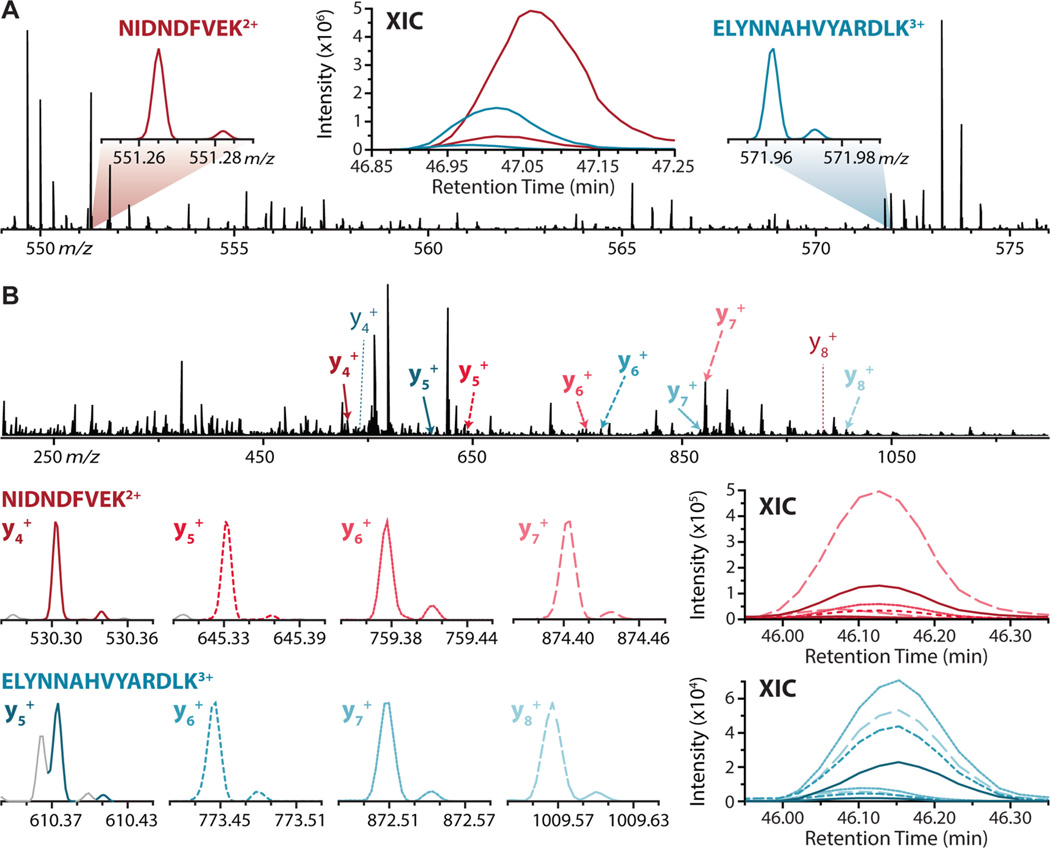
An example of DDA and DIA NeuCode quantification is depicted in Figure 2. This figure highlights two peptide precursors (NIDNDFVEK2+ and ELYNNAHVYARDLK2+) that fall within the same 27 Th bin (549–576 Th) and coelute over the same 24 s retention time window. Figure 2a shows the MS1 spectra (averaged over the 24 s elution window) and XICs resulting from DDA analysis of the two precursors. Ratios of 9.99 and 6.45 were recorded for each, respectively.
Figure 2.
NeuCoDIA (MS2 quantification) generates fragment ion extracted ion chromatograms (XICs) that are comparable to those attained for the same parent ions using data-dependent acquisition (MS1 quantification): (A) MS1 spectra (240k resolution) attained during data-dependent analysis of the 10:1 yeast mixed-ratio sample and averaged over the 24 s that the peptides NIDNDFVEK2+ and ELYNNAHVYARDLK3+ were eluting. XICs reflect MS1 precursor ion intensities for NeuCode pairs over time (5-point boxcar average). (B) MS2 spectra (120k resolution, 27 Th isolation window centered on m/z 562.5) attained during NeuCoDIA analysis of the 10:1 yeast mixed-ratio sample and averaged over the 24 s that the precursors in part a (NIDNDFVEK2+ and ELYNNAHVYARDLK3+) were eluting. XICs reflect MS2 fragment ion intensities for fragment ion pairs over time (5-point boxcar average).
Figure 2b illustrates the equivalent NeuCoDIA analysis of the two peptides in Figure 2a. MS2 scans resulting from the isolation and fragmentation of the 549–576 Th window were averaged over the comparable 24 s retention time range during which the precursors of interest were eluting to generate the MS2 spectrum shown. Not only are the NeuCode doublets resolved for each fragment ion found, but quantification is consistent between fragments generated from the same precursor. Ratios of 11.43 and 11.48 were obtained for the respective peptides, highlighting the sensitivity of NeuCoDIA for low-abundance precursors.
Validation Using C2C12 Myogenesis Sample
To test the utility of NeuCoDIA in the context of complex biological sample analysis, we next evaluated the proteomic changes that occur during the differentiation of cultured C2C12 mouse myoblasts into myotubes. A major change that occurs during the differentiation of proliferative progenitor cells, such as myoblasts, to quiescent postmitotic cells, such as myotubes, is a widespread reprogramming of cellular metabolism.17 Normal proliferating cells (as well as cancer cells) rely heavily on the consumption of glucose (aerobic glycolysis) and glutamine (glutaminolysis) for energy generation and for the facilitation of cell growth and division via anabolic pathways (such as lipid, nucleic acid, and protein biosynthesis). Conversely, differentiated cells rely increasingly on the catabolism of fatty acids (through beta-oxidation, the tricarboxylic acid (TCA) cycle, and oxidative phosphorylation (OxPhos)) to generate energy for essential cellular processes.18,19
Mouse-derived C2C12 cells were grown in media supplemented with either 13C6 15N2-heavy lysine (+8.0142 Da) or D8-heavy lysine (+8.0502 Da). Myoblast cells labeled with D8-heavy lysine were harvested upon full incorporation of the NeuCode label, while those labeled with 13C6 15N2-heavy lysine were allowed to differentiate into myotubes over 5 additional days. The labeled myoblasts and myotubes were mixed in a 1:1 ratio by mass. Myogenesis samples were prepared and analyzed in biological duplicate using the NeuCoDIA method described above with the following precursor ranges: 300–400 Th, 400–500 Th, 500–600 Th, 600–700 Th, 700–800 Th, 800–900 Th, 900–1 000 Th, 1 000–1 400 Th. Note that the final experiment in this data set covered a 400 Th window and isolated/fragmented four 102 Th precursor m/z windows in its DIA scan sequence. Overall, NeuCoDIA analysis quantified 17 280 (94.0%) and 17 176 (93.0%) of the identified myogenic peptides, corresponding to 3 884 (96.3%) and 3 804 (96.5%) proteins, respectively (Supplemental Figure 5 in the Supporting Information).
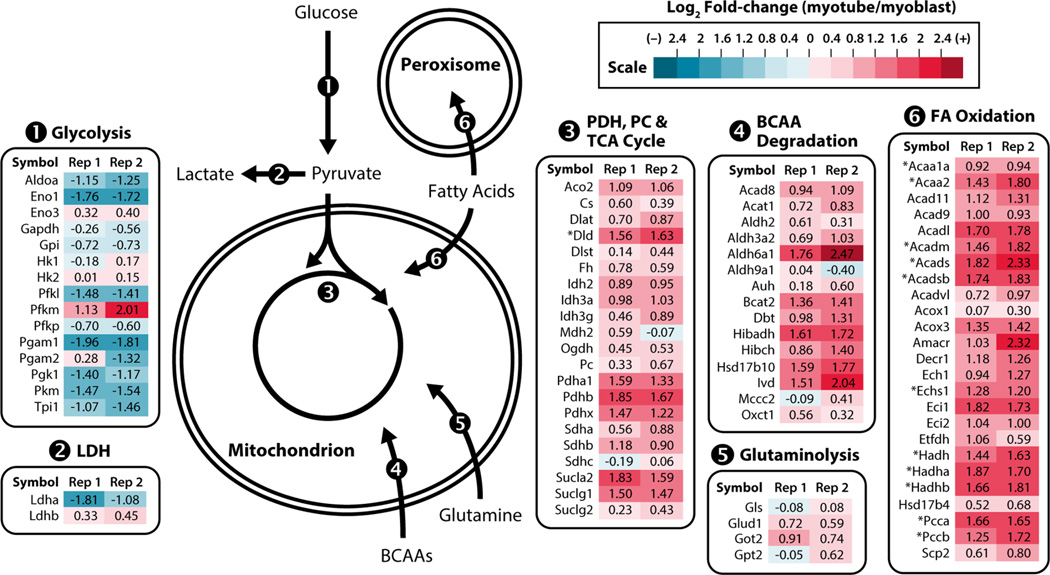
Consistent with a metabolic switch from aerobic glycolysis to mitochondrial respiration during differentiation, our analysis revealed an overall decrease in the abundance of enzymes involved in glycolysis and lactate production, coincident with an increase in enzymes of the TCA cycle and fatty acid oxidation (Figure 3). We have previously shown that myoblast differentiation also results in an overall increase in the levels of OxPhos proteins20 and, taken together, these protein abundance changes suggest that the rewiring of cellular metabolism during myogenic differentiation is, in part, due to the regulation of enzyme expression. Additionally, we found an increase in the abundance of enzymes involved in the catabolism of the branched chain amino acids (BCAAs) leucine, valine, and isoleucine (Figure 3). Increases in BCAA and fatty acid oxidation have also been observed during both adipogenesis21 and in muscle in response to exercise.22 Increased BCAA degradation could also be important for myotube metabolism, as this pathway both contributes to energy production through generation of NADH and FADH2 and provides TCA cycle intermediates (anaplerosis).23 Specifically, the succinyl-CoA generated from the oxidation of valine and isoleucine could ensure an adequate supply of oxaloacetate for the acetyl-CoA generated from fatty acid oxidation to gain entrance to the TCA cycle.
Figure 3.
NeuCoDIA reveals coordinated changes in protein expression of major cellular catabolic pathways during myogenic differentiation. Protein expression fold changes from two biological replicates are displayed. * indicates enzymes that are/are predicted to be involved in BCAA degradation (LDH, lactate dehydrogenase; PDC, pyruvate dehydrogenase complex; PC, pyruvate carboxylase; TCA, tricarboxylic acid; FA, fatty acid; BCAA, branched chain amino acid).
CONCLUSIONS
We have demonstrated how NeuCode SILAC enables multiplexed quantification from DIA methods. The effective nature of this technique is supported both by the accuracy and precision attained during NeuCoDIA analysis of mixed-ratio yeast samples and by the dynamic peptide changes revealed through NeuCoDIA analysis of myogenesis samples. The reduced noise level and charge-state associated with MS2 peptide fragments (as opposed to MS1 peptide precursors) lowers the resolution requirements for NeuCode quantification, enabling the observation and resolution of isotopologues that cannot be readily distinguished in MS1 scans. This opens up the possibility of moving to higher plexes (such as 4-plex and 6-plex) without significantly increasing the spectral complexity of the already complex SWATH-type MS2 scan. Although we only considered DIA quantification in this study, we believe that the identification of peptides from DIA analyses will not be deterred by NeuCode multiplexing; if anything, the multiplexed C-terminal ion peaks intrinsic to NeuCode may even facilitate identification from SWATH-MS analyses.10 We therefore establish that the utility of NeuCode is not limited to DDA applications but shows promise for the extension of isotope-based quantification to DIA strategies as well.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health Grant GM080148 to J.J.C. C.E.M. was supported by an NLM training grant to the Computation and Informatics in Biology and Medicine Training Program (Grant NLM T15LM007359). J.W.R. acknowledges support from a National Institutes of Health Molecular Biosciences Training Program (Grant 2T32GM007215-36).
Footnotes
ASSOCIATED CONTENT
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Venable JD, Dong M-Q, Wohlschlegel J, Dillin A, Yates JR. Nat. Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 2.Geiger T, Cox J, Mann M. Mol. Cell. Proteomics. 2010;9:2252–2261. doi: 10.1074/mcp.M110.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bern M, Finney G, Hoopmann MR, Merrihew G, Toth MJ, MacCoss MJ. Anal. Chem. 2009;82:833–841. doi: 10.1021/ac901801b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Mol. Cell. Proteomics. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins BC, Gillet LC, Rosenberger G, Röst HL, Vichalkovski A, Gstaiger M, Aebersold R. Nat. Methods. 2013;10:1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Hüttenhain R, Surinova S, Gillet LC, Mouritsen J, Brunner R, Navarro P, Aebersold R. Proteomics. 2013;13:1247–1256. doi: 10.1002/pmic.201200417. [DOI] [PubMed] [Google Scholar]
- 7.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Mol. Cell. Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebert AS, Merrill AE, Bailey DJ, Still AJ, Westphall MS, Strieter ER, Pagliarini DJ, Coon JJ. Nat. Methods. 2013;10:332–334. doi: 10.1038/nmeth.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merrill AE, Hebert AS, MacGilvray ME, Rose CM, Bailey DJ, Bradley JC, Wood WW, El Masri M, Westphall MS, Gasch AP, Coon JJ. Mol. Cell. Proteomics. 2014;13:2503–2512. doi: 10.1074/mcp.M114.040287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards AL, Vincent CE, Guthals A, Rose CM, Westphall MS, Bandeira N, Coon JJ. Mol. Cell. Proteomics. 2013;12:3812–3823. doi: 10.1074/mcp.M113.028951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bamberger C, Pankow S, Park SKR, Yates JR. J. Proteome Res. 2014;13:1494–1501. doi: 10.1021/pr401035z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wenger CD, Phanstiel DH, Lee M, Bailey DJ, Coon JJ. Proteomics. 2011;11:1064–1074. doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Mueller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach J-J, Cox J, Horning S, Mann M, Makarov A. Mol. Cell. Proteomics. 2012;11:O111.013698. doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senko MW, Remes PM, Canterbury JD, Mathur R, Song Q, Eliuk SM, Mullen C, Earley L, Hardman M, Blethrow JD, Bui H, Specht A, Lange O, Denisov E, Makarov A, Horning S, Zabrouskov V. Anal. Chem. 2013;85:11710–11714. doi: 10.1021/ac403115c. [DOI] [PubMed] [Google Scholar]
- 15.Schaub TM, Hendrickson CL, Horning S, Quinn JP, Senko MW, Marshall AG. Anal. Chem. 2008;80:3985–3990. doi: 10.1021/ac800386h. [DOI] [PubMed] [Google Scholar]
- 16.Senko MWR, Philip M, Canterbury JD, Mathur R, Song Q, Eliuk SM, Lange O, Denisov E, Makarov A, Specht A, Zabrouskov V. Improving Data Dependent MSn Performance with a Multitasking Mass Spectrometer. Bremen, Germany: Thermo Fisher Scientific: San Jose, CA; Thermo Fisher Scientific GmbH; 2013. [Google Scholar]
- 17.Agathocleous M, Harris WA. Trends Cell Biol. 2013;23:484–492. doi: 10.1016/j.tcb.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Vander Heiden MG, Cantley LC, Thompson CB. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward PS, Thompson CB. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincent CE, Rensvold JW, Westphall MS, Pagliarini DJ, Coon JJ. Anal. Chem. 2012;85:2079–2086. doi: 10.1021/ac302156t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedishvili NY, Popov KM, Jaskiewicz JA, Harris RA. Arch. Biochem. Biophys. 1994;315:317–322. doi: 10.1006/abbi.1994.1506. [DOI] [PubMed] [Google Scholar]
- 22.Shimomura Y, Murakami T, Nakai N, Nagasaki M, Harris RA. J. Nutr. 2004;134:1583S–1587S. doi: 10.1093/jn/134.6.1583S. [DOI] [PubMed] [Google Scholar]
- 23.Owen OE, Kalhan SC, Hanson RW. J. Biol. Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.