Human Msd1/SSX2IP is a component of centriolar satellites and essential for microtubule anchoring to the centrosome. Anchoring defects lead to abnormal accumulation of centriolar components at centriolar satellites, which interferes with centriole assembly. Loss of Msd1/SSX2IP produces supernumerary centriole precursors specifically in tumor cells.
Abstract
Centriolar satellites are numerous electron-dense granules dispersed around the centrosome. Mutations in their components are linked to various human diseases, but their molecular roles remain elusive. In particular, the significance of spatial communication between centriolar satellites and the centrosome is unknown. hMsd1/SSX2IP localizes to both the centrosome and centriolar satellites and is required for tethering microtubules to the centrosome. Here we show that hMsd1/SSX2IP-mediated microtubule anchoring is essential for proper centriole assembly and duplication. On hMsd1/SSX2IP knockdown, the centriolar satellites become stuck at the microtubule minus end near the centrosome. Intriguingly, these satellites contain many proteins that normally localize to the centrosome. Of importance, microtubule structures, albeit not being anchored properly, are still required for the emergence of abnormal satellites, as complete microtubule depolymerization results in the disappearance of these aggregates from the vicinity of the centrosome. We highlighted, using superresolution and electron microscopy, that under these conditions, centriole structures are faulty. Remarkably, these cells are insensitive to Plk4 overproduction–induced ectopic centriole formation, yet they accelerate centrosome reduplication upon hydroxyurea arrest. Finally, the appearance of satellite aggregates is cancer cell specific. Together our findings provide novel insights into the mechanism of centriole assembly and microtubule anchoring.
INTRODUCTION
Centrosome functions are important for a wide range of cellular processes, including the cell cycle, cell motility, ciliogenesis, and development. Over the past decade, it has become evident that the centrosome plays a multifaceted role in these processes; nonetheless, its canonical function as a microtubule-organizing center is still generally regarded to be crucial. The centrosome consists of a pair of centrioles associated with surrounding pericentriolar material (PCM; Bornens, 2002; Azimzadeh and Marshall, 2010; Nigg and Stearns, 2011; Gonczy, 2012). In addition, numerous electron-dense granules 70–100 nm in size, referred to as centriolar satellites, exist around the centrosome (Kubo et al., 1999). Proteomics and systematic genome-wide analysis suggest that the human centrosome comprises 250–300 different components, representing an almost complete catalogue (Neumann et al., 2010; Balestra et al., 2013; Jakobsen et al., 2013). The number of known centriolar satellite components has continued to increase, from ∼10 in 2011 (Barenz et al., 2013) to ∼30 in 2014 (Tollenaere et al., 2014), with more probably remaining to be identified. Compared to the knowledge of the centrosome and centriole, our understanding of the structural architecture and molecular roles of centriolar satellites is still limited, although there is strong evidence that centriolar satellites are involved in centrosome-mediated processes such as mitotic spindle assembly, ciliogenesis, neurogenesis, and autophagy (Nachury et al., 2007; Kim et al., 2008; Lopes et al., 2011; Stowe et al., 2012; Barenz et al., 2013; Tang et al., 2013). Of note, the perturbation of centriolar satellite function is causative in a variety of human diseases (Nachury et al., 2007; Kim et al., 2008; Lopes et al., 2011).
Centriolar satellites dynamically localize along the microtubule cytoskeleton that emanates from the centrosome (Kubo et al., 1999). One proposed role for centriolar satellites is the transport of centriolar/centrosomal components from the cytoplasm to the centrosome via microtubules in a dynein-mediated manner (Dammermann and Merdes, 2002; Barenz et al., 2011). However, the questions of which components are transported, how these molecules are properly delivered to the centriolar/centrosomal region, and the physiological significance of this delivery system remain to be resolved.
Among the numerous known components of centriolar satellites, PCM1 was the first molecule to be identified and plays a major structural role (Balczon et al., 1994; Kubo et al., 1999; Dammermann and Merdes, 2002). In line with this notion, PCM1 particles still localize around the centrosome after the depletion of several satellite components (Staples et al., 2012; Lee and Stearns, 2013). The depletion of certain other components alternatively causes the distribution pattern of PCM1 to become more dispersed (Lopes et al., 2011; Kim and Rhee, 2012). Conversely, more concentrated distributions may be induced, including colocalization with the centrosome, where PCM1 is usually not detected (Lopes et al., 2011; Stowe et al., 2012). How these differences in PCM1 localization patterns affect the function of centriolar satellites is not well understood.
The conserved coiled-coil protein Msd1 was originally identified as a mitosis-specific component of the fission yeast spindle pole body (SPB; the fungal equivalent of the animal centrosome), which is essential for anchoring the minus end of the spindle microtubule to the SPB (Toya et al., 2007). Intriguingly, we and others have shown that orthologues in humans, frogs, and zebrafish (referred to as SSX2IP; Breslin et al., 2007) localize to the centrosome and in addition are components of centriolar satellites (Barenz et al., 2013; Hori et al., 2014), with vertebrate Msd1/SSX2IP colocalizing and physically interacting with PCM1. Knockdown of Msd1/SSX2IP leads to microtubule-anchoring defects, accompanied by the disorganization of interphase microtubules, compromised centrosome maturation, spindle misorientation, and defective ciliogenesis (Barenz et al., 2013; Hori et al., 2014; Klinger et al., 2014).
In the present study, we address the cellular consequences resulting from defective microtubule anchoring after hMsd1/SSX2IP depletion. In particular, we focus our analysis on the structural integrity of the centriole and centrosome. We present evidence showing the dramatic effect on centriole architecture and function associated with hMsd1/SSX2IP depletion.
RESULTS
hMsd1/SSX2IP depletion leads to formation of abnormal aggregates of centriolar satellites with reduced motility around the centrosome
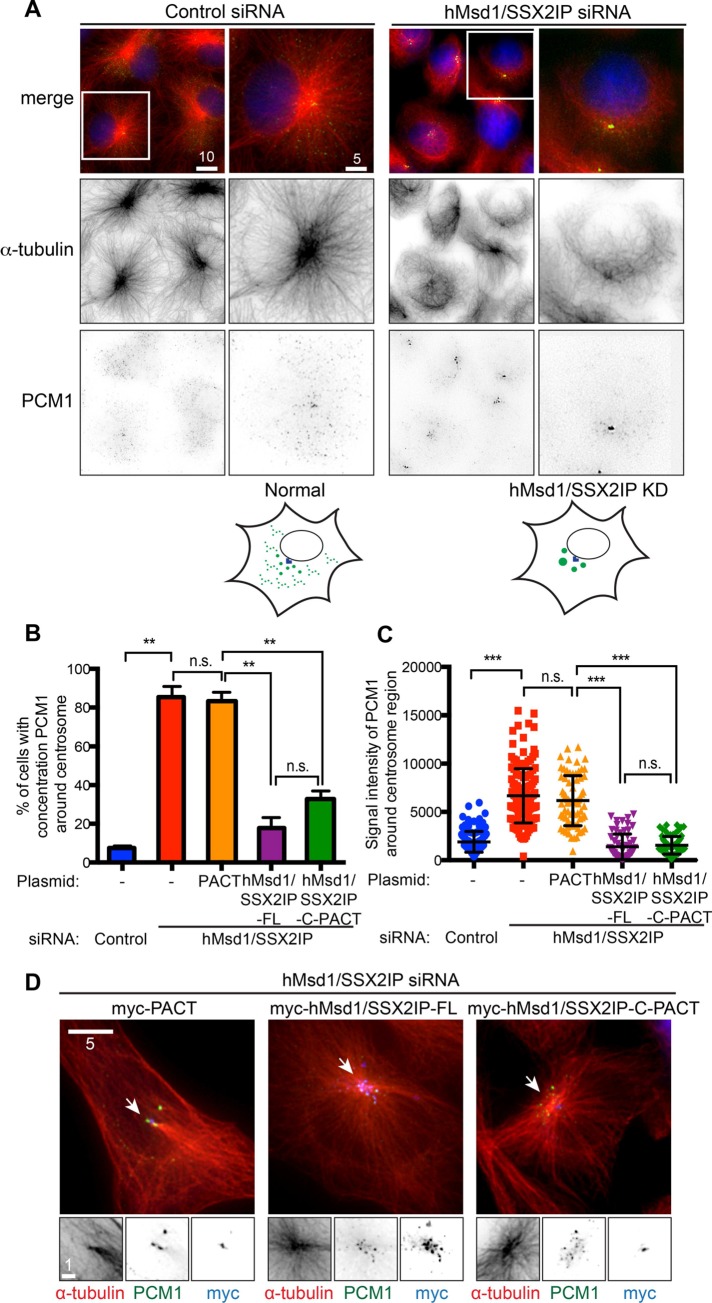
Although PCM1 is deemed to be a structural platform that forms centriolar satellites (Kubo et al., 1999; Dammermann and Merdes, 2002), its precise cellular localization pattern varies to a large degree, depending on which satellite component is depleted (see Introduction). To examine the contribution of hMsd1/SSX2IP to the localization and structural integrity of centriolar satellites, we performed immunofluorescence microscopy using an anti-PCM1 antibody after hMsd1/SSX2IP small interfering RNA (siRNA)–mediated knockdown in U2OS cells (Hori et al., 2014; see Supplemental Figure S1A for the efficient depletion of hMsd1/SSX2IP by siRNA). This analysis showed that whereas PCM1 particles were widely distributed around and away from the centrosome in control cells, hMsd1/SSX2IP-depleted cells displayed dense PCM1 aggregates close to the centrosome in addition to dispersed smaller dots (Figure 1A). These aggregates were not specific to PCM1, as another centriolar satellite component, Cep290 (Kim et al., 2008; Lopes et al., 2011), was also included, indicating that the structures retained centriolar satellite integrity (Supplemental Figure S1, B–D). Quantification of immunofluorescence signal for PCM1 around the centrosomal region indicated that the depletion of hMsd1/SSX2IP resulted in a significant increase in signal intensity (Figure 1, B and C) without changing total protein levels (Hori et al., 2014). This phenotype was attributable to hMsd1/SS2XIP depletion rather than RNA interference (RNAi)–mediated nonspecific outcomes, as the simultaneous introduction of siRNA-resistant full-length hMsd1/SSX2IP construct fully rescued the dispersal of centriolar satellites (Figure 1, B and C). These results show that the absence of hMsd1/SSX2IP affects the localization patterns but not the quantitative abundance of centriolar satellites.
FIGURE 1:
Microtubule anchoring of hMsd1/SSX2IP to the centrosome is essential for the scattered localization of PCM1. (A) Microtubule disorganization upon hMsd1/SSX2IP depletion leads to PCM1 aggregation. U2OS cells were transfected with control or hMsd1/SSX2IP siRNA and immunostained after 48 h with anti–α-tubulin (red) and anti-PCM1 antibodies (green). Regions outlined by squares in the top row are enlarged in the bottom two rows. DAPI staining (blue) is also shown in merged images (top). Scale bars, 10 μm, 5 μm. Schematics for PCM1 localization patterns are depicted below. (B, C) Quantification. The percentage of cells displaying PCM1 aggregation (B) and PCM1 signal intensities around the centrosome (C) were quantified. Twenty-five–pixel squares around the centrosome were measured. U2OS cells were treated with hMsd1/SSX2IP siRNA and simultaneously transfected with plasmids containing myc-PACT, myc-tagged full-length hMsd1/SSX2IP (hMsd1/SSX2IP-FL), or myc-PACT connected to the C-terminal half of hMsd1/SSX2IP (hMsd1/SSX2IP-C-PACT). Data represent mean ± SD (B, >300 cells derived from three independent experiments, n = 3; C, >100 cells, n = 3). Statistical analysis was performed using two-tailed unpaired Student's t tests. **p < 0.001, ***p < 0.0001; n.s., not significant. (D) PCM aggregates upon hMsd1/SSX2IP depletion stem from defects in microtubule anchoring of the centrosome. Cells were stained with antibodies against myc (blue), PCM1 (green), and α-tubulin (red). Enlarged images corresponding to regions marked with arrowheads (top) are shown at the bottom. Scale bars, 5 μm, 1 μm (enlarged images).
hMsd1/SSX2IP depletion leads to the disorganization of interphase radial microtubule arrays (Hori et al., 2014). Previous work showed that the centrosomal-targeting motif (the PACT domain; Gillingham and Munro, 2000) connected to the C-terminal region of hMsd1/SSX2IP (hMsd1/SSX2IP-C-PACT), which lacks the siRNA target site and is therefore RNAi resistant (Hori et al., 2014), was enough to rescue this microtubule defect. Accordingly, we examined whether PCM1 aggregation was reversed by the introduction of hMsd1/SSX2IP-C-PACT. Whereas the PACT domain alone showed abnormal aggregation of PCM1 around the centrosome, expression of hMsd1/SSX2IP-C-PACT, as well as siRNA-resistant full-length hMsd1/SS2XIP, in hMsd1/SSX2IP-depleted cells suppressed this defect and prevented microtubule disorganization (Figure 1, B–D). Because hMsd1/SSX2IP-C-PACT, but not PACT-less hMsd1/SSX2IP-C, rescues microtubule-anchoring defects (this study; Hori et al., 2014), the centrosomal region is critical for hMsd1/SSX2IP function. However, it is formally possible that a small faction of this protein localizing to other places also contributes to the suppression of microtubule disorganization and the emergence of aggregated satellites. In any case, the results indicate that PCM1 aggregation stems from microtubule abnormalities.
Satellite aggregates upon hMsd1/SSX2IP depletion are dependent on the existence of microtubules despite their disorganization
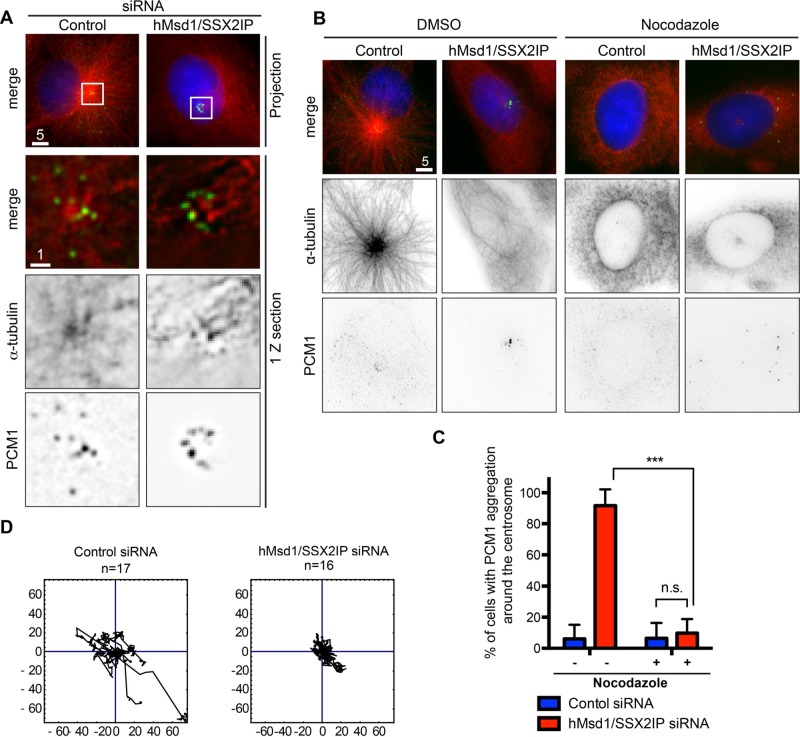
We noticed that upon hMsd1/SSX2IP depletion, the majority of PCM1 aggregates localized to the end of microtubules that are released from the centrosome, whereas PCM1 dots were broadly dispersed on microtubule arrays in control cells (Figure 2A). If the appearance of satellite aggregates around the centrosome is due to disorganized microtubules arising from their anchoring defects, the elimination of microtubules should lead to the dispersion of these aggregates. To examine this proposition, we treated cells depleted of hMsd1/SSX2IP with microtubule-depolymerizing nocodazole. Markedly, numerous PCM1 aggregates became completely scattered away from the centrosome (Figure 2, B and C, and Supplemental Figure S2).
FIGURE 2:
hMsd1/SSX2IP knockdown compromises dynamic motility of PCM1 and results in the aggregation that is dependent on microtubule structures. (A) PCM1 aggregates localize to the end of microtubules. Regions outlined by squares are enlarged, in which images of only one z-section are shown. Scale bars, 5 μm, 1 μm. (B) Microtubule depolymerization leads to the dispersed localization of PCM1 in hMsd1/SSX2IP-depleted cells. U2OS cells were transfected with control or hMsd1/SSX2IP siRNA and treated with DMSO (left) or nocodazole (right) for 2 h. Cells were then fixed and immunostained with antibodies against PCM1 (green) and α-tubulin (red). DNA was stained with DAPI (blue). Scale bars, 5 μm. (C) Quantification of cells displaying PCM1 aggregation around the centrosome. Data represent the mean ± SD (>150 cells, n = 3). ***p < 0.0001; n.s., not significant. (D) The trajectory of PCM1 signals. U2OS cells were treated with control or hMsd1/SSX2IP siRNA and further transfected with EGFP-PCM1 after 24 h. At 24 h after the second transfection, the cells were observed and time-lapse imaging performed (n = 17 in control siRNA cells; n = 16 in hMsd1/SSX2IP siRNA cells).
A previous report showed that PCM1 particles move dynamically toward the centrosome, where radial microtubule arrays are used as trafficking routes (Kubo et al., 1999). Consistent with microtubule disorganization, tracking the trajectory of GFP-tagged PCM1 by live imaging showed that hMsd1/SSX2IP depletion resulted in substantially attenuated PCM1 motility when compared with control cells (Figure 2D). Taking the results together, we consider that the anchoring of microtubule minus ends to the centrosome via hMsd1/SSX2IP is essential for dynamic motility of PCM1 along microtubules. In the absence of hMsd1/SSX2IP's anchoring function, centriolar satellites accumulate at the microtubule ends that are released from the centrosome.
hMsd1/SSX2IP-mediated microtubule anchoring is required for recruitment of centrin to the centriole
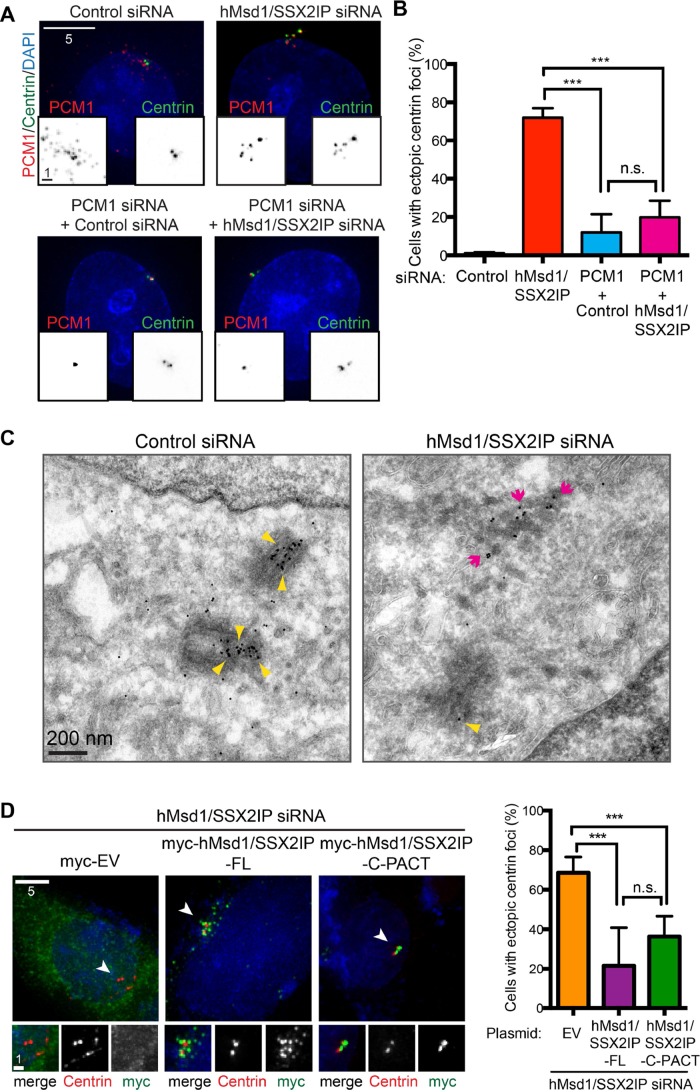
It was previously suggested that one of the cellular functions of centriolar satellites involves the transport of centrosomal/centriolar components from the cytoplasm to the centrosome via microtubules (Kubo et al., 1999; Dammermann and Merdes, 2002; Nachury et al., 2007; Barenz et al., 2011). However, except under nonphysiological conditions such as ectopic overproduction of PCM1 (Dammermann and Merdes, 2002), localization of these components to the centriolar satellites is normally undetectable, possibly because the individual structures are too small and/or the velocity of transport is too fast for detection. Given that centriolar satellites abnormally accumulate as aggregates in close proximity to the centrosomal region with reduced motility (Figure 2, A and D), we next examined the localization of centrin, which is known to colocalize with PCM1 aggregates when induced by ectopic overproduction of PCM1-truncated mutants (Dammermann and Merdes, 2002). Intriguingly, we found that the number of centrin dots per cell was increased in hMsd1/SSX2IP-depleted HeLa cells stably expressing centrin–green fluorescent protein (GFP) and furthermore that the majority (>90%, >300 cells) of these centrin signals colocalized with PCM1 (Figure 3, A and B). In clear contrast, under control siRNA-treated conditions, we always observed no more than four dots of centrin (Figure 3A). It is notable that the appearance of ectopic centrin foci is not attributable to stably expressed centrin-GFP; anti-centrin labeling also showed centrin amplification in normal U2OS and HeLa cells (Supplemental Figure S3). To address whether the appearance of these extra centrin dots was dependent on the integrity of centriolar satellites, we performed double depletions of PCM1 and hMsd1/SSX2IP, and it was evident that supernumerary centrin dots disappeared with this treatment (Figure 3, A and B), suggesting that the extra centrin dots reside on centriolar satellites. We envision that the abnormal aggregation of PCM1 particles impedes the proper transport of centrin toward the centriole, leading to accumulation of centrin molecules on centriolar satellites.
FIGURE 3:
hMsd1/SSX2IP depletion leads to the formation of supernumerary centrin foci that are dependent on PCM1. (A) Extra centrin foci appear upon hMsd1/SSX2IP depletion in a PCM1-dependent manner. HeLa cells stably expressing centrin-GFP were cotransfected with control, hMsd1/SSX2IP, control and PCM1, or hMsd1/SSX2IP and PCM1 siRNAs. At 48 h after transfection, cells were fixed and immunostained with anti-GFP (green) and anti-PCM1 antibodies (red). DNA was stained with DAPI (blue). Scale bars, 5 μm, 1 μm (bottom squares). (B) Quantification of cells containing ectopic centrin foci. Data represent the mean ± SD (>300 cells, n = 3). ***p < 0.001; n.s., not significant. (C) Representative images of immuno-EM using an anti-GFP antibody in HeLa cells stably expressing centrin-GFP. Note that gold particles overlap with the electron-dense granules that represent centriolar satellites in hMsd1/SSX2IP-depleted cells (magenta arrows). Gold particles at the centriole (yellow arrowheads) in hMsd1/SSX2IP-depleted cells (n = 49) were fewer than with control cells (n = 61). Scale bar, 200 nm. (D) The centrosome-targeted C-terminal half of hMsd1/SSX2IP suppressed the formation of ectopic centrin foci. U2OS cells were cotransfected with hMsd1/SSX2IP siRNA and plasmids containing myc alone, siRNA-resistant, full-length myc-hMsd1/SSX2IP (myc-hMsd1/SSX2IP-FL), or myc-PACT connected C-terminal half of hMsd1/SSX2IP (myc-hMsd1/SSX2IP-C-PACT). Left, cells were stained with antibodies against myc (green) and centrin-2 (red). DNA was stained with DAPI (blue). Regions marked with arrowheads (top) are enlarged at the bottom. Scale bars, 5 μm, 1μm (bottom, enlarged images). Right, quantification of cells displaying ectopic centrin foci (>200 cells, n = 3). ***p < 0.0001, n.s., not significant.
To visualize abnormal centrin dots at the ultrastructural level, we performed immuno–electron microscopy (EM) in control and hMsd1/SSX2IP-depleted cells. This analysis highlighted numerous electron-dense granules around the centrioles in hMsd1/SSX2IP-depleted cells, and, of greater importance, anti-GFP/protein A–conjugated gold labeling of some of the accumulated granules was evident (Figure 3C, magenta arrows, n = 61 for control siRNA and 49 for hMsd1/SSX2IP siRNA). We did not observe overduplicated centrosomes in either control or hMsd1/SSX2IP-depleted cells. In addition, although gold particles also localized as expected to the lumen of authentic centrioles in control and hMsd1/SSX2IP-depleted cells, the overall intensity of labeling was slightly reduced in hMsd1/SSX2IP-depleted cells (Figure 3D, yellow arrowheads).
Consistent with the notion that defects in microtubule anchoring are the primary reason for accumulation of extra centrin dots upon hMsd1/SSX2IP depletion, the introduction of siRNA-resistant full-length hMsd1/SS2XIP or forced targeting of the C-terminal hMsd1/SSX2IP (hMsd1/SSX2IP-C-PACT) was capable of suppressing this phenotype (Figure 3D). Taking the results collectively, we suggest that hMsd1/SSX2IP-mediated microtubule anchoring is important for the proper delivery of centrin to the centriole via centriolar satellites.
A subset of centriolar/centrosomal components accumulates in centriolar satellites upon hMsd1/SSX2IP depletion
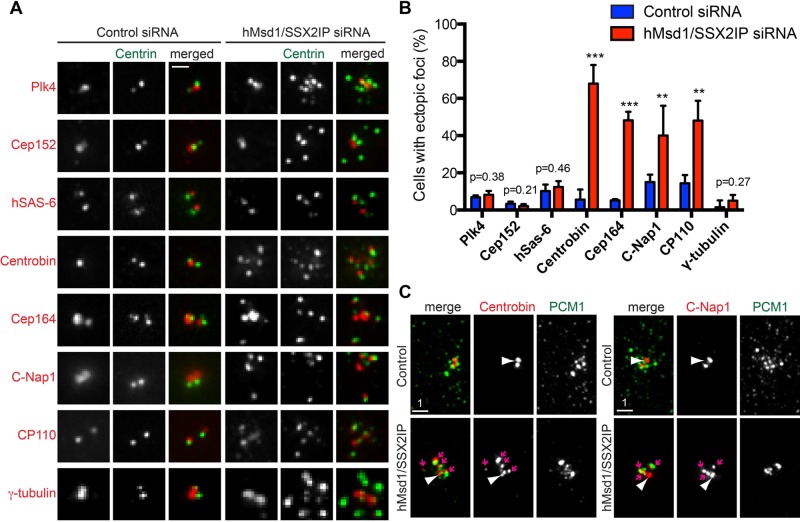
Because centrin is arguably not the sole protein transported to the centrosome via centriolar satellites (Dammermann and Merdes, 2002; Nachury et al., 2007; Kim and Rhee, 2012), we next systematically examined the localization of other centriole/centrosome components after hMsd1/SSX2IP knockdown. Remarkably, a cohort of components, including centrobin (normally localizing to the daughter centriole; Zou et al., 2005), Cep164 (the distal appendage; Graser et al., 2007), C-Nap1 (the proximal end; Fry et al., 1998), and CP110 (the distal end; Chen et al., 2002), also accumulated at ectopic centrin foci (Figure 4, A and B). Furthermore, centrobin and c-NAP also colocalized with PCM1 aggregates (Figure 4C).
FIGURE 4:
A subset of centriolar/centrosomal components accumulates as aggregates around the centrosome and colocalize with PCM1. (A) Some centriolar/centrosomal components were amplified at the pericentriolar region. HeLa cells stably expressing centrin-GFP were transfected with control or hMsd1/SSX2IP siRNA, and 48 h after transfection, cells were fixed and immunostained with antibodies against individual proteins indicated (red) and centrin-GFP (green). (B) Quantification of cells containing ectopic foci of each centriolar/centrosomal component. Data represent the mean ± SD (>200 cells, n = 3). ***p < 0.0001, **p < 0.001. (C) Ectopic foci containing centrobin (left) or C-Nap1 (right) colocalize with PCM1 aggregates in hMsd1/SSX2IP-depleted cells. Cells were stained with antibodies against PCM1 (green) and centrobin or C-Nap1 (red). Scale bar, 1 μm.
In stark contrast, Plk4 (the proximal end; Kleylein-Sohn et al., 2007), Cep152 (the proximal end; Cizmecioglu et al., 2010; Hatch et al., 2010), hSAS-6 (procentriole; Strnad et al., 2007), and γ-tubulin (the centrosome and pericentriolar material; Stearns et al., 1991) did not accumulate at the ectopic foci; these proteins instead localized normally as one or two spots (Figure 4, A and B). Overall it appeared that centriolar/centrosomal proteins that are normally recruited during the initial stages of centriole duplication, such as those involved in the formation of the procentriole, did not accumulate as supernumerary dots, whereas components recruited in later stages, including the elongation and maturation steps, did accumulate (Bornens, 2002; Azimzadeh and Marshall, 2010; Nigg and Stearns, 2011; Gonczy, 2012). It is therefore possible that centriole components required for the elongation and/or maturation steps are transported to the centrosome in a centriolar satellite– and microtubule-mediated manner, but additional parallel pathways may also be used to deliver other components to the centriole/centrosome.
Centriole components stuck on centriolar satellites lead to defective assembly of intact centriole structures
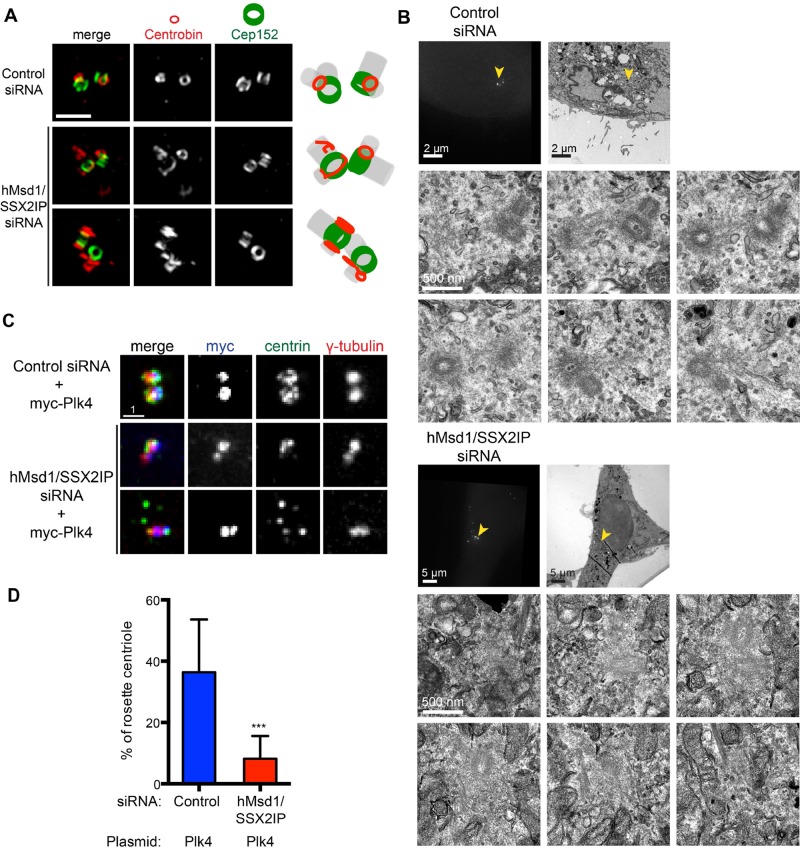
The observation that several centriolar/centrosomal components do not properly localize upon hMsd1/SSX2IP depletion, moving instead to noncentrosomal centriolar satellite sites, led us to the idea that the structural integrity of authentic centrioles might be compromised as a result of the shortage of properly delivered components. To address this point, we undertook three different yet complementary approaches. First, we performed superresolution immunofluorescence imaging of centriole components using three-dimensional structured-illumination microscopy (3D-SIM). Consistent with its normal localization, Cep152 formed two ring-like structures (Lawo et al., 2012; Sonnen et al., 2012) in both control and hMsd1/SSX2IP siRNA–treated cells (Figure 5A). By contrast, the localization of centrobin was somewhat irregular. In control cells, centrobin formed two smaller rings juxtaposed with those of Cep152. In hMsd1/SSX2IP-depleted cells however, we rarely observed two symmetrical rings. Instead, at least one of the centrioles displayed irregular/filamentous staining, sometimes perpendicular in relation to the Cep152 rings (Figure 5A, bottom two rows).
FIGURE 5:
Centriole structures are abnormal in hMsd1/SSX2IP-depleted cells. (A) Superresolution microscopy (OMX) images. Representative images show two normal ring structures of Cep152 in either control (top) or after hMsd1/SSX2IP depletion (bottom two rows), whereas centrobin displays abnormal non-ring structures in hMsd1/SSX2IP-depleted cells. In control cells, centrobin forms smaller rings juxtaposed with Cep152 rings (>20 cells, n = 2). Right, schematics depicting the localization patterns of centrobin and Cep152 together with predicted centriole structures (gray). Scale bar, 1 μm. (B) CLEM images. HeLa cells stably expressing centrin-GFP were transfected with control or hMsd1/SSX2IP siRNA and processed for CLEM analysis. Top row, correlative images of fluorescence (left) and EM (right). Bottom two rows, serial section images corresponding to the areas marked with yellow arrowheads in the top row (control siRNA n = 6, hMsd1/SSX2IP siRNA n = 12). Note that distinctive electron-dense centriole structures were more difficult to identify in the hMsd1/SSX2IP-depleted cells. Scale bars, 2 μm, 5 μm (top), 500 nm (bottom two rows). (C) Loss of hMsd1/SSX2IP suppresses the appearance of a rosette-like arrangement of extra centrins induced by myc-Plk4 overproduction. U2OS cells were transfected with control or hMsd1/SSX2IP siRNA, and plasmids containing myc-connected Plk4 constructs were further transfected after 24 h. Cells were stained with antibodies against myc (blue), centrin-2 (green), and γ-tubulin (red). Representative images of rosette formation in control siRNA cells (top) and two examples in hMsd1/SSX2IP siRNA cells (bottom two rows). One (middle) displays two to four centrin foci, whereas the other (bottom) exhibits the dispersed appearance of extra centrin foci, the phenotype reminiscent of hMsd1/SSX2IP depletion. Scale bar, 1 μm. (D) Quantification of cells containing a rosette-like arrangement of extra centrins. Data represent the mean ± SD (>200 cells, n = 2). ***p < 0.0001.
Second, we performed correlative light and electron microscopy (CLEM) in combination with serial sectioning of HeLa cells stably expressing centrin-GFP (Figure 5B). Whereas we observed intact centriole structures with typical orthogonal configuration in control cells (n = 6), ∼42% of centrioles in hMsd1/SSX2IP-depleted cells displayed abnormalities (n = 12); of interest, centriole structures were obscure, and the relative density of the pericentriolar region in Msd1-depleted cells was often increased compared with control cells (Figure 5B).
Finally, we ectopically overproduced Plk4 in hMsd1/SSX2IP-depleted cells to induce extra centriole assembly. Plk4 is a master regulator of centriole copy number; overproduction leads to centriole overduplication, whereas depletion leads to defects in centriole duplication (Kleylein-Sohn et al., 2007). We reasoned that if centriolar assembly was compromised by hMsd1/SSX2IP knockdown, overproduction of Plk4 might not be able to produce ectopic centrioles. Whereas Plk4 overproduction resulted in a characteristic rosette-like arrangement of additional centrin dots around the parental centriole in >35% of control cells (Figure 5, C and D), cells depleted of hMsd1/SSX2IP failed to do so; either two centrin dots (Figure 5C, middle row) or several disconnected centrin dots were observed (Figure 5C, bottom two rows), a phenotype reminiscent of hMsd1/SSX2IP depletion (see Figure 3A). Taking these results together, we propose that hMsd1/SSX2IP is critical for the assembly of proper centriole structures and suggest that faulty centriole structures stem from the microtubule-anchoring defect associated with hMsd1/SSX2IP loss, causing accumulation of several centriole/centrosome components at centriolar satellites and preventing normal delivery to the centriolar region.
Depletion of hMsd1/SSX2IP promotes centrosome reduplication under hydroxyurea-arrest conditions
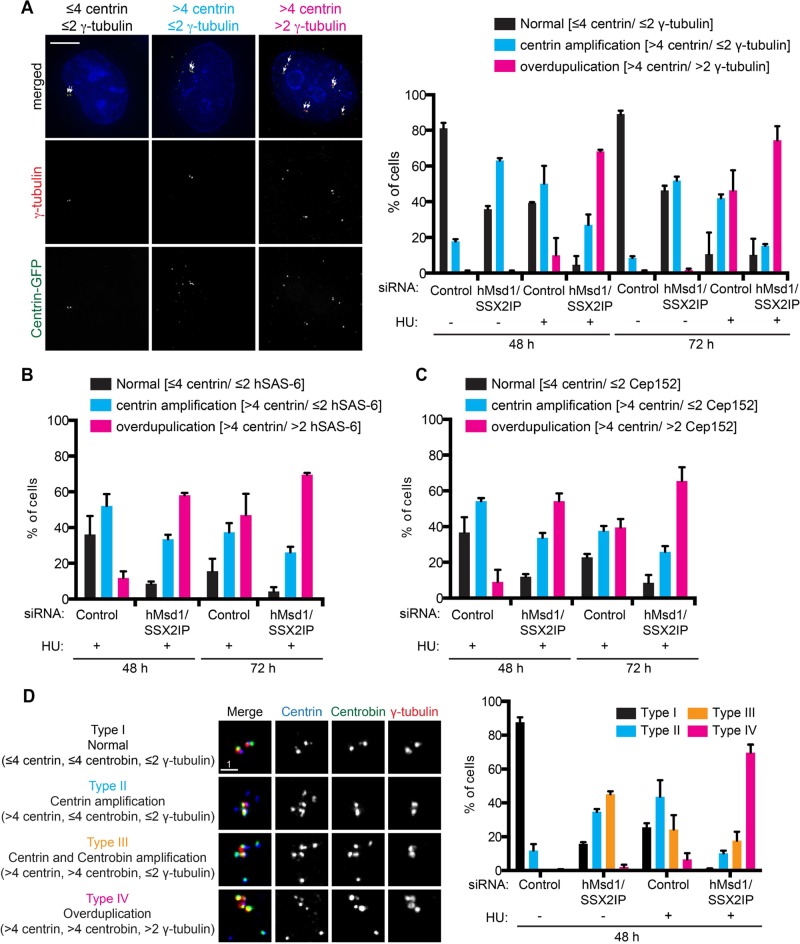
It is established that upon prolonged hydroxyurea (HU) treatment, centrosome reduplication occurs in certain types of cell lines, including CHO and U2OS (Balczon et al., 1995; Prosser et al., 2009; Collins et al., 2010; Kuriyama et al., 2007). Under these conditions, centriolar satellites are transiently used as precursors consisting of incomplete reduplication intermediates (Prosser et al., 2009; Collins et al., 2010). As shown earlier, upon hMsd1/SSX2IP knockdown, centriolar proteins accumulate in centriolar satellites. We suspected that hMsd1/SSX2IP knockdown might accelerate the timing of centrosome reduplication when combined with HU treatment. To test this hypothesis, we transfected U2OS cells stably expressing centrin-GFP with control or hMsd1/SSX2IP siRNA in the presence or absence of HU, and 48 h and 72 h after treatment, we observed centrin and the γ-tubulin signals. As shown in Figure 6A, at the 48-h time point, HU treatment alone poorly induced centrosome reduplication (defined as cells containing more than two and four γ-tubulin and centrin dots, respectively); only 10% of the whole population displayed reduplication. Strikingly, simultaneous hMsd1/SSX2IP knockdown increased this percentage to 68%. By 72 h, the percentage of cells displaying reduplication rose to 46% (HU alone) and 74% (HU and hMsd1/SSX2IP knockdown). Note that hMsd1/SSX2IP knockdown alone resulted in ectopic accumulation of only centrin foci, and generally not γ-tubulin, even after 72 h of treatment.
FIGURE 6:
hMsd1/SSX2IP depletion promotes centrosome overduplication in HU-arrested U2OS cells. (A–C) HU was added to U2OS cells stably expressing centrin-GFP and simultaneously transfected with control or hMsd1/SSX2IP siRNA. At 48 and 72 h after transfection, cells were fixed and stained with antibodies against γ-tubulin (A), hSAS-6 (B), or Cep152 (C). (A, left) Images displaying three representative types of cells: ≤4 centrin/≤2 γ-tubulin (denoted as normal), >4 centrin/≤2 γ-tubulin (centrin amplification), and >4 centrin/>2 γ-tubulin (overduplication). Arrows mark centrin dots that colocalize with γ-tubulin. Scale bars, 5 μm. Quantification of the percentage of cells representing the three types of centrin/γ-tubulin (A, right), hSAS-6 (B), or Cep152 (C) patterns. Data represent the mean ± SD (>200 cells, n = 2). For immunofluorescence images corresponding to B and C, see Supplemental Figure S4, A and B, respectively. (D) HU was added to U2OS cells stably expressing centrin-GFP and simultaneously transfected with control or hMsd1/SSX2IP siRNA. At 48 h after transfection, cells were fixed and stained with antibodies against centrin-2 (blue), centrobin (green), and γ-tubulin (red). Left, the four representative types of cells: type I, ≤4 centrin, ≤4 centrobin, ≤2 γ-tubulin (normal); type II, >4 centrin, ≤4 centrobin, ≤2 γ-tubulin (centrin amplification), type III, >4 centrin, >4 centrobin, ≤2 γ-tubulin (centrin and centrobin amplification); and type IV, >4 centrin, >4 centrobin, >2 γ-tubulin (overduplication). Right, quantification of the percentage of cells representing the four types of centrin/centrobin/γ-tubulin patterns. Data represent the mean ± SD (>200 cells, n = 2). Note that in type III, most, if not all, amplified centrin foci overlapped with those of centrobin, whereas in type IV, this is also the case for γ-tubulin extra foci.
To substantiate the notion of intact centrosome reduplication that was judged by only the presence of γ-tubulin (Figure 6, A and B), we further performed immunofluorescence microscopy with other antibodies in addition to anti–γ-tubulin. Both hSAS-6 and Cep152, which do not show ectopic accumulation upon hMsd1/SSX2IP depletion alone (Figure 4, A and B), were amplified much earlier under conditions of both hMsd1/SSX2IP depletion and HU treatment (Figure 6, B and C, and Supplemental Figure S4, A and B).
Previous studies using HU-treated CHO and U2OS cells showed that under S phase arrest, centriolar satellites act as precursors for reduplicating centrosomes; these satellites contain a group of centriolar/centrosomal components in addition to centrin (Prosser et al., 2009; Collins et al., 2010). Intrigued by these preceding studies, and given that several centriolar/centrosomal proteins, including centrin, coaccumulate in centriolar satellites upon hMsd1/SSX2IP depletion (Figure 4), we immunostained hMsd1/SSX2IP-depleted cells with antibodies against centrobin and CP110 at 48 h under HU treatment. Of interest, both proteins colocalized with the extra centrin foci (Figure 6D and Supplemental Figure S4C). Even control siRNA–treated cells treated with HU displayed coaccumulation, albeit to a lesser extent. Under all conditions examined, the emergence of extra centrin foci preceded those of centrobin and CP110, which were followed by recruitment of γ-tubulin, hSAS-6, and Cep152. This suggests the temporal regulation of the delivery and/or recruitment of centriolar/centrosomal components during centrosome reduplication (Prosser et al., 2009). The result further supports the proposition that centrin-containing centriolar satellites are assembly intermediates for centrosome reduplication under HU treatment, of which the kinetics of their emergence is accelerated upon hMsd1/SSX2IP knockdown.
The appearance of extra centrin foci in hMsd1/SSX2IP-depleted cells is cancer cell specific
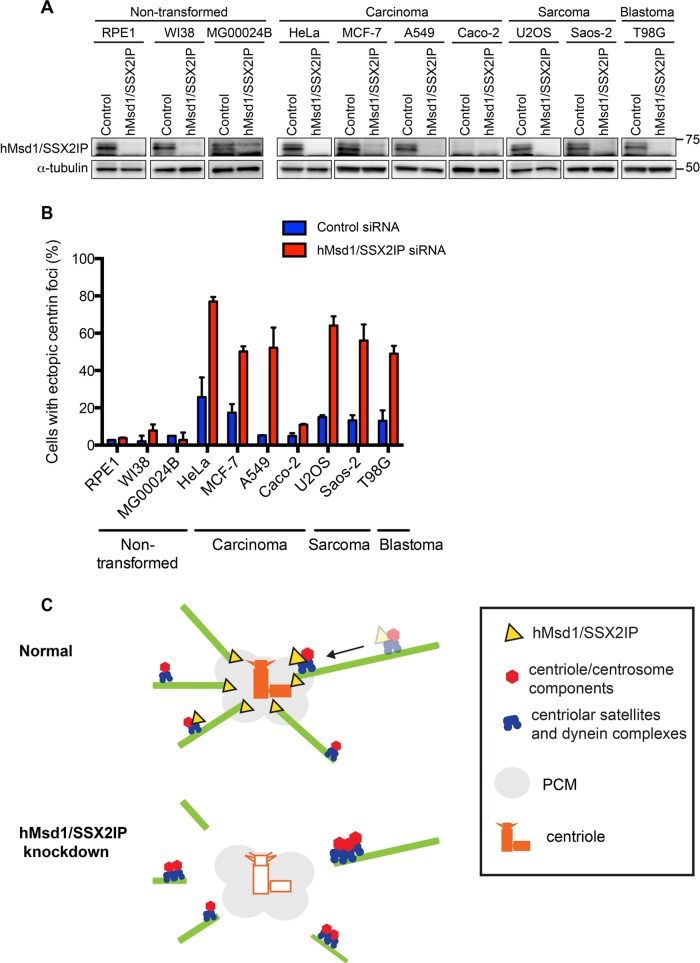
During the course of experiments using hTERT-immortalized RPE1 cells (human retinal pigment epithelial cells) to examine the requirement of hMsd1/SSX2IP in ciliogenesis (Hori et al., 2014), we realized that unlike the other two cell types (HeLa and U2OS cells), extra centrin dots did not emerge, despite the negative effect of hMsd1/SSX2IP depletion on microtubule organization and ciliogenesis. Total protein levels of hMsd1/SSX2IP were very similar, if not identical, when comparing RPE1 and HeLa or U2OS cells, and the efficiency of protein depletion was also indistinguishable (Figure 7A). One obvious difference among these three cell lines is that RPE1 is nontransformed, whereas HeLa and U2OS cells are derived from cancer patients. Given that the function and regulation of the centrosome are intimately associated with tumorigenesis (Godinho and Pellman, 2014), we next tested the consequence of hMsd1/SSX2IP depletion in a panel of human nontransformed and cancer cell lines. These included nontransformed cell lines W138 (lung fibroblast) and MG00024B and cancer cell lines MCF-7 (breast cancer), A549 (epithelial lung adenocarcinoma), Caco-2 (epithelial colorectal adenocarcinoma), Saos-2 (bone cancer), and T98G (glioblastoma). As shown in Figure 7A, global protein expression levels of hMsd1/SSX2IP were comparable, except for CaCo-2 cells, where expression was substantially reduced. Intriguingly, upon successful depletion of hMsd1/SSX2IP, ectopic centrin foci were only observed in the cancer cell lines (Figure 7B). We conclude therefore that nontransformed and transformed cells respond very differently to hMsd1/SSX2IP depletion, the abnormal expression of centrin foci increasing in cancer cell lines in the absence of hMsd1/SSX2IP function.
FIGURE 7:
Centrin amplification upon hMsd1/SSX2IP knockdown is cancer cell specific. (A) Nontransformed (RPE1, W138, MG00024B), carcinoma (HeLa, MCF-7, A549, Caco-2), sarcoma (U2OS, Saos-2), or blastoma cell lines (T98G) were treated with control or hMsd1/SSX2IP siRNA. Immunoblotting with an anti-hMsd1/SSX2IP antibody is shown for each cell line with α-tubulin used as a loading control. (B) Quantification of cells containing ectopic centrin foci. Data represent the mean ± SD (>150 cells, n = 2). (C) A scheme depicting the transport and accumulation of centriolar components in control and hMsd1/SSX2IP-depleted cells. Top, under normal conditions, hMsd1/SSX2IP (yellow) localizes to centriolar satellites (blue) and around the centrosome (gray area), and is responsible for anchoring the minus end of microtubules to the centrosome. Centriolar satellites transport a cohort of centriolar/centrosomal components (red) toward the centrosome via microtubules. This process is essential to the assembly of intact centriole structures (orange). Bottom, in the absence of hMsd1/SSX2IP, particularly in cancer cells, microtubules are no longer tethered to the centrosome. Consequently, centriolar satellites carrying centriolar/centrosomal components become stuck at the end of the microtubules, leading to faulty assembly of centrioles (outlined in orange). Note that our work indicates that in the absence of hMsd1/SSX2IP-mediated microtubule anchoring, particularly in cancer cells, other independent pathways responsible for centriole assembly, such as diffusion-based protein–protein interaction, are not sufficient to build centrioles properly. At the moment, however, we cannot explicitly conclude whether the sole reason for faulty centriole assembly is attributed to the defects in the release of the centriolar/centrosomal proteins from centriolar satellites. By contrast, in nontransformed cells, microtubule disorganization is observed; however, the role of hMsd1/SSX2IP in centriole assembly is cryptic, and alternate compensatory pathways may be exploited in these cells.
DISCUSSION
In this work, we uncovered a critical role of hMsd1/SSX2IP, a conserved microtubule-anchoring protein, in centriole assembly and duplication. On hMsd1/SSX2IP knockdown, a subset of centriolar/centrosomal components colocalize with centriolar satellites and accumulate around the centrosome in the form of discrete aggregates. Of importance, cells depleted of hMsd1/SSX2IP displayed defective centriole structures that were unresponsive to Plk4 overproduction, which normally induces ectopic centriole formation, whereas HU treatment accelerated centrosome reduplication. Intriguingly, the emergence of supernumerary centrin foci was cancer cell specific. Thus hMsd1/SSX2IP is crucial for the formation of intact centrioles, yet it plays opposing roles under different centrosome reduplication conditions.
The proper tethering of interphase microtubules to the centrosome via hMsd1/SSX2IP is essential for assembly and duplication of centrioles
It has been suggested that the microtubule cytoskeleton is important for the assembly and duplication of the centriole and centrosome (Barenz et al., 2011; Tollenaere et al., 2014). However, this has never been explicitly demonstrated, which can be primarily attributed to the lack of suitable tools by which to address the issue. Here we took advantage of the hMsd1/SSX2IP-knockdown system, which specifically disrupts the anchoring of microtubules to the centrosome yet retains microtubule structures and the integrity of centriolar satellites. To our knowledge, this is the first report demonstrating that a substantial number of centriolar/centrosome components become stuck on trafficking centriolar satellites when microtubule anchoring to the centrosome is disrupted. Furthermore, to analyze the structural integrity of centrioles, we implemented 3D-SIM and EM imaging. These analyses unequivocally demonstrated the structural abnormalities associated with hMsd1/SSX2IP depletion, showing faulty incorporation of centriole components and imperfect centriole architecture (depicted in Figure 7C).
In addition to the centriole deficiencies associated with hMsd1/SSX2IP depletion, Plk4 overproduction fails to induce ectopic centriole assembly in hMsd1/SSX2IP-depleted cells. We envisage two possible scenarios for this defect. One is that as the structure of the parental centrioles is impaired, they cannot produce extra centrioles around the proximal end (i.e., a rosette arrangement; Kleylein-Sohn et al., 2007). Another possibility is that as the subset of proteins required for centriole assembly is stuck in the vicinity of the centrosome as aggregates, they are incapable of assembling into centrioles as building blocks. At present, we cannot distinguish between these two situations, although they are not mutually exclusive. However, if the former possibility were the case, Plk4 overproduction might be expected to induce extra centriole formation via a de novo pathway independent of template parental centrioles (Khodjakov et al., 2002; Loncarek and Khodjakov, 2009). Because in our case this did not occur, we consider the latter scenario to be more feasible. It is also consistent with the idea that centriolar satellites serve as storage sites for cargo proteins (Tollenaere et al., 2014). More work is needed to clarify this point.
Biogenesis of the centriole and centrosome requires intricate pathways for delivery of individual components
A number of recent studies indicated that the assembly of the centriole and centrosome may not depend solely on diffusion-based protein–protein interactions. Instead, multiple delivery systems seem to be involved. For instance, the γ-tubulin complex components are transported via the Rab11-endosome pathway during mitosis (Hehnly and Doxsey, 2014), whereas pericentrin is responsible for the transport of several other components to the mitotic centrosome in a dynein-mediated manner (Chen et al., 2014). Furthermore, immune response–related LGALS3BP is involved in the delivery of pericentriolar material components (Fogeron et al., 2013). Our results demonstrated the critical role of hMsd1/SSX2IP and centriolar satellites in the transport of a subset of centriolar/centrosomal components, but the spectrum of transport cargo molecules differs from preceding studies. Intriguingly, emerging evidence suggests that even within individual centriolar satellite particles, protein compositions are not uniform; instead, multiple satellites each containing different sets of cargo are likely to be responsible for the transport of a myriad of centriolar/centrosomal components (Dammermann and Merdes, 2002; Nachury et al., 2007; Stowe et al., 2012; Firat-Karalar et al., 2014). One future direction would therefore be to elucidate the complex delivery systems implemented by centriolar satellites and the identification of the specific roles of each satellite component in cargo transport.
hMsd1/SSX2IP-mediated microtubule anchoring in cancer cells
We showed here a striking correlation of the phenotypic manifestation upon hMsd1/SSX2IP depletion between nontransformed and cancer cell lines. Because microtubule anchoring is equally impaired in both cell types (RPE1 and U2OS; Hori et al., 2014), it is likely that the difference lies in the centriole assembly processes. It is widely accepted that centriolar/centrosomal anomalies are coupled to tumorigenesis, but these aberrations are generally detrimental for cell division in normal cells (Godinho and Pellman, 2014). Recent studies show that in addition to hMsd1/SSX2IP, multiple molecules and pathways are involved in the suppression of supernumerary centriole formation. These include Cep76 (Tsang et al., 2009), LGALS3BP (Fogeron et al., 2013), Cep131/Azi1 (Staples et al., 2012), and CCDC14 (Firat-Karalar et al., 2014). Of interest, the depletion of these molecules has a variety of effects in nontransformed and cancer cells, often varying within specific lines (Staples et al., 2012; Fogeron et al., 2013). Cancer cells are likely to endure defective centriole duplication, possibly by exploiting alternate pathways. Among molecules exerting inhibitory roles in centriole amplification, Cep131/Azi1 and CCDC14 are components of centriolar satellites (Staples et al., 2012; Firat-Karalar et al., 2014). Functional relationships between these two molecules and hMsd1/SSX2IP need to be addressed in the future.
In conclusion, centriolar satellites and hMsd1/SSX2IP play a crucial role in ensuring centriole integrity via their microtubule-anchoring functions. Under conditions that lead to centriole/centrosome reduplication, hMsd1/SSX2IP plays both positive and negative roles, depending on the system investigated. Of importance, cancer cells are more susceptible to the emergence of supernumerary centrin dots in the absence of normal hMsd1/SSX2IP function. Development of small-molecule inhibitors against hMsd1/SSX2IP would provide novel cancer therapeutics in conjunction with drug treatment such as HU, which may specifically kill cancer cells.
MATERIALS AND METHODS
Cell cultures
U2OS, HeLa, and hTERT-RPE1 cells were maintained as described previously (Hori et al., 2014). MCF-7, A549, Caco-2, Saos-2, T98G, Wl38, and MG00024B cells were cultured in high-glucose DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum at 37°C in the presence of 5–10% CO2. HeLa and U2OS cells stably expressing GFP-centrin (kindly provided by Michel Bornens, Institut Curie, Paris, France) were cultured in DMEM supplemented with 10% fetal bovine serum and 0.5 mg/ml G418.
Plasmid DNA transfection
Plasmid DNA transfections were conducted as described previously (Hori et al., 2014) using Lipofectamine 2000 (Invitrogen). To generate RNAi-resistant, myc-tagged hMsd1/SSX2IP constructs, the full-length hMsd1/SSX2IP (hMsd1/SSX2IP-FL) or C-terminal half of hMsd1/SSX2IP connected to the PACT domain (Gillingham and Munro, 2000) (hMsd1/SSX2IP-C-PACT) was inserted into the pcDNA-myc plasmid. Nucleotide sequences replaced in the RNAi-resistant version of hMsd1/SSX2IP were previously described (Hori et al., 2014). pEGFP-hPCM1 was a gift from Song-Hai Shi (Memorial Sloan Kettering Cancer Center, New York, NY). pcDNA-myc-Plk4 was a gift from Erich Nigg (Biozentrum, University of Basel, Basel, Switzerland). pmCherry–α-tubulin was obtained from Addgene.
RNA interference
RNAi experiments were performed as described previously (Hori et al., 2014). Double-strand RNA at 40 nM was used for each experiment with the RNAiMAX transfection reagent (Invitrogen), and cells were fixed 48 h after transfection unless otherwise stated. Double-stranded siRNA oligonucleotides used for hMsd1/SSX2IP and PCM1 depletion were previously described (Hori et al., 2014).
Antibodies
For immunofluorescence microscopy, the following antibodies were used: chicken anti-myc (1:300, A-21281; Molecular Probes, Eugene, OR), rabbit anti-SSX2IP (1:150, HPA027306; Sigma-Aldrich, Gillingham, United Kingdom), rabbit anti-PCM1 (1:300, sc67204; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti–γ-tubulin (1:250, T5192; Sigma-Aldrich), mouse anti–α-tubulin (1:250, T9026; Sigma-Aldrich), rabbit anti–centrin-2 (1:400, sc27793R; Santa Cruz Biotechnology), mouse anti-Plk4 (1:200, H00010733-B01; Abnova, Taipei, Taiwan), rabbit anti-Cep152 (1:400, A302-480A; Bethyl Laboratories, Montgomery, TX), rabbit anti-hSAS-6 (1:100, sc98506; Santa Cruz Biotechnology), mouse anti-centrobin (1:100, H00116840-B01P; Abnova), rabbit anti-Cep164 (1:200, 45330002; Novus Biologicals, Littleton, CO), mouse anti–C-Nap1 (1:100, 611374; BD Biosciences, San Jose, CA), and rabbit anti-CP110 (1:100, ab99337; Abcam, Cambridge, MA). Secondary antibodies were Alexa Fluor 488–coupled anti-rabbit, Alexa Fluor 594–coupled anti-rabbit, Alexa Fluor 594–coupled anti-mouse, Alexa Fluor 488–coupled anti-mouse, Alexa Fluor 488–coupled anti-chicken, and Cy3-coupled anti-mouse antibodies (all used at 1:1500). For immunoblotting, rabbit anti-SSX2IP (1:1000) and mouse anti–α-tubulin (1:5000) antibodies were used.
Immunofluorescence, superresolution microscopy, and image analysis
Immunofluorescence microscopy was performed as described previously (Hori et al., 2014). Briefly, cells were fixed with methanol at −20°C for 5 min and washed in phosphate-buffered saline (PBS). After blocking in 3% bovine serum albumin (BSA) for 1 h at room temperature, cells were incubated with primary and then secondary antibodies. DNA was visualized by the addition of 4,6-diamidino-2-phenylindole (DAPI; Vector Lab, Burlingame, CA). After incubation of the coverslips at room temperature for 20 min, the samples were mounted on slides. During time-lapse imaging, cells were kept at 34–37°C by a chamber heater. For microtubule depolymerization experiments, cells were treated with dimethyl sulfoxide (DMSO) or 20 μM nocodazole for 2 h before immunofluorescence microscopy.
Images were taken using an Olympus IX71 wide-field inverted epifluorescence microscope with Olympus PlanApo 60×/numerical aperture (NA) 1.4 or UApo 40×/NA 1.35, oil immersion objectives. DeltaVision image acquisition software (softWoRx 3.3.0; Applied Precision) equipped with CoolSnap-HQ digital charge-coupled device (CCD) camera or Cascade electron multiplying CCD 512B camera (Roper Scientific) was used. The sections of images were compressed into a two-dimensional (2D) projection using the DeltaVision maximum intensity algorithm. Deconvolution was applied before generating the 2D projection. Images were taken as 64 sections along the z-axis at 0.2-μm intervals. Captured images were processed with Photoshop CS3, version 10.0 (Adobe).
Superresolution microscopy was performed using a structured-illumination microscopy system (DeltaVision OMX V3; Applied Precision, Issaquah, WA). A 100×/1.4 NA oil objective (Olympus, Tokyo, Japan) was used with 488- and 593-nm laser illumination and standard excitation and emission filter sets. We applied 125-nm z-steps to acquire raw images, which we reconstructed in 3D using SoftWoRx software (Applied Precision). Image analysis was performed using Photoshop.
Quantification and fluorescence signal intensity measurement
For fluorescence signal intensity measurement, fluorescence signals were quantified using maximum intensity, after subtracting background signals in the vicinity of the fluorescent spot. SoftWoRx software was used for analysis. To quantify the number of centrin-GFP foci, at least 200 cells were counted in each sample, independently, twice, from which SDs and p values were calculated.
Electron microscopy techniques
Cells grown on gridded coverslips were fixed in 4% paraformaldehyde in 0.1 M phosphate buffer (PB), followed by secondary fixation in 1.5% glutaraldehyde/2% paraformaldehyde in 0.1 M PB for 60 min. The coverslips were then processed using 1.5% potassium ferricyanide/1% osmium tetroxide and 1% tannic acid in 0.05 M PB to enhance contrast before dehydration and embedding in epoxy resin. The cells of interest were identified by correlating the grid reference/cell pattern on the surface of the block with fluorescence images. Serial ultrathin sections were collected through the entire extent of the cells of interest and were viewed using an electron microscope (FEI Tecnai G2 Spirit BioTWIN with Gatan Orius CCD camera [FEI, Eindhoven, The Netherlands]). Serial images were adjusted for brightness and contrast using Photoshop and stacked and aligned using Amira (Visage Imaging, Berlin, Germany).
Cells processed for cryosectioning and immunolabeling were fixed in 4% formaldehyde, or 4% formaldehyde with 0.1% glutaraldehyde, in 0.1 M PB. After fixation and embedding in 12% gelatin, blocks of 1 mm3 were trimmed and cryoprotected in 2.3 M sucrose at 4°C, ready for mounting onto pins and snap freezing (Slot and Geuze, 2007). For cryosectioning (Tokuyasu, 1973), 70-nm-thick serial sections were cut at −120°C and collected onto Formvar-coated finder grids or slot grids using a wire loop filled with 1% methyl cellulose/1.15 M sucrose in PBS.
For immunolabeling, excess pick-up solution was removed by floating the grids in 2% gelatin at 37°C for 30 min before washing in 0.01 M PBS containing 0.1% glycine, blocking in 1% BSA in PBS, and incubating in anti-GFP primary antibody (1:50; ab6556; Abcam) in 1% BSA/PBS for 2 h at room temperature. After washing in 0.1% BSA/PBS, the grids were incubated in protein A conjugated to 10-nm gold particles (1:60; CMC, Utrecht, the Netherlands) for 30 min. After washes in PBS, the antisera were fixed with 1% glutaraldehyde/PBS and washed in distilled water.
For correlative microscopy, grids were mounted on microscope slides and coverslipped using buffered glycerol, pH 7.4. Images of GFP fluorescence were acquired using an epifluorescence microscope (Zeiss, Cambridge, United Kingdom) and used for locating GFP- positive structures in the TEM. After image acquisition, the grids were washed in distilled water and contrasted using 2% uranyl oxalate and 1.8% methyl cellulose/0.6% uranyl acetate (Tokuyasu, 1978). The labeled sections were examined in the transmission electron microscope, and images correlating to GFP positive structures were acquired.
Centrosome duplication assay
For HU treatment, U2OS cells stably expressing centrin-GFP were transfected with control or hMsd1/SSX2IP siRNA and simultaneously treated with 5 mM hydroxyurea (HU; Sigma-Aldrich) to arrest cells at S phase and promote centrosome amplification. At 48 and 72 h, cells were fixed and stained with antibodies against γ-tubulin, hSAS-6, Cep152, centrobin, or CP110, as well as with DAPI, to visualize the centrosome/centriole and chromosome, respectively.
To examine Plk4 overproduction, U2OS cells were transfected with control or hMsd1/SSX2IP siRNA, and after 24 h, plasmids containing myc-Plk4 (pcDNA-myc-Plk4) were further transfected. Cells were fixed 24 h after the second transfection.
Tracking the trajectory of PCM1 motility
U2OS cells were treated with control or hMsd1/SSX2IP siRNA for 24 h and then further transfected with pEGFP-PCM1 and pmCherry–α-tubulin plasmids. At 18 h after the second transfection, live imaging was started. Movie streams were acquired at a frame rate of 10 frames/s. Subsequent particle tracking analysis was carried out using AQM 6.0 Kinetic Acquisition Manager software (Siemens Medical Solutions, Forchheim, Germany). Briefly, the diffusion trajectories of single particles were determined by connecting the spots corresponding to individual time points. The data processing and fitting were carried out using Mathematica (Wolfram Research, Champaign, IL).
Statistical data analysis
All data represent the mean of multiple experiments ± SD (Figures 1, B and C, 2C, 3, B and D, 4B, 5D, 6, A–D, and 7B and Supplemental Figures S1, C and D and S4C). Experiment sample numbers and number of replicates used for statistical testing are reported in the corresponding figure legends. All p values are from two-tailed unpaired Student's t tests. Unless otherwise stated, we followed this key for asterisk placeholders for p values in the figures: ***p < 0.0001, **p < 0.001, *p < 0.01.
Supplementary Material
Acknowledgments
We thank Michel Bornens, Fanni Gergely, Sean Munro, Erich Nigg, Wei Shao, and Song-Hai Shi for their generous gift of reagents used in this study. We are grateful to Anne Vaahtokari for help with the OMX microscope. We are grateful to Risa Mori for critical reading of the manuscript. A.H. was supported by a fellowship from the Daiichi-Sankyo Foundation. T.T. and L.C. were supported by Cancer Research UK. Note that The Francis Crick Institute is a consortium of six of the UK's scientific academic organizations—the Medical Research Council (National Institute for Medical Research), Cancer Research UK (London Research Institute), the Wellcome Trust, University College London, Imperial College London, and King's College London—founded on April 1, 2015.
Abbreviations used:
- CLEM
correlative light and electron microscopy
- DAPI
4,6-diamidino-2-phenylindole
- RNAi
RNA interference
- siRNA
small interfering RNA
- SPB
spindle pole body
- 3D-SIM
three-dimensional structured-illumination microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1561) on April 1, 2015.
The experiments were designed by A.H. and T.T., A.H. performed the majority of the experiments and data analysis, and C.P. and L.C. performed electron microscopy. A.H. and T.T. wrote the article with suggestions from the other authors.
The authors declare no conflict of interest arising from this work.
REFERENCES
- Azimzadeh J, Marshall WF. Building the centriole. Curr Biol. 2010;20:R816–R825. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE. PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J Cell Biol. 1994;124:783–793. doi: 10.1083/jcb.124.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra FR, Strnad P, Fluckiger I, Gonczy P. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev Cell. 2013;25:555–571. doi: 10.1016/j.devcel.2013.05.016. [DOI] [PubMed] [Google Scholar]
- Barenz F, Inoue D, Yokoyama H, Tegha-Dunghu J, Freiss S, Draeger S, Mayilo D, Cado I, Merker S, Klinger M, et al. The centriolar satellite protein SSX2IP promotes centrosome maturation. J Cell Biol. 2013;202:81–95. doi: 10.1083/jcb.201302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenz F, Mayilo D, Gruss OJ. Centriolar satellites: busy orbits around the centrosome. Eur J Cell Biol. 2011;90:983–989. doi: 10.1016/j.ejcb.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Breslin A, Denniss FA, Guinn BA. SSX2IP: an emerging role in cancer. Biochem Biophys Res Commun. 2007;363:462–465. doi: 10.1016/j.bbrc.2007.09.052. [DOI] [PubMed] [Google Scholar]
- Chen CT, Hehnly H, Yu Q, Farkas D, Zheng G, Redick SD, Hung HF, Samtani R, Jurczyk A, Akbarian S, et al. A unique set of centrosome proteins requires pericentrin for spindle-pole localization and spindle orientation. Curr Biol. 2014;24:2327–2334. doi: 10.1016/j.cub.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins ES, Hornick JE, Durcan TM, Collins NS, Archer W, Karanjeet KB, Vaughan KT, Hinchcliffe EH. Centrosome biogenesis continues in the absence of microtubules during prolonged S-phase arrest. J Cell Physiol. 2010;225:454–465. doi: 10.1002/jcp.22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Rauniyar N, Yates JR, 3rd, Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol. 2014;24:664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogeron ML, Muller H, Schade S, Dreher F, Lehmann V, Kuhnel A, Scholz AK, Kashofer K, Zerck A, Fauler B, et al. LGALS3BP regulates centriole biogenesis and centrosome hypertrophy in cancer cells. Nat Commun. 2013;4:1531. doi: 10.1038/ncomms2517. [DOI] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof Y-D, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled- coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Pellman D. Causes and consequences of centrosome abnormalities in cancer. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130452. doi: 10.1098/rstb.2013.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol. 2012;13:425–435. doi: 10.1038/nrm3373. [DOI] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Doxsey S. Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev Cell. 2014;28:497–507. doi: 10.1016/j.devcel.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Ikebe C, Tada M, Toda T. Msd1/SSX2IP-dependent microtubule anchorage ensures spindle orientation and primary cilia formation. EMBO Rep. 2014;15:175–184. doi: 10.1002/embr.201337929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen L, Schroder JM, Larsen KM, Lundberg E, Andersen JS. Centrosome isolation and analysis by mass spectrometry-based proteomics. Methods Enzymol. 2013;525:371–393. doi: 10.1016/B978-0-12-397944-5.00018-3. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krishnaswami SR, Gleeson JG. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Rhee K. CEP90 is required for the assembly and centrosomal accumulation of pericentriolar satellites, which is essential for primary cilia formation. PLos One. 2012;7:e48196. doi: 10.1371/journal.pone.0048196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Klinger M, Wang W, Kuhns S, Barenz F, Drager-Meurer S, Pereira G, Gruss OJ. The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone. Mol Biol Cell. 2014;25:495–507. doi: 10.1091/mbc.E13-09-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Terada Y, Lee KS, Wang CL. Centrosome replication in hydroxyurea-arrested CHO cells expressing GFP-tagged centrin2. J Cell Sci. 2007;120:2444–2453. doi: 10.1242/jcs.008938. [DOI] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- Lee JY, Stearns T. FOP is a centriolar satellite protein involved in ciliogenesis. PLoS One. 2013;8:e58589. doi: 10.1371/journal.pone.0058589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol Cells. 2009;27:135–142. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes CA, Prosser SL, Romio L, Hirst RA, O'Callaghan C, Woolf AS, Fry AM. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J Cell Sci. 2011;124:600–612. doi: 10.1242/jcs.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, Straatman KR, Fry AM. Molecular dissection of the centrosome overduplication pathway in S-phase-arrested cells. Mol Cell Biol. 2009;29:1760–1773. doi: 10.1128/MCB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2:2480–2491. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples CJ, Myers KN, Beveridge RD, Patil AA, Lee AJ, Swanton C, Howell M, Boulton SJ, Collis SJ. The centriolar satellite protein Cep131 is important for genome stability. J Cell Sci. 2012;125:4770–4779. doi: 10.1242/jcs.104059. [DOI] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. γ-Tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Stowe TR, Wilkinson CJ, Iqbal A, Stearns T. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol Biol Cell. 2012;23:3322–3335. doi: 10.1091/mbc.E12-02-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, Zhong Q. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013;502:254–257. doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT. A technique for ultracryotomy of cell suspensions and tissues. J Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT. A study of positive staining of ultrathin frozen sections. J Ultrastruct Res. 1978;63:287–307. doi: 10.1016/s0022-5320(78)80053-7. [DOI] [PubMed] [Google Scholar]
- Tollenaere MA, Mailand N, Bekker-Jensen S. Centriolar satellites: key mediators of centrosome functions. Cell Mol Life Sci. 2014;72:11–23. doi: 10.1007/s00018-014-1711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya M, Sato M, Haselmann U, Asakawa K, Brunner D, Antony C, Toda T. γ-Tubulin complex-mediated anchoring of spindle microtubules to spindle-pole bodies requires Msd1 in fission yeast. Nat Cell Biol. 2007;9:646–653. doi: 10.1038/ncb1593. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Spektor A, Vijayakumar S, Bista BR, Li J, Sanchez I, Duensing S, Dynlacht BD. Cep76, a centrosomal protein that specifically restrains centriole reduplication. Dev Cell. 2009;16:649–660. doi: 10.1016/j.devcel.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, Gao Q. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol. 2005;171:437–445. doi: 10.1083/jcb.200506185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.