The yeast Rim8/Art9 α-arrestin, involved in ambient pH signaling, is regulated through multisite phosphorylation of the hinge region by the plasma membrane–associated casein kinase 1. This modification prevents its stable association with the pH sensor protein Rim21 at the plasma membrane and thereby inhibits signal transduction at acidic pH.
Abstract
α-Arrestins play a key role as trafficking adaptors in both yeast and mammals. The yeast Rim8/Art9 α-arrestin mediates the recruitment of endosomal sorting complex required for transport (ESCRT) to the seven-transmembrane protein Rim21 in the ambient pH signaling RIM pathway. ESCRT is believed to function as a signaling platform that enables the proteolytic activation of the Rim101 transcription factor upon external alkalization. Here we provide evidence that the pH signal promotes the stable association of Rim8 with Rim21 at the plasma membrane. We show that Rim8 is phosphorylated in a pH-independent but Rim21-dependent manner by the plasma membrane–associated casein kinase 1 (CK1). We further show that this process involves a cascade of phosphorylation events within the hinge region connecting the arrestin domains. Strikingly, loss of casein kinase 1 activity causes constitutive activation of the RIM pathway, and, accordingly, pH signaling is activated in a phosphodeficient Rim8 mutant and impaired in the corresponding phosphomimetic mutant. Our results indicate that Rim8 phosphorylation prevents its accumulation at the plasma membrane at acidic pH and thereby inhibits RIM signaling. These findings support a model in which CK1-mediated phosphorylation of Rim8 contributes to setting a signaling threshold required to inhibit the RIM pathway at acidic pH.
INTRODUCTION
Adaptor proteins of the arrestin clan play an important role in the regulation of the intracellular trafficking of membrane receptors, transporters, or channels (Becuwe et al., 2012a). These proteins share a common structure called the arrestin fold and can be divided into three subfamilies: visual/β-arrestins, α-arrestins, and Vps26 (Alvarez, 2008). Visual/β-arrestins are restricted to the animal kingdom, and their main role is to regulate seven-transmembrane (7TM)- spanning receptor desensitization, endocytosis, and signaling (Shukla et al., 2011). α-Arrestins and Vps26 are present in all eukaryotes, except plants, which possess only Vps26 (Alvarez, 2008). α-Arrestins regulate the trafficking of membrane transporters/receptors, whereas Vps26 is a component of the retromer complex mediating endosome-to-Golgi retrograde transport (Shi et al., 2006; Lin et al., 2008; Nikko et al., 2008; Nikko and Pelham, 2009; Hatakeyama et al., 2010; Nabhan et al., 2010; O'Donnell et al., 2010; Becuwe et al., 2012b; Merhi and Andre, 2012; Han et al., 2013; Karachaliou et al., 2013; Puca et al., 2013; Wu et al., 2013; Alvaro et al., 2014; Smardon and Kane, 2014).
The Saccharomyces cerevisiae Rim8/Art9 protein (PalF in Aspergillus nidulans) is unique among α-arrestins in that it shares with β-arrestins the ability to function as signal transducer of 7TM protein signals (Herranz et al., 2005; Herrador et al., 2010). Rim8 functions as an adaptor between the 7TM protein Rim21 and endosomal sorting complex required for transport (ESCRT) in the pH-sensing RIM signaling pathway (Herrador et al., 2010). Rim21 is the putative sensor component of the pH-sensing machinery that also includes 7TM and 3TM proteins Dfg16 and Rim9 (Obara et al., 2012). According to the current model, Rim8-mediated recruitment of ESCRT provides a signaling platform that enables the proteolytic activation of the Rim101 transcription factor upon external alkalization (Maeda, 2012). In this process, the scaffold protein Rim20 links the ESCRT subunit Snf7 to Rim101 and Rim13, the putative protease involved in Rim101 cleavage (Xu and Mitchell, 2001; Boysen and Mitchell, 2006; Subramanian et al., 2012). Although earlier work suggested that Rim101 cleavage occurs at the endosome after endocytosis of the pH sensor machinery (Boysen and Mitchell, 2006; Subramanian et al., 2012), recent studies in both A. nidulans and S. cerevisiae instead support the idea that the overall process takes place at the plasma membrane (Galindo et al., 2012; Obara and Kihara, 2014).
α-Arrestins, like β-arrestins, are regulated by phosphorylation and ubiquitination (Becuwe et al., 2012a). We previously found that Rim8 monoubiquitination by the Rsp5 E3 ubiquitin ligase contributes to the recruitment of ESCRT (Herrador et al., 2010, 2013). In the present study, we investigated the mechanism of Rim8 activation, focusing on how Rim8 function is modulated by phosphorylation. Our results indicate that Rim8 is regulated through multisite phosphorylation of the interdomain hinge region by the plasma membrane–associated casein kinase 1. We present evidence that Rim8 phosphorylation prevents its accumulation at the plasma membrane at acidic pH and therefore inhibits RIM signaling. Our findings support a model in which casein kinase 1 functions as a molecular brake that inhibits signal transduction in the absence of stimulus.
RESULTS
Rim8 interacts with Rim21 at the plasma membrane
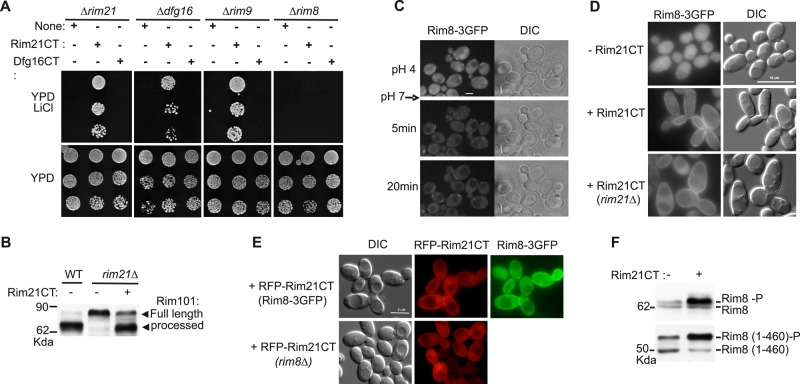
We previously reported that Rim8 interacts with a C-terminal fragment (C-tail) of the 7TM protein Rim21 in two-hybrid assays (Herrador et al., 2010). However, the interaction in the natural context of cellular membranes between Rim8 and the different components of the pH-sensing machinery had not been tested. We used the mating-based split-ubiquitin system (mbSUS) designed for membrane proteins (Obrdlik et al., 2004) to test for interaction between Rim8 and transmembrane proteins Rim21, Dfg16, and Rim9 (Figure 1A). Using this assay, we found that Rim8 interacts with Rim21 but not with its paralogue Dfg16 or with the 3TM protein Rim9 (Figure 1B). In additional, Dfg16 C-tail, in contrast to Rim21 C-tail, does not bind to Rim8 in two-hybrid assays (Figure 1C). A substitution (S86P) in the arrestin N-domain of the Rim8 homologue in A. nidulans, PalF, strongly impairs its interaction with the 7TM protein PalH (Herranz et al., 2005). The corresponding substitution in Rim8 (S76P) also disrupts its interaction with Rim21 C-tail without preventing Rim8 binding to the ESCRT subunit Vps23 (Figure 1C). By using lithium sensitivity as readout for the activity of the RIM pathway (Xu and Mitchell, 2001), we further showed that this mutation impairs RIM signaling (Figure 1D). This result, together with mbSUS data, supports the idea that Rim21 C-tail binding to Rim8 plays a major role in recruiting Rim8 to the pH-sensing machinery.
FIGURE 1:
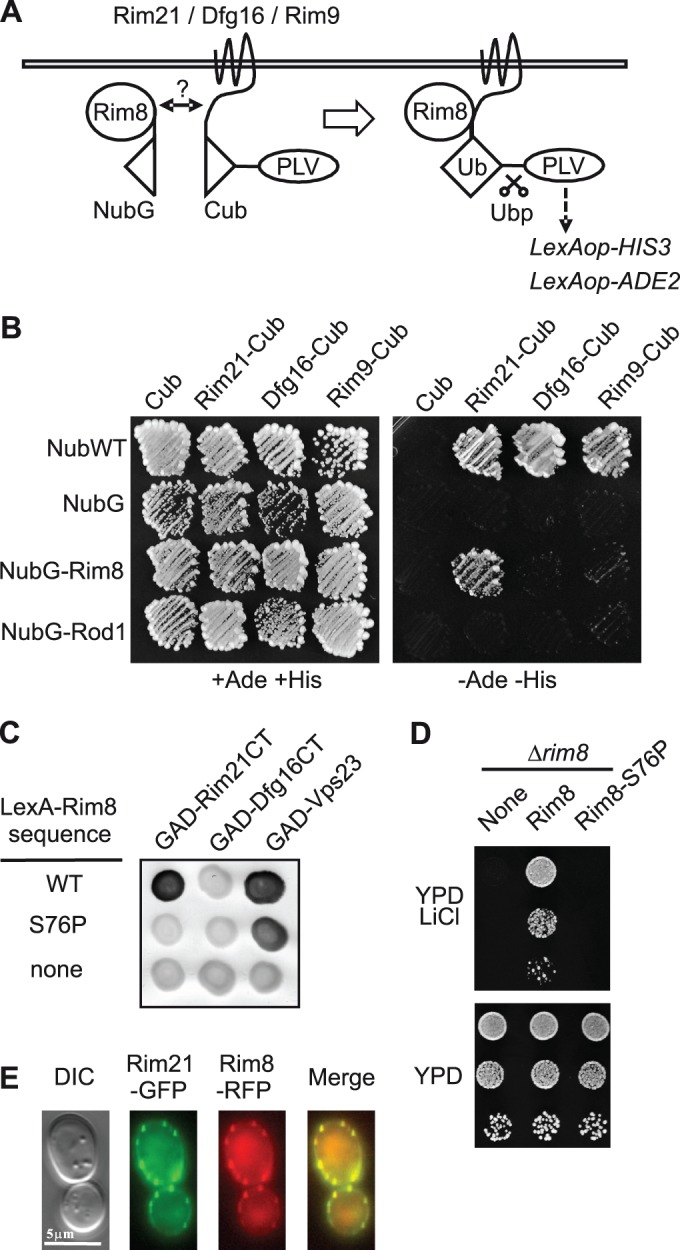
Interaction and colocalization of Rim8 with Rim21. (A) Schema of the mbSUS basic principle for detecting interaction between Rim8 and components of the pH-sensing machinery. The ubiquitin N-terminal fragment (NubG) carries the I13G substitution, which prevents nonspecific binding to the ubiquitin C-terminal fragment (Cub). Interaction between the two fusion proteins results in reconstitution of the split-ubiquitin heterodimer (Ub). Ubiquitin recognition and cleavage by a deubiquitinase (Ubp) leads to liberation of a transcription factor (PLV), which activates the HIS3 and ADE2 reporter genes. (B) mbSUS assay to test the interaction between Rim8 and Rim21, Dfg16, or Rim9. Diploid yeast cells coexpressing the indicated NubG- and Cub-PLV fused proteins were grown for 3 d on SD medium without adenine and histidine (–Ade –His) in order to detect interacting proteins (right) or on the same medium supplemented with adenine and histidine (+Ade +His) used as a control plate (left). Unfused NubG and NubG-Rod1 were used as negative controls and unfused NubWT as positive control. (C) Two-hybrid interactions between GAD fusions to Rim21 C-tail (Rim21CT), Dfg16 C-tail (Dfg16CT), or Vps23 and LexA fusions to Rim8 or a S76P mutant derivative. Positive interactions were revealed by β-galactosidase lift filter assays. (D) Phenotypic analysis of the Rim8-S76P mutant. Y04414 (rim8∆) was transformed with pHA-Rim8, a S76P mutant derivative, or the vector control pRS313 (none). Transformants were grown to stationary phase on SD medium, and serially diluted samples were spotted on YPD or YPD containing 200 mM LiCl. (E) Colocalization of Rim8 and Rim21. OVY137 (ADH1p-RIM21-GFP, ADH1p-RIM8-RFP, ADH1p-DFG16, ADH1p-RIM9) was grown to mid log phase in YPD medium and resuspended in SD medium. Cells were examined by fluorescence and differential interference contrast (DIC) microscopy.
We then examined the possible colocalization of Rim8 with Rim21. Because Rim21-GFP fluorescence was undetectable when expressed at endogenous levels, and since Rim21 levels are dependent on the other components of the pH-sensing machinery (Obara et al., 2012), we constructed a strain that co-overexpresses Rim21, Dfg16, and Rim9, as well as Rim8. To avoid possible artifacts of gene overexpression from multicopy plasmids, we expressed untagged or functional green fluorescent protein (GFP) and red fluorescent protein (RFP) fusion proteins under the control of the ADH1 promoter from genomically integrated constructs. In these conditions, Rim21-GFP and Rim8-RFP colocalize in discrete foci at the plasma membrane (Figure 1E). These findings provide additional evidence of Rim8-Rim21 association and support the idea that this interaction occurs at the plasma membrane.
Rim21 is required for pH-independent phosphorylation of Rim8
Previous studies suggest that α-arrestins are activated by dephosphorylation (Macgurn et al., 2011; Becuwe et al., 2012b; Merhi and Andre, 2012; O'Donnell et al., 2013). Therefore we proceeded to examine whether pH-dependent activation of the RIM pathway correlates with changes in the phosphorylation status of Rim8. On Western blots, Rim8 migrates as a major band and two minor bands of lower and higher mobility (Figure 2A, lane 1). We previously showed that the faint upper band corresponds to the monoubiquitinated protein (Herrador et al., 2010). This pattern remained largely unchanged after exposure to neutral-alkaline pH, although a small decrease in protein level was observed (Figure 2A, lane 2). Treatment with λ phosphatase resulted in a shift of the major band to the faster-migrating species and also increased the mobility of the ubiquitinated protein (Figure 2B, top). Taken together, these results indicate that both ubiquitinated and nonubiquitinated forms of Rim8 are largely phosphorylated at either acidic or neutral-alkaline pH.
FIGURE 2:
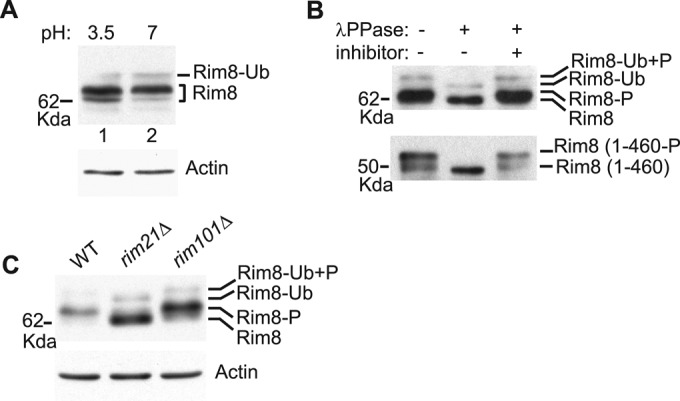
Phosphorylation of Rim8. (A) Effect of ambient pH on Rim8 electrophoretic mobility. Protein extracts from strain OVY247 (RIM8-3HA) prepared before and after a shift from pH 3.5 to 7 were prepared by glass bead disruption and immunoblotted with anti-HA (top) or anti-actin (bottom) antibodies. (B) Phosphatase assay. Y04414 (rim8∆) was transformed with pHA-Rim8 (top) or a C-terminal truncated derivative (residues 1–460; bottom), grown to mid log phase, and shifted to pH 7.5. Anti-HA–immunoprecipitated protein extracts were treated with λ phosphatase (λPPase) with or without phosphatase inhibitor and immunoblotted with anti-HA antibody. Positions of the different Rim8 forms (P, phosphorylated; Ub, ubiquitinated) are indicated. (C) Rim21-dependent phosphorylation of Rim8. OVY247 (RIM8-3HA), OVY270 (rim21∆ RIM8-3HA), and OVY271 (rim101∆ RIM8-3HA) were grown to mid log phase (final pH 3.5), and protein extracts were immunoblotted with anti-HA (top) or anti-actin (bottom) antibodies.
We then tested whether Rim8 phosphorylation is dependent on Rim21. Of interest, we found that Rim8 phosphorylation is strongly reduced in a rim21Δ mutant but not in a rim101Δ mutant, thus demonstrating that phosphorylation of Rim8 is dependent on Rim21 but independent of Rim101 processing (Figure 2C). In contrast, and in accordance with previous work (Herrador et al., 2010), neither Rim21 nor Rim101 is required for Rim8 ubiquitination. We also note that Rim8 levels are increased in the rim mutant strains, in agreement with previous findings that RIM8 is repressed in a negative feedback loop by the Rim101 transcription factor (Lamb and Mitchell, 2003).
Overexpression of Rim21 C-tail activates the RIM pathway and increases both Rim8 membrane localization and phosphorylation
Because Rim8 binding to Rim21 plays an essential role in RIM signaling (Figure 1D), we tested whether overexpression of the Rim21 C-tail fragment can bypass the requirement for the pH-sensing machinery in the activation of the RIM pathway. Lithium growth tests indicate that overexpression of Rim21 C-tail, but not Dfg16 C-tail, restores RIM signaling in rim21Δ, dfg16Δ, and rim9Δ mutants but not in a rim8Δ mutant (Figure 3A). Accordingly, Rim21 C-tail overexpression restores Rim101 processing in a rim21Δ mutant (Figure 3B). Thus overexpression of Rim21 C-tail can activate the RIM pathway in the absence of any component of the pH-sensing machinery but still in a Rim8-dependent manner.
FIGURE 3:
Effect of Rim21 C-tail overexpression on RIM signaling and Rim8 localization and phosphorylation. (A) Phenotypic analysis of rim mutant strains overexpressing Rim21 or Dfg16 C-tails (CT). Y01150 (rim21∆), Y11806 (dfg16∆), Y06196 (rim9∆), and Y04414 (rim8∆) were transformed with pADH1-Flag-Rim21CT, pADH1-Flag-Dfg16CT, or empty vector (none). Growth tests in the presence of LiCl were as described for Figure 1D. Immunoblot analysis of transformants expressing Rim21CT and Dfg16CT confirmed the expression of both proteins (unpublished data). (B) Effect of Rim21 C-tail overexpression on Rim101 processing. OVY24 (wild type [WT] RIM101-HA) and OVY43 (rim21∆ RIM101-HA) were transformed with pADH1-Flag-Rim21CT (+) or empty vector (–). After a shift to pH 7.5, protein extracts were prepared and immunoblotted with anti-HA antibody. (C) Effect of ambient pH on Rim8 localization. OVY172 (RIM8-3GFP) was grown to mid log phase in YPD medium and then injected into a microfluidics device in SD medium containing 0.1 M HEPES, pH 4. Cells were then perfused with SD medium containing 0.1 M HEPES, pH 7, and examined by fluorescence and DIC microscopy before and after the change of medium at the indicated time. (D) Rim8 localization in cells overexpressing Rim21 C-tail. OVY172 (RIM8-3GFP) and OVY304 (rim21∆ RIM8-3GFP) were transformed with pADH1-Flag-Rim21CT or empty vector as indicated on the left. Cells were grown to mid log phase on SD medium (final pH 3.5) and examined by fluorescence and DIC microscopy. (E) Colocalization of Rim8 and overexpressed Rim21 C-tail. OVY172 (RIM8-3GFP) and OVY31(rim8∆) were transformed with pADH1-RFP-Rim21CT, grown to mid log phase on SD medium (final pH 3.5), and examined by fluorescence and DIC microscopy. (F) Effect of Rim21 C-tail overexpression on Rim8 phosphorylation. Y04414 was cotransformed with pHA-Rim8 (top) or a Rim8 truncated derivative (residues 1–460; bottom) and pADH1-Flag-Rim21CT (+) or empty vector (–). Transformants were grown to mid log phase (final pH 3.5), and protein extracts were prepared and immunoblotted with anti-HA antibody.
A recent study showed that the activation of the RIM pathway correlates with an accumulation of Rim8 at the plasma membrane (Obara and Kihara, 2014). We confirmed these results by using time-lapse microscopy and a microfluidics system to monitor the subcellular localization of a functional Rim8-3GFP fusion expressed from its chromosomal locus. At acidic pH, Rim8-3GFP was mainly present in the cytosol, although some punctuate structures of weak intensity were also detected (Figure 3C; see also Figure 3D). However, within 5 min after a shift from pH 4 to 7, Rim8-3GFP was recruited to bright, discrete foci at the plasma membrane, and these foci were still detected 30 min after the shift (Figure 3C and Supplemental Figure 3C.movie).
We then examined whether Rim21 C-tail overexpression affects Rim8 localization. Overexpressed Rim21 C-tail markedly increases Rim8-3GFP localization at the plasma membrane and produces an elongated cell phenotype in both wild-type and rim21Δ mutant strains (Figure 3D). We further showed that overexpressed RFP-Rim21C-tail colocalizes with Rim8-3GFP at the plasma membrane and is also detected at the cell surface in a ∆rim8 mutant (Figure 3E). These results suggest that Rim21 C-tail can associate with the plasma membrane to mediate the recruitment of Rim8. In addition, we found that the amount of phosphorylated Rim8 or a C-terminal-truncated derivative increase in cells overexpressing Rim21 C-tail (Figure 3F). Thus accumulation of Rim8 at the plasma membrane correlates with an increase of Rim8 phosphorylation.
The interdomain connector (hinge) of Rim8 is phosphorylated by the plasma membrane–associated casein kinase 1
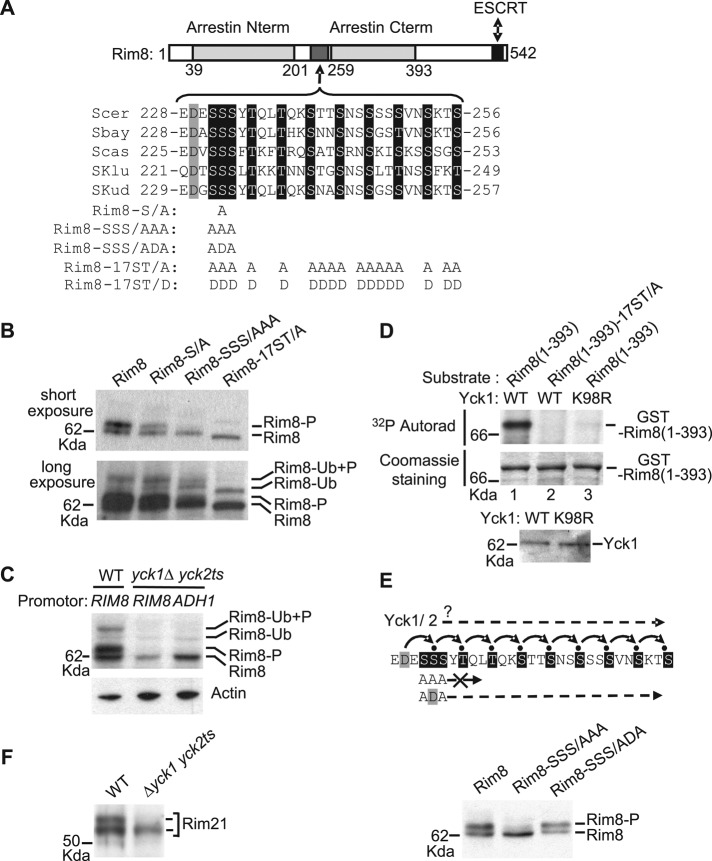
Rim8 has two arrestin domains that interact with Rim21, and a C-terminal region that contains the ubiquitination site (K521) and binds to ESCRT (Figure 4A; Herrador et al., 2010). A C-terminal-truncated derivative of Rim8 (residues 1–460) is still phosphorylated (Figure 2B, bottom), indicating that major phosphorylation sites reside in the N-terminal part of the protein, which contains the arrestin domains. Rim8 contains a Ser/Thr-rich region (residues 231–256) between the two arrestin domains (17 Ser/Thr residues from a total of 26; Figure 4A). The corresponding interdomain connector in β-arrestins is referred to as the “hinge” region and plays a key role in the conformational changes of these proteins upon 7TM protein binding (Vishnivetskiy et al., 2002). We then tested whether this Ser/Thr-rich region in Rim8 is phosphorylated. Alanine substitutions of S232, SSS(231–233), or the 17 Ser/Thr residues present in the Ser/Thr-rich region led to a progressive disappearance of the phosphorylated forms of both ubiquitinated and nonubiquitinated Rim8 (Figure 4B). These results indicate that the Ser/Thr-rich region in the interdomain connector of Rim8 is responsible for Rim8 phosphorylation. In addition, it shows that phosphorylation is not a prerequisite for Rim8 ubiquitination.
FIGURE 4:
Casein kinase 1–mediated phosphorylation of Rim8 interdomain connector (hinge). (A) Conservation of a cluster of Ser/Thr residues in the hinge region of Rim8 orthologues in the Saccharomyces clade. Top, schematic representation (to scale) of Rim8, showing the positions of the arrestin N-terminal and C-terminal PFAM domains, the ESCRT binding region, and the Ser/Thr cluster. The alignment (ClustalW) shows the conserved Asp (gray shading) and Ser/Thr (black shading) residues in S. cerevisiae (Scer), Saccharomyces bayanus (Sbar), Saccharomyces castellii (Scas), Saccharomyces kluyveri (Sklu), and Saccharomyces kudriavzevii (Skud). The position and amino acid substitutions for each Rim8 mutant are shown below. (B) Phosphorylation of Rim8 hinge region. Y04414 (rim8∆) was transformed with pHA-Rim8 or the mutant derivatives pHA-Rim8-S/A, pHA-Rim8-SSS/AAA, and pHA-Rim8-17ST/A. After a shift to pH 7.5, protein extracts were prepared by glass bead disruption, immunoblotted with anti-HA, and visualized after short (top) or long (bottom) autoradiography exposure to detect ubiquitinated species. Position of phosphorylated (P) and/or ubiquitinated (Ub) Rim8 is indicated. Note that the Rim8-17ST/A mutant migrates with slightly higher mobility than wild- type Rim8. (C) Casein kinase 1–dependent phosphorylation of Rim8. LRB341 (WT) and LRB362 (yck1∆ yck2ts) were transformed with pHA-Rim8 or pADH1-HA-Rim8, expressing HA-Rim8 under its own promoter or ADH1 promoter, respectively. Transformants were grown at a semipermissive temperature for yck2ts (30ºC). After a shift to pH 7.5, protein extracts were prepared by glass bead disruption and immunoblotted with anti-HA (top) or anti-actin (bottom) antibodies. (D) In vitro phosphorylation of Rim8 hinge region by casein kinase 1. Top, in vitro phosphorylation assay of GST-Rim8(1-393), or the corresponding mutant derivative GST-Rim8(1-393)-17ST/A, by affinity-purified His-tagged Yck1 or inactive Yck1-K98R. The 32P incorporation and protein levels were determined by autoradiography (top) and Coomassie staining (bottom), respectively. Bottom, immunoblot analysis of purified His-tagged Yck1 and Yck1-K98R with anti-His antibody (10% of the reaction). (E) Sequential phosphorylation of Rim8. Top, model for the cascade of CK1-mediated phosphorylations of Rim8. Small black circles represent phosphate groups. Bottom, Y04414 (rim8∆) was transformed with pHA-Rim8 or the mutant derivatives pHA-Rim8-SSS/AAA and pHA-Rim8-SSS/ADA. Protein extracts were prepared by glass bead disruption and immunoblotted with anti-HA antibody. (F) Same assay as in C with Rim21. LRB341 (WT) and LRB362 (yck1∆ yck2ts) were transformed with pRim21-HA, and protein extracts were immunoblotted with anti-HA antibody.
Of note, 13 of the 17 Ser/Thr residues in the Ser/Thr-rich region fit the plasma membrane–associated casein kinase 1 (Yck1/2) target-site consensus (D/E-X-X-S/T and S/T-X-X-S/T; Flotow et al., 1990; Knippschild et al., 2005). We then examined whether Rim8 is phosphorylated in a yck1Δ yck2ts temperature-sensitive mutant at the semipermissive temperature of 30ºC. Although Rim8 protein levels are lower in this strain, the phosphorylated species are undetectable, even when Rim8 is overexpressed under the control of the ADH1 promoter (Figure 4C). Therefore in vivo phosphorylation of Rim8 is dependent on casein kinase 1. To show further that Rim8 is a substrate of casein kinase 1, we used C-terminal-truncated Rim8 (residues 1–393, fused to glutathione S-transferase (GST) and purified from Escherichia coli) as substrate in an in vitro phosphorylation assay of Yck1 (fused to a heptahistidine tag and purified from yeast). Yck1 catalyzed the phosphorylation of this substrate (Figure 4D, lane 1) and phosphorylation was prevented by either removing the 17 Ser/Thr residues in the Rim8 hinge region (Figure 4D, lane 2) or inactivating Yck1 catalytic activity (Yck1-K98R; Figure 4D, lane 3). Thus, together with in vivo data, these results indicate that this cluster of serines and threonines is a major site of Rim8 phosphorylation by casein kinase 1.
Casein kinase 1 optimal consensus sites require either priming phosphorylation or a phosphomimetic residue (Asp/Glu) at the P-3 position (Flotow et al., 1990; Knippschild et al., 2005). Multiple sequence alignment of Rim8 orthologues in the Saccharomyces clade shows that most of the potential casein kinase 1 phosphorylation sites are conserved and form a stretch of nine adjacent repeats of the S/T-X-X-S/T motif, preceded by an Asp residue at the P-3 position of the first motif (Figure 4A). This striking pattern suggests that these S/T residues are phosphorylated sequentially and that the initial phosphorylation event does not require priming phosphorylation (Figure 4E, top). Such a mechanism would explain why elimination of the SSS(231–233) triplet is almost sufficient to prevent Rim8 phosphorylation (Figure 4B). Reintroduction of a phosphomimetic Asp residue at position 232 in this mutant restores Rim8 phosphorylation (Figure 4E, bottom), thus showing that the strong effect of the SSS(231-233)AAA substitution is mainly due to the inhibition of the phosphorylation cascade. This finding indicates that multisite phosphorylation of the Rim8 hinge involves a cascade of casein kinase 1 (CK1)–mediated phosphorylations.
A previous study reported that Rim21 is also phosphorylated, with a concomitant shift in gel mobility (Obara et al., 2012). We found that Rim21 migrates as a doublet, which is reduced to a single band in the yck1Δ yck2ts mutant. These results thus suggest that Rim21 phosphorylation is also dependent on casein kinase 1 activity.
Casein kinase 1 negatively modulates the RIM signaling pathway
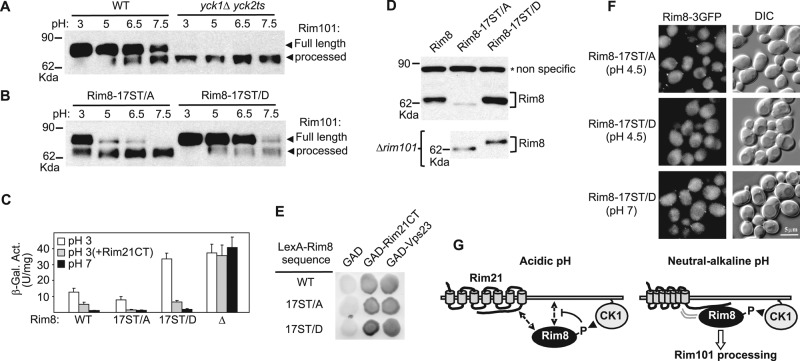
Our findings implicate casein kinase 1 as a potential new player in the pH-sensing RIM signaling pathway. We then tested whether RIM signaling is altered in a yck1Δ yck2ts mutant at the semipermissive temperature of 30ºC. Strikingly, we found that Rim101 processing is constitutive and pH independent in this mutant (Figure 5A), thus indicating that casein kinase 1 is a negative regulator of the RIM signaling pathway.
FIGURE 5:
Regulation of the RIM signaling pathway by casein kinase 1. (A) Constitutive Rim101 processing in casein kinase 1 mutant cells. OVY243 (WT) and OVY143 (yck1∆ yck2ts) were grown at the indicated pH values and at a semipermissive temperature for yck2ts (30ºC). Protein extracts were immunoblotted with anti-HA antibody to detect Rim101-HA. (B–F) Effect of phosphomimetic or phosphodeficient mutations in RIM8 on RIM signaling and Rim8 function. (B) Same experiment as in A with OVY244 (RIM8-17ST/A) and OVY245 (RIM8-17ST/D). (C) Y04414 (rim8∆) was transformed with the plasmid pLGn+3xNRE22D carrying a CYC-NRE-lacZ reporter and pFlag-Rim8 (WT), the mutant derivative pFlag-Rim8-17ST/A (17ST/A) or pFlag-Rim8-17ST/D (17ST/D), or the empty vector (∆). When indicated, cells were also cotransformed with pADH1-Rim21CT(L). Values are average β-galactosidase activity for four to seven transformants grown to mid log phase at pH 3 or 7. (D) Top, Y04414 (rim8∆) was transformed with pFlag-Rim8 or the mutant derivative pFlag-Rim8-17ST/A or pFlag-Rim8-17ST/D and grown to mid log phase (final pH 3.5), and protein extracts were immunoblotted with anti-Flag antibody. Bottom, Y00936 (rim101∆) was transformed with pHA-Rim8-17ST/A or pHA-Rim8-17ST/D and grown to mid log phase (final pH 3.5), and protein extracts were immunoblotted with anti-HA antibody. (E) Two-hybrid assay as in Figure 1C between GAD fusions to Rim21 C-tail (Rim21CT) or Vps23 and wild-type LexA-HA-Rim8 or the indicated mutant derivatives. (F) OVY264 (RIM8(17ST/A)-3GFP) and OVY265 (RIM8(17ST/D)-3GFP) were grown to mid log phase in YPD medium (final pH 5.8) and resuspended in SD medium at pH 4.5 or 7. Cells were examined by fluorescence and DIC microscopy. (G) Model for transduction of the pH signal from Rim21 to Rim8 (see the text for a description). Dashed arrows and arrowheads indicate weak interactions and phosphorylation events, respectively.
We further examined whether the lack of Rim8 phosphorylation in the yck1Δ yck2ts mutant accounts for the constitutive activation of the RIM pathway. Rim101 processing is partially constitutive in the phosphodeficient mutant Rim8-17ST/A, whereas this effect is not observed in the phosphomimetic mutant Rim8-17ST/D, in which the 17 Ser/Thr residues of the interdomain connector have been substituted with aspartate (Figure 5B). These results indicate that the casein kinase 1 mutant phenotype can be explained, at least partially, by a lack of phosphorylation of Rim8.
To corroborate these findings, we used the Rim101-dependent reporter CYC1-NRE-lacZ (Rothfels et al., 2005). At acidic pH (pH 3), repression of this reporter by Rim101 is increased in the Rim8-17ST/A mutant, in agreement with overactivation of the RIM pathway, and decreased in the Rim8-17ST/D mutant, indicating that basal signaling is impaired in these conditions (Figure 5C). Because RIM8 itself is repressed by Rim101 in a negative feedback loop (Lamb and Mitchell, 2003), overactivation of the RIM pathway would be expected to cause reduced expression of RIM8. Indeed, the 17ST/A mutation leads to a decrease of Rim8 levels, whereas the 17ST/D mutation has the opposite effect (Figure 5D, top). This difference is not observed in a Δrim101 mutant (Figure 5D, bottom), fitting with the idea that it results from interference with RIM signaling. Taken together, these data demonstrate that Rim8 phosphorylation by casein kinase 1 negatively modulates the function of this protein at acidic pH.
As expected, we found that neutral pH activates the RIM pathway and fully represses the CYC1-NRE-lacZ reporter in a wild-type strain (Figure 5C). Rim21 Ctail overexpression in wild-type cells grown at acidic pH also activates the RIM pathway, albeit only partly, as shown by the partial repression of the CYC1-NRE-lacZ reporter (Figure 5C). In both cases, and in agreement with the results obtained in the analysis of Rim101 processing at pH 7.5 (Figure 5B), the activation of the RIM pathway is not impaired by the phosphomimetic mutation Rim8-17ST/D (Figure 5C). These data suggest that Rim8 phosphorylation is inhibitory only at acidic pH and that an alternative mechanism overrides this inhibition at neutral-alkaline pH.
We then tested whether phosphorylation of Rim8 disrupts its interaction with Rim21 C-tail or the ESCRT subunit Vps23. However, two-hybrid assays show that both phosphomimetic and phosphodeficient Rim8 mutants strongly interact with either protein (Figure 5E). By contrast, we found that these mutations have a differential effect on the localization of Rim8 at acidic pH. Numerous foci are detected with the phosphodeficient mutant Rim8-17ST/A, whereas no foci are observed with the phosphomimetic mutant Rim8-17ST/D (Figure 5F). These results support the idea that Rim8 phosphorylation prevents its accumulation in foci at the plasma membrane at acidic pH. In contrast, and in agreement with the results described, the Rim8-17ST/D mutation does not prevent the formation of foci at neutral pH, thus confirming that Rim8 phosphorylation is inhibitory only at acidic pH (Figure 5F).
DISCUSSION
Previous work indicates that Rim21 is the sensor component of the pH-sensing machinery, whereas Rim9 and Dfg16 play an auxiliary role (Obara et al., 2012). Our results also support an essential role for Rim21 in the transmission of the pH signal: Rim21 interacts with Rim8 in both split-ubiquitin and two-hybrid assays, and overexpression of the C-terminal cytoplasmic tail of Rim21 is sufficient to activate the RIM pathway. In addition, we showed that a mutation that disrupts the interaction between Rim8 and Rim21 C-tail blocks RIM signaling. By co-overexpressing the different components of the pH-sensing machinery, we showed that Rim8 and Rim21 colocalize in discrete foci at the plasma membrane. These data are consistent with previous findings that both Rim8 and Rim21 are visualized as punctate foci at the plasma membrane and that Rim8 foci are dependent on Rim21 (Obara et al., 2012; Obara and Kihara, 2014). In addition, and as recently reported (Obara and Kihara, 2014), we found that Rim8 accumulates at the plasma membrane after exposure to neutral-alkaline pH. By contrast, Rim21 foci are detected in the absence of pH signal (Obara et al., 2012). Taken together, these results support the idea that the pH signal elicits the recruitment of Rim8 to Rim21 at the plasma membrane. Strikingly, we observed that Rim21 C-tail overexpression at acidic pH could partly mimic the effect of neutral-alkaline pH and activate the RIM pathway. This finding raises the possibility that activation of this pathway involves a conformational change of Rim21 that enhances the accessibility of the C-tail to Rim8. We also found that overexpressed Rim21 C-tail could mediate the recruitment of Rim8 to the plasma membrane, suggesting that this region of Rim21 possesses some lipid-binding activity.
We showed both in vivo and in vitro that a Ser/Thr-rich region in the interdomain hinge of Rim8 is multisite phosphorylated by the plasma membrane–associated casein kinase 1 (CK1). Of note, a recent study showed that β-arrestin-2 hinge is phosphorylated by mitogen-activated protein kinase to regulate G protein–coupled receptor intracellular trafficking (Khoury et al., 2014). We present evidence that the Ser/Thr residues in Rim8 hinge are phosphorylated sequentially rather than independently of one another. In addition, the presence of acidic residues at the P-3 position of each of the Ser residues SSS(231–233) (Figure 4A) supports the idea that the initial phosphorylation event does not require priming phosphorylation by another kinase. In fact, we found that Rim8 accumulation at the plasma membrane in cells overexpressing Rim21 C-tail correlates with enhanced Rim8 phosphorylation, consistent with the hypothesis that plasma membrane localization is the main determinant of Rim8 phosphorylation by casein kinase 1. Intriguingly, we observed that Rim21 C-tail overexpression also results in an elongated cell phenotype reminiscent of that caused by loss of casein kinase 1 activity (Robinson et al., 1993). This effect suggests that the prominent plasma membrane localization of Rim21 C-tail and Rim8 in these cells may titer out casein kinase 1.
Our results identified casein kinase 1 as a negative regulator of the RIM pathway that inhibits signal transduction at acidic pH. Previous studies established that both glucose- and amino acid–sensing mechanisms also involve casein kinase 1 (Moriya and Johnston, 2004; Liu et al., 2008), which thus stands as a key player in plasma membrane signaling events. By using phosphomimetic and phosphodeficient Rim8 mutants, we showed that CK1-mediated phosphorylation of Rim8 contributes to the inhibition of the RIM pathway at acidic pH. These findings, together with those of previous studies (O'Donnell, 2012), suggest that phosphoinhibition is a common regulatory mechanism for α-arrestins. Our results indicate that the nonphosphorylatable Rim8 mutant, unlike the casein kinase 1 mutant, only confers partial constitutive activation of the RIM pathway. Although in vivo phosphorylation of this mutant is undetectable, we cannot discard the possibility that additional CK1 phosphorylation sites in Rim8 contribute to this difference. Alternatively, casein kinase 1 may phosphorylate other components of the RIM pathway. The change in Rim21 electrophoretic mobility in a casein kinase 1 mutant points toward this protein as a likely candidate.
Unexpectedly, we found that Rim8 is phosphorylated in a pH-independent but Rim21-dependent manner. Rim8 phosphorylation is strongly reduced in a rim21Δ mutant and markedly enhanced upon overexpression of Rim21 C-tail. These results support the idea that even at acidic pH, Rim21 interacts with Rim8 to induce its phosphorylation. However, Rim8 foci are barely detectable in these conditions, suggesting that this interaction is transient, albeit sufficient to trigger Rim8 phosphorylation by the plasma membrane–associated casein kinase 1. Rim8 phosphorylation after Rim21 binding could induce Rim8 release from the plasma membrane. In keeping with this hypothesis, we found that a Rim8 phosphodeficient mutant accumulates in foci at the plasma membrane at acidic pH. Of note, a similar mechanism has been proposed for the Art1 α-arrestin: the transient association between Art1 and the Npr1 protein kinase at the plasma membrane would trigger Art1 phosphorylation, thereby limiting its plasma membrane association (Macgurn et al., 2011). Phosphorylation of the Rod1/Art4 α-arrestin by the Snf1 protein kinase has also been proposed to block its association with the trans-Golgi membrane (Becuwe and Leon, 2014). The mechanism, by which Rim8 phosphorylation prevents its accumulation at the plasma membrane at acidic pH and therefore inhibits RIM signaling, remains unknown. Two-hybrid assays suggest that Rim8 phosphorylation does not impair the interaction with Rim21 C-tail. However, these assays may not reflect the in vivo situation, and multisite phosphorylation of the interdomain hinge could affect its flexibility, which appears to play an important role in the conformational change of visual arrestin upon binding to rhodopsin (Vishnivetskiy et al., 2002). On the other hand, previous work reported that β-arrestins bind phospholipids, and it has been proposed that this interaction may provide, together with the 7TM protein, a multipoint attachment of arrestin to the plasma membrane (Gaidarov et al., 1999). By analogy, one might speculate that Rim8, like β-arrestins, associates with phospholipids at the plasma membrane and that introduction of multiple negative charges by the phosphorylation cascade could disrupt such interaction.
Our results indicate that Rim8 is largely phosphorylated at both acidic and neutral-alkaline pH. Thus Rim8 regulation is significantly different in diverse fungal species, since homologues in A. nidulans and Candida albicans are phosphorylated in response to the pH signal (Herranz et al., 2005; Gomez-Raja and Davis, 2012). Although earlier studies indicated that most α-arrestins are activated by dephosphorylation (O'Donnell, 2012), recent reports showed that a single α-arrestin can be regulated by different mechanisms (Crapeau et al., 2014; Ghaddar et al., 2014). For instance, dephosphorylation of the Bul α-arrestins is required to mediate the endocytosis of the yeast general amino acid permease Gap1 in response to internal amino acids but not for substrate-induced endocytosis of Gap1, which appears to depend on a conformational change within the permease (Ghaddar et al., 2014). The latter mechanism may also apply to the RIM pathway: as discussed earlier, the pH sensor protein Rim21 could undergo a conformational change in response to neutral-alkaline pH that increases the accessibility of Rim21 C-tail and overrides the inhibitory effect of Rim8 phosphorylation by casein kinase 1. In keeping with this hypothesis, phosphomimetic mutations in Rim8 are sufficient to block the weak RIM signaling activity at acidic pH but do not prevent the activation of the RIM pathway at neutral-alkaline pH or in cells overexpressing Rim21 C-tail.
On the basis of these and other results, we propose a model for the activation of the RIM pathway (Figure 5G). At acidic pH, Rim21 adopts an inactive conformation in which the Rim21 C-tail is only partially accessible. Transient binding of Rim8 to Rim21 C-tail triggers Rim8 phosphorylation by CK1, which could lock Rim8 in an inactive conformation and/or prevent Rim8 binding to lipids, precluding its stable association with the plasma membrane. At neutral-alkaline pH, Rim21 undergoes a conformational change that increases the accessibility of Rim21 C-tail. The strong interaction of Rim8 with Rim21 overrides the CK1-mediated inhibition and stabilizes its association with the plasma membrane. Rim8 ubiquitination appears to be an early event, since ubiquitinated Rim8 is phosphorylated in a Rim21-dependent manner. We previously showed that the ubiquitinated form of Rim8 is found in complex with the ESCRT-I subunit Vps23 (Herrador et al., 2010, 2013), suggesting that Rim8-bound ESCRT-I complex can be recruited to the plasma membrane. The stable association of Rim8 with the plasma membrane would then promote the assembly of ESCRT and the associated Rim101 processing machinery, resulting in Rim101 cleavage and activation.
MATERIALS AND METHODS
Strains and genetic methods
The S. cerevisiae strains used in this study are shown in Table 1. OVY24, OVY227, and OVY143 expressing hemagglutinin (HA)-tagged RIM101 were constructed by transforming the strains Y00000, Y01150, Y11806, and LRB362 with a fragment of plasmid pKR41, as previously described (Rothfels et al., 2005). OVY243, OVY244, and OVY245 are derivatives of OVY24 in which the RIM8 gene has been replaced by Flag-tagged RIM8 (Herrador et al., 2010) or mutant derivatives containing multiple Ser/Thr-to-Ala (17ST/A) or -Asp (17ST/D) substitutions. OVY172, OVY264, and OVY265 were constructed using the same procedure with the Y00000 strain and a derivative of pRim8-GFP (Herrador et al., 2010) containing a triple-GFP tag. OVY304 is a derivative of OVY172 in which the RIM21 gene has been deleted. OVY247 was constructed by gene replacement of the RIM8 gene by HA-tagged RIM8 (Herrador et al., 2010) in Y00000, and OVY270 and OVY271 are derivatives of OVY247 in which the RIM21 and RIM101 genes have been deleted, respectively. OVY137 was constructed by genomic integration of pRS303, pRS304, pRS305, and pRS306 derivatives (Sikorski and Hieter, 1989), expressing Rim8-RFP, Rim9, Rim21-GFP, and Dfg16 under the control of the ADH1p promoter at the HIS3, TRP1, LEU2, and URA3 locus of W303-1A, respectively. CTY10.5d was used for two-hybrid experiments. Standard genetic methods were followed, and yeast cultures were grown in yeast extract/peptone/dextrose (YPD) medium or synthetic dextrose (SD) medium lacking appropriate amino acids to maintain selection for plasmids (Rose et al., 1990). To test for lithium sensitivity of rim mutant strains (Xu and Mitchell, 2001), serial dilutions of cultures were spotted onto YPD plates containing 200 mM LiCl.
TABLE 1:
Yeast strains used in this study.
| Strain | Genotype | Reference/source |
|---|---|---|
| LRB341 | MATa his3 leu2 ura3-52 | Panek et al. (1997) |
| LRB362 | MATa his3 leu2 ura3-52 yck1-1::ura3 yck2-2ts | Panek et al. (1997) |
| Y00000 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 | EUROSCARF |
| Y04414 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 rim8::kanMX4 | EUROSCARF |
| Y01150 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 rim21::kanMX4 | EUROSCARF |
| Y11806 | MATα his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 dfg16::kanMX4 | EUROSCARF |
| Y06196 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 rim9::kanMX4 | EUROSCARF |
| OVY24 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 RIM101-HA::LEU2 | This study |
| OVY43 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 rim21::kanMX4 RIM101-HA::LEU2 | This study |
| OVY143 | MATa his3 leu2 ura3-52 yck1-1::ura3 yck2-2ts RIM101-HA::LEU2 | This study |
| THY.AP4 | MATa ura3 leu2 lexA::lacZ::trp1 lexA::HIS3 lexA::ADE2 | Obrdlik et al. (2004) |
| THY.AP5 | MATα URA3 leu2 trp1 his3 loxP::ade2 | Obrdlik et al. (2004) |
| CTY10.5d | Mata ade2-101 his3-Δ200 leu2-Δ1 trp1-Δ901gal4 gal80 URA3::lexAop-lacZ | Bartel et al., 1993) |
| OVY137 | Mata ura3-1 his3-11 leu2-3_11; trp1Δ2 ade2-1 can1-100 ADH1p-RIM9::TRP1 ADH1p-RIM8-RFP::HIS3 ADH1p-DFG16::URA3 ADH1p-RIM21-GFP::LEU2 | This study |
| OVY172 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 RIM8-3GFP | This study |
| OVY243 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 FLAG-RIM8 RIM101-HA::LEU2 | This study |
| OVY244 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 FLAG-RIM8-17ST/A RIM101-HA::LEU2 | This study |
| OVY245 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 FLAG-RIM8-17ST/D RIM101-HA::LEU2 | This study |
| OVY304 | MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 rim21::kanMX4 RIM8-3GFP | This study |
EUROSCARF, European Saccharomyces cerevisiae Archive for Functional analysis, Frankfurt, Germany.
Plasmids
Plasmids and vectors used in this study are listed in Table 2. The mbSUS vectors were prepared as described previously (Obrdlik et al., 2004). pADH1-Flag-Rim21CT and pADH1-RFP-Rim21CT were constructed by cloning the Rim21 C-tail (residues 327–533) in the polylinker of a derivative of pWS93 (Song and Carlson, 1998) in which the triple-HA tag had been replaced by a triple-Flag epitope or the mCherry RFP sequence. The same procedure was used to clone the Dfg16 C-tail (residues 426–619) in pADH1-Flag-Dfg16CT and construct pADH1-Flag-Rim21CT(L), except that the parental vector is a derivative of pSK134 (Vincent and Carlson, 1999) containing a triple-Flag epitope. Dfg16 C-tail and C-terminal truncated Rim8 were cloned in pACT2 and pGEX-5X-1 to give pGAD-Dfg16CT and pGST-Rim8(1-393). respectively. pLexA-HA-Rim8 and pADH1-HA-Rim8 were constructed by cloning a cassette containing the triple HA-tagged RIM8 ORF from pHA-Rim8 (Herrador et al., 2010) in pLexA(1-202)+PL and a pRS313 derivative containing the ADH1 promotor and terminator, respectively. The Yck1 open reading frame was cloned in a pYEX4T-1 (Clontech, Palo Alto, CA) derivative in which GST was replaced by a heptahistidine tag. pRim21-HA, encoding C-terminal HA tagged Rim21 under the control of its native promoter, is a derivative of pRS315. Rim8 and Yck1 missense and truncation mutations were obtained by using mutagenic PCR. pLGn+3xNRE22D carries a CYC1-NRE-lacZ reporter (Rothfels et al., 2005).
TABLE 2:
Plasmids used in this study.
| Plasmid | Expressed protein; vector | Experiment | Reference/source |
|---|---|---|---|
| pNXgate-Rim8 | NubG-3HA-Rim8; pNXgate | mbSUS | This study |
| pNXgate-Rod1 | NubG-3HA-Rod1; pNXgate | mbSUS | This study |
| MetYCgate-Rim21 | Cub-PLV-Rim21; pMetYCgate | mbSUS | This study |
| MetYCgate-Dfg16 | Cub-PLV-Dfg16; pMetYCgate | mbSUS | This study |
| MetYCgate-Rim9 | Cub-PLV-Rim9; pMetYCgate | mbSUS | This study |
| pLexA-HA-Rim8 | LexA-HA-Rim8; pLexA(1-202)+PL | Two-hybrid | Herrador et al. (2010) |
| pLexA-HA-Rim8-S76P | LexA-HA-Rim8-S76P; pLexA(1-202)+PL | Two-hybrid | This study |
| pGAD-Rim21CT | GAD-Rim21(327-533); pACT2 | Two-hybrid | Herrador et al. (2010) |
| pGAD-Dfg16CT | GAD-Dfg16(426-619); pACT2 | Two-hybrid | This study |
| pGAD-Vps23 | GAD-Vps23; pACT2 | Two-hybrid | Herrador et al. (2010) |
| pHA-Rim8 | HA-Rim8; pRS313 | Western blot | Herrador et al. (2010) |
| pHA-Rim8-K521R | HA-Rim8-K521R; pRS313 | Western blot | Herrador et al. (2010) |
| pHA-Rim8-S76P | HA-Rim8-S76P; pRS313 | Western blot | This study |
| pHA-Rim8(1-460) | HA-Rim8(1-460); pRS313 | Western blot | This study |
| pHA-Rim8(1-460)-S76P | HA-Rim8(1-460)-S76P; pRS313 | Western blot | This study |
| pHA-Rim8-1S/A | HA-Rim8-S232A; pRS313 | Western blot | This study |
| pHA-Rim8-3S/A | HA-Rim8-SSS(231-233)AAA; pRS313 | Western blot | This study |
| pHA-Rim8-17ST/A | HA-Rim8-17ST(231-256)A; pRS313 | Western blot | This study |
| pHA-Rim8-17ST/D | HA-Rim8-17ST(231-256)D; pRS313 | Western blot | This study |
| pFlag-Rim8 | Flag-Rim8; pRS313 | Western blot | Herrador et al. (2010) |
| pFlag-Rim8-17ST/A | Flag-Rim8-17ST(231-256)A; pRS313 | Western blot | This study |
| pFlag-Rim8-17ST/D | Flag-Rim8-17ST(231-256)D; pRS313 | Western blot | This study |
| pADH1-HA-Rim8 | HA-Rim8; pRS313 | Western blot | This study |
| pADH1-Flag-Rim21CT | Flag-Rim21(327-533); pWS93 derivative | Western blot | This study |
| pADH1-Flag-Rim21CT(L) | Flag-Rim21(327-533); pSK134 derivative | β-Galactosidase assays | This study |
| pADH1-RFP-Rim21CT | RFP-Rim21(327-533); pWS93 derivative | Fluorescence microscopy | This study |
| pADH1-Flag-Dfg16CT | Flag-Dfg16(426-619); pWS93 derivative | Western blot | This study |
| pGST-Rim8(1-393) | GST-Rim8(1-393); pGEX-5X-1 | In vitro phosphorylation | This study |
| pGST-Rim8(1-393)-17ST/A | GST-Rim8(1-393)-17ST(231-256)A; pGEX-5X-1 | In vitro phosphorylation | This study |
| pCUP1-His7-Yck1 | His7-Yck1; pYEX4T-1 derivative | In vitro phosphorylation | This study |
| pCUP1-His7-Yck1-K98R | His7-Yck1-K98R; pYEX4T-1 derivative | In vitro phosphorylation | This study |
| pRim21-HA | Rim21-HA; pRS315 | Western blot | This study |
Vectors: pNubWT-2, pNXgate, and pMetYCgate (Obrdlik et al., 2004); pLexA(1-202)+PL (Ruden et al., 1991); pACT2 (Clontech); pRS313 and pRS315 (Sikorski and Hieter, 1989); pGEX-5X-1 (GE Healthcare); pYEX4T-1 (Clontech), pWS93 (Song and Carlson, 1998); pSK134 (Vincent and Carlson, 1999).
β-Galactosidase assays
Two-hybrid interactions were detected by filter lift assays with X-gal, as described previously (Yang et al., 1992). For analysis of the CYC1-NRE-lacZ reporter, transformants were grown in a selective SD medium titrated to pH 3 with HCl or buffered with 0.1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) at pH 7, and β-galactosidase activity was measured with protein extracts prepared by cell disruption with glass beads and normalized to the protein concentration of the extract (Guarente, 1983).
Split-ubiquitin assays
The mbSUS strains and plasmids used in this study are shown in Tables 1 and 2, respectively. The assay was carried out as described previously (Obrdlik et al., 2004), and interactions were detected via growth selection on SD medium lacking histidine and adenine.
Immunoblot
Yeast cells were grown until mid log phase in selective SD medium (final pH 3.5) and, when indicated, shifted for 30 min to neutral-alkaline conditions (pH 7–7.5) with KOH to activate the RIM pathway (Herrador et al., 2010). For growth at different pH values, mid log cultures were diluted fivefold into fresh SD medium buffered with 0.1 M HEPES at the appropriate pH and grown for 5 h. Protein extracts were prepared by the NaOH/trichloroacetic acid (TCA) lysis method (Volland et al., 1994) or, when indicated, by cell disruption with glass beads in IP buffer (50 mM HEPES, pH 7.5/150 mM NaCl/0.1% Triton X-100/1 mM dithiothreitol [DTT]/10% glycerol/5 mM N-ethylmaleimide/Complete protease inhibitor mixture [Roche, Basel, Switzerland]; Vincent and Carlson, 1999), followed by TCA precipitation. Immunoprecipitation experiments with anti-HA (Roche) affinity matrix were performed as previously reported (Herrador et al., 2010), and λ phosphatase treatment of Rim8-HA immunoprecipitates was carried out as described before (Herranz et al., 2005). Total (20 μg or 1 × 107 cell equivalents for NaOH/TCA lysis) and immunoprecipitated extracts were analyzed by 7.5% SDS–PAGE and immunoblotting with monoclonal anti-Flag (M2; Sigma, St. Louis, MO), anti-HA (3F10; Roche), or anti-actin (C4; MP Biomedicals, Irvine, CA) antibodies. Antibodies were detected by enhanced chemiluminescence with ECLPlus reagents (GE Healthcare, Piscataway, NJ).
Protein kinase assays
In vitro kinase experiments were performed essentially as reported before (Moriya and Johnston, 2004). Recombinant GST-Rim8(1–393) was affinity purified from E. coli BL21 (Novagen, Madison, WI) with glutathione–Sepharose 4B beads (Pharmacia), as described previously (Vincent et al., 2003), and GST fusion proteins bound to beads were resuspended in 150 μl of kinase buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2). His-tagged Yck1 and Yck1K98R were purified from Y00000 transformants grown to mid log phase in SD medium. Cells were broken with glass beads in IP buffer (without DTT and EDTA), and His-tagged proteins were purified with Talon metal affinity matrix (Clontech), following the same procedure as for immunoprecipitation experiments, and then eluted with 50 μl of kinase buffer containing 150 mM imidazole. His-tagged proteins were detected by immunoblotting with an anti-His antibody. Protein kinase assays were performed by mixing 5 μl of purified GST-fusion substrate attached to beads, 5 μl of purified His-tagged kinase, 0.5 μCi of [γ-32P]ATP (Perkin Elmer, Waltham, MA), and 40 μl of kinase buffer. After 30 min incubation at room temperature, beads were washed with STE buffer (10 mM Tris-HCl, pH8.0/1 mM EDTA/150 mM NaCl) containing 0.1% Triton X-100 and 0.5 M NaCl and then washed with STE buffer with 0.1% Triton X-100. GST-fusion substrate was eluted by boiling in SDS sample buffer and separated by SDS–PAGE. Total and phosphorylated proteins were detected by Coomassie staining and autoradiography, respectively.
Fluorescence microscopy
Yeast cells expressing GFP- and RFP-tagged proteins were grown to mid log phase either in SD medium (final pH 3.5) to select for plasmids or in YPD medium before being resuspended in fresh SD medium at pH 4.5 or, when indicated, titrated at pH 7 with KOH. GFP and RFP localization in live cultures was visualized by fluorescence microscopy as previously described (Herrador et al., 2010). For the microfluidics experiment (Figure 3C), OVY172 cells grown to mid log phase in YPD were injected in a microfluidics chamber (ref. YO4C; CellASIC, Hayward, CA), using the Microfluidic Perfusion Platform (ONIX) driven with the interface software ONIX-FG-SW (CellASIC). Cells were resuspended in SD medium containing 0.1M HEPES, pH 4, and trapped and maintained in a uniform plane. Cells were then perfused with 0.1 M SD medium containing 0.1 M HEPES, pH 7, and imaged. Normal growth conditions were reproduced by adjusting the ambient temperature at 30°C with a thermostated chamber and flowing cells with the indicated culture medium at 3 psi. The microfluidics device was coupled to a DMI6000 (Leica, Wetzlar, Germany) microscope equipped with an oil immersion Plan Apochromat 100×/numerical aperture 1.4 objective, a QuantEM cooled electron-multiplying charge-coupled device camera (Photometrics, Tucson, AZ), and a spinning-disk confocal system CSU22 (Yokogawa, Tokyo, Japan). Rim8-3GFP was visualized with a GFP filter 535AF45. Images were acquired with MetaMorph 7 software (Molecular Devices, Sunnyvale, CA).
Supplementary Material
Acknowledgments
We thank L. Robinson for strains LRB341 and LRB362. This work was supported by Grant BFU2008-02005 from the Spanish Ministerio de Ciencia e Innovación and by the Fondation ARC pour la Recherche sur le Cancer (SFI20121205762 to S.L. and DOC20130606445 to M.B.). A.H. is the recipient of a Spanish National Research Council JAE Predoctoral Fellowship.
Abbreviations used:
- CK1
casein kinase 1
- ESCRT
endosomal sorting complex required for transport
- 7TM
seven transmembrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1552) on April 7, 2015.
REFERENCES
- Alvarez CE. On the origins of arrestin and rhodopsin. BMC Evol Biol. 2008;8:222. doi: 10.1186/1471-2148-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro CG, O'Donnell AF, Prosser DC, Augustine AA, Goldman A, Brodsky JL, Cyert MS, Wendland B, Thorner J. Specific alpha-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol Cell Biol. 2014;34:2660–2681. doi: 10.1128/MCB.00230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel PL, Chien C, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley DA, editor. Cellular Interactions in Development: A Practical Approach, Oxford, UK: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- Becuwe M, Herrador A, Haguenauer-Tsapis R, Vincent O, Leon S. Ubiquitin-mediated regulation of endocytosis by proteins of the arrestin family. Biochem Res Int. 2012a;2012:242764. doi: 10.1155/2012/242764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M, Leon S. Integrated control of transporter endocytosis and recycling by the arrestin-related protein Rod1 and Rsp5. Elife. 2014;3:e03307. doi: 10.7554/eLife.03307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M, Vieira N, Lara D, Gomes-Rezende J, Soares-Cunha S, Casal M, Haguenauer-Tsapis R, Vincent O, Paiva S, Leon S. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J Cell Biol. 2012b;196:247–259. doi: 10.1083/jcb.201109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen J.H, Mitchell AP. Control of Bro1-domain protein Rim20 Localization by external pH ESCRT machinery and the Saccharomyces cerevisiae Rim101 pathway. Mol Biol Cell. 2006;17:1344–1353. doi: 10.1091/mbc.E05-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crapeau M, Merhi A, Andre B. Stress conditions promote yeast Gap1 permease ubiquitylation and down-regulation via the arrestin-like Bul and Aly proteins. J Biol Chem. 2014;289:22103–22116. doi: 10.1074/jbc.M114.582320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem. 1990;265:14264–14269. [PubMed] [Google Scholar]
- Gaidarov I, Krupnick JG, Falck JR, Benovic JL, Keen JH. Arrestin function in G protein-coupled receptor endocytosis requires phosphoinositide binding. EMBO J. 1999;18:871–881. doi: 10.1093/emboj/18.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo A, Calcagno-Pizarelli AM, Arst HN, Jr, Peñalva MA. An ordered pathway for the assembly of fungal ESCRT-containing ambient pH signalling complexes at the plasma membrane. J Cell Sci. 2012;125:1784–1795. doi: 10.1242/jcs.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaddar K, Merhi A, Saliba E, Krammer EM, Prevost M, Andre B. Substrate-induced ubiquitylation and endocytosis of yeast amino acid permeases. Mol Cell Biol. 2014;34:4443–4463. doi: 10.1128/MCB.00699-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Raja J, Davis DA. The beta-arrestin-like protein Rim8 is hyperphosphorylated and complexes with Rim21 and Rim101 to promote adaptation to neutral-alkaline pH. Eukaryot Cell. 2012;11:683–693. doi: 10.1128/EC.05211-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Han SO, Kommaddi RP, Shenoy SK. Distinct roles for beta-arrestin2 and arrestin-domain-containing proteins in beta2 adrenergic receptor trafficking. EMBO Rep. 2013;14:164–171. doi: 10.1038/embor.2012.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama R, Kamiya M, Takahara T, Maeda T. Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol Cell Biol. 2010;30:5598–5607. doi: 10.1128/MCB.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrador A, Herranz S, Lara D, Vincent O. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol Cell Biol. 2010;30:897–907. doi: 10.1128/MCB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrador A, Leon S, Haguenauer-Tsapis R, Vincent O. A mechanism for protein monoubiquitination dependent on a trans-acting ubiquitin binding domain. J Biol Chem. 2013;288:16206–16211. doi: 10.1074/jbc.C113.452250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz S, Rodriguez JM, Bussink HJ, Sanchez-Ferrero JC, Arst HN, Jr, Peñalva MA, Vincent O. Arrestin-related proteins mediate pH signaling in fungi. Proc Natl Acad Sci USA. 2005;102:12141–12146. doi: 10.1073/pnas.0504776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachaliou M, Amillis S, Evangelinos M, Kokotos AC, Yalelis V, Diallinas G. The arrestin-like protein ArtA is essential for ubiquitination and endocytosis of the UapA transporter in response to both broad-range and specific signals. Mol Microbiol. 2013;88:301–317. doi: 10.1111/mmi.12184. [DOI] [PubMed] [Google Scholar]
- Khoury E, Nikolajev L, Simaan M, Namkung Y, Laporte SA. Differential regulation of endosomal GPCR/beta-arrestin complexes and trafficking by MAPK. J Biol Chem. 2014;289:23302–23317. doi: 10.1074/jbc.M114.568147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Mitchell AP. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:677–686. doi: 10.1128/MCB.23.2.677-686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Liu Z, Thornton J, Spirek M, Butow RA. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component Ptr3. Mol Cell Biol. 2008;28:551–563. doi: 10.1128/MCB.00929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgurn JA, Hsu PC, Smolka MB, Emr SD. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 2011;147:1104–1117. doi: 10.1016/j.cell.2011.09.054. [DOI] [PubMed] [Google Scholar]
- Maeda T. The signaling mechanism of ambient pH sensing and adaptation in yeast and fungi. FEBS J. 2012;279:1407–1413. doi: 10.1111/j.1742-4658.2012.08548.x. [DOI] [PubMed] [Google Scholar]
- Merhi A, Andre B. Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14–3-3-dependent control of the Bul arrestin-like adaptors. Mol Cell Biol. 2012;32:4510–4522. doi: 10.1128/MCB.00463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci USA. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Pan H, Lu Q. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 2010;11:605–611. doi: 10.1038/embor.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E, Pelham HR. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 2009;10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E, Sullivan JA, Pelham HR. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 2008;9:1216–1221. doi: 10.1038/embor.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell AF. The running of the Buls: control of permease trafficking by alpha-arrestins Bul1 and Bul2. Mol Cell Biol. 2012;32:4506–4509. doi: 10.1128/MCB.01176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell AF, Apffel A, Gardner RG, Cyert MS. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol Biol Cell. 2010;21:3552–3566. doi: 10.1091/mbc.E10-07-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell AF, Huang L, Thorner J, Cyert MS. A calcineurin-dependent switch controls the trafficking function of alpha-arrestin Aly1/Art6. J Biol Chem. 2013;288:24063–24080. doi: 10.1074/jbc.M113.478511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Kihara A. Signaling events of the Rim101 pathway occur at the plasma membrane in a ubiquitination-dependent manner. Mol Cell Biol. 2014;34:3525–3534. doi: 10.1128/MCB.00408-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Yamamoto H, Kihara A. Membrane protein Rim21 plays a central role in sensing ambient pH in Saccharomyces cerevisiae. J Biol Chem. 2012;287:38473–38481. doi: 10.1074/jbc.M112.394205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA. 2004;101:12242–12247. doi: 10.1073/pnas.0404467101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek HR, Stepp JD, Engle HM, Marks KM, Tan PK, Lemmon SK, Robinson LC. Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J. 1997;16:4194–4204. doi: 10.1093/emboj/16.14.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca L, Chastagner P, Meas-Yedid V, Israel A, Brou C. Alpha-arrestin 1 (ARRDC1) and beta-arrestins cooperate to mediate Notch degradation in mammals. J Cell Sci. 2013;126:4457–4468. doi: 10.1242/jcs.130500. [DOI] [PubMed] [Google Scholar]
- Robinson LC, Menold MM, Garrett S, Culbertson MR. Casein kinase I-like protein kinases encoded by YCK1 and YCK2 are required for yeast morphogenesis. Mol Cell Biol. 1993;13:2870–2881. doi: 10.1128/mcb.13.5.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1990. [Google Scholar]
- Rothfels K, Tanny JC, Molnar E, Friesen H, Commisso C, Segall J. Components of the ESCRT pathway DFG16 and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:6772–6788. doi: 10.1128/MCB.25.15.6772-6788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruden DM, Ma J, Li Y, Wood K, Ptashne M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature. 1991;350:250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- Shi H, Rojas R, Bonifacino JS, Hurley JH. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nat Struct Mol Biol. 2006;13:540–548. doi: 10.1038/nsmb1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK, Xiao K, Lefkowitz RJ. Emerging paradigms of beta-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem Sci. 2011;36:457–469. doi: 10.1016/j.tibs.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon AM, Kane PM. Loss of vacuolar H+-ATPase activity in organelles signals ubiquitination and endocytosis of the yeast plasma membrane proton pump Pma1p. J Biol Chem. 2014;289:32316–32326. doi: 10.1074/jbc.M114.574442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Carlson M. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 1998;17:5757–5765. doi: 10.1093/emboj/17.19.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Woolford CA, Desai JV, Lanni F, Mitchell AP. Cis- and trans-acting localization determinants of pH response regulator Rim13 in Saccharomyces cerevisiae. Eukaryot Cell. 2012;11:1201–1209. doi: 10.1128/EC.00158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Carlson M. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 1999;18:6672–6681. doi: 10.1093/emboj/18.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Rainbow L, Tilburn J, Arst HN, Jr, Peñalva MA. YPXL/I is a protein interaction motif recognized by aspergillus PalA and its human homologue AIP1/Alix. Mol Cell Biol. 2003;23:1647–1655. doi: 10.1128/MCB.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Hirsch JA, Velez MG, Gurevich YV, Gurevich VV. Transition of arrestin into the active receptor-binding state requires an extended interdomain hinge. J Biol Chem. 2002;277:43961–43967. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- Volland C, Urban-Grimal D, Geraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]
- Wu N, Zheng B, Shaywitz A, Dagon Y, Tower C, Bellinger G, Shen CH, Wen J, Asara J, McGraw TE, et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell. 2013;49:1167–1175. doi: 10.1016/j.molcel.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Mitchell AP. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J Bacteriol. 2001;183:6917–6923. doi: 10.1128/JB.183.23.6917-6923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hubbard EJ, Carlson M. A protein kinase substrate identified by the two-hybrid system. Science. 1992;257:680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.