Abstract
Recently various pathways of human telomere (ht) DNA folding into G-quadruplexes and of ligand binding to these structures have been proposed. However, the key issue as to the nature of forces driving the folding and recognition processes remains unanswered. In this study, structural changes of 22-mer ht-DNA fragment (Tel22), induced by binding of ions (K+, Na+) and specific bisquinolinium ligands, were monitored by calorimetric and spectroscopic methods and by gel electrophoresis. Using the global model analysis of a wide variety of experimental data, we were able to characterize the thermodynamic forces that govern the formation of stable Tel22 G-quadruplexes, folding intermediates, and ligand-quadruplex complexes, and then predict Tel22 behavior in aqueous solutions as a function of temperature, salt concentration, and ligand concentration. On the basis of the above, we believe that our work sets the framework for better understanding the heterogeneity of ht-DNA folding and binding pathways, and its structural polymorphism.
Introduction
Guanine-rich DNA sequences in the presence of cations can fold into four-stranded structures called G-quadruplexes. The existence of potential quadruplex sequences in key regions of the eukaryotic genome, including the immunoglobulin heavy chain switch region, promoter regions, ribosomal DNA, oncogenes, and telomeres, suggests that they may play an important role in the mechanism and control of several cellular processes (1–3). Therefore, G-quadruplexes are relevant targets of small molecules that can potentially modulate their biological functions, gene expression, and protein synthesis (4,5).
Quadruplex topologies may differ in glycosidic bond angles, strand orientation, connecting loop regions, and molecularity leading to conformational heterogeneity of G-quadruplex structures. This is well exemplified by guanine-rich human telomeric (ht) repeat sequences, which are capable of adopting multiple topologies. For example, monomeric ht quadruplexes containing the core sequence d(AGGG(TTAGGG)3) (Tel22) can adopt several distinct quadruplex topologies. X-ray crystallography reveals that in the presence of K+ ions, Tel22 shows all-parallel strand orientation (6) while in K+ solutions it adopts, according to NMR and other biophysical techniques, a (3+1) hybrid-type topology (denoted as HK+) (7–10). By contrast, in Na+ solutions Tel22 adopts a conformation with antiparallel strand orientation (denoted as ANa+) (11). As shown recently, binding of ligands capable of inducing binding-coupled conformational transitions of G-quadruplexes may be an additional cause of the observed ht-quadruplex polymorphism (12–16).
Detailed computational and experimental (thermodynamic, kinetic) investigation of folding and ligand binding pathways of quadruplex DNA has begun very recently (17–22). These studies leave no doubt that in solution, the Tel22 folding/unfolding process may consist of several structural steps, meaning that such solutions should be considered as equilibrium mixtures of Tel22 molecules in their folded (Q), unfolded (U), and intermediate (I) states. Because in the reported studies of ligand binding to Tel22, as a rule, the involvement of Tel22 folding intermediates has been neglected, one may expect the reported thermodynamics of binding to be incomplete. Thus, in addition to investigating the nature and the relative importance of forces that govern folding of Tel22 and its recognition by ligands, we aimed to estimate how important in controlling these binding events may be the involvement of the Tel22 intermediates. We believe that such an approach will lead to an improvement in our understanding of the heterogeneity observed in the ht-quadruplex folding and binding processes, and of the complex interplay between the enthalpic and entropic contributions to the free energy changes accompanying these events.
Materials and Methods
Sample preparation
HPLC pure oligonucleotide 5′-AGGGTTAGGGTTAGGGTTAGGG-3′ (Tel22) was obtained from Midland Chemical (Midland, MI). The buffer solutions used in our experiments consisted of 20 mM cacodylic acid, 1 mM EDTA, and various concentrations of Na+ or K+ ions. NaOH (KOH) was added to cacodylic acid to reach pH = 6.9. Then, NaCl (KCl) was added to obtain the desired concentration of Na+ (K+) ions (100 or 200 mM Na+ (K+) in cacodylic buffer). DNA was first dissolved in water and then extensively dialyzed against the buffer using a Float-A-Lyser dialysis tube (molecular mass cutoff 500–1000 Da; Spectrum Laboratories, Piscataway, NJ). The starting solution of oligonucleotide was first heated up to 95°C in an outer thermostat for 5 min to make sure that all DNA transforms into the unfolded form, and subsequently cooled down to 5°C at the cooling rate of 0.05°C min−1 to allow DNA to adopt quadruplex structure(s). It is then used in the experiments. Concentration of the DNA in the buffer solution was determined at 25°C spectrophotometrically using a Cary 100 BIO UV/Visible Spectrophotometer (Varian, Cary, NC) equipped with a thermoelectric temperature controller. Tel22 concentrations at 25°C were obtained from the melting curves monitored at wavelength λ = 260 nm. For the extinction coefficient of Tel22 unfolded form at 25°C, we used the value ε260 = 228,500 M−1 cm−1 estimated from the nearest-neighbor data of Cantor et al. (23).
The two bisquinolinium derivatives, Phen-DC3 (M = 848.12 g mol−1; Fig. S1 in the Supporting Material), and 360A-Br (M = 780.90 g mol−1; Fig. S1), are poorly soluble in aqueous solution, therefore they were first dissolved in DMSO and then transferred into buffer solution. The lowest possible content of the DMSO in the buffer solution sufficient for ligand solubility (μM range) was 3%. Therefore, all quantities of the ligand and Tel22 were dissolved in buffer solution with 3% DMSO. Concentrations of the ligands were determined by measuring absorbance at 25°C (εPhen-DC3, 350 nm = 6200 M−1 cm−1 and ε360A-Br, 370 nm = 5980 M−1 cm−1).
Circular dichroism spectroscopy
Circular dichroism (CD) spectra of DNA and ligand-DNA complexes were recorded at 25°C in a 1.0 cm cuvette in the wavelength range between 215 and 350 nm. CD titrations were conducted at 25°C by titrating DNA solution (cDNA ≈ 70 μM) into a 600 μL ligand solution (cL ≈ 20 μM). Ellipticity, Θ, was measured at 293 nm in a 1.0 cm cuvette with signal averaging time of 30 s and 5 nm bandwidth. Temperature dependence of CD spectra of Tel22 was collected between 215 and 320 nm in a 0.25 mm cuvette with a signal averaging time of 10 s and 5 nm bandwidth. Experiments were performed using a CD spectrophotometer model No. 62A DS (Aviv Biomedical, Lakewood, NJ) equipped with a thermoelectric temperature controller.
Fluorimetry
Fluorimetry (FL) titrations were conducted at 25°C by titrating DNA solution (cDNA ≈ 80 μM) into a 1000 μL 360A-Br ligand solution (cL ≈ 10 μM). Emission spectra were recorded between 340 and 570 nm (excitation wavelength λex = 330 nm) in a 1.0 cm cuvette with scanning speed of 50 nm min−1. Phen-DC3 exhibits very weak induced fluorescence, therefore we were unable to obtain reliable experimental data for further model analysis of its binding. Experiments were performed using a Cat. No. LS 55 luminescence spectrometer (Perkin Elmer, Waltham, MA) equipped with a thermally controlled cell holder.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were performed between 15 and 35°C by titrating a solution of DNA (cDNA ≈ 80 μM) into a ligand solution (cL ≈ 10 μM, V = 1.386 mL) using a VP-ITC isothermal titration calorimeter from Microcal (Northampton, MA). The area under the peak after each injection of DNA solution was obtained by integration of the raw signal, corrected for the corresponding heat of dilution (blank titration of DNA into buffer solution) and expressed per mole of added DNA per injection, to give the enthalpy of interaction, (ΔHT).
Differential scanning calorimetry
Differential scanning calorimetry (DSC) experiments were performed at the DNA concentration of ∼0.3 mM in the temperature range between 1 and 95°C at the heating and cooling rate of 1.0°C min−1. The corresponding baseline (buffer-buffer) thermograms were subtracted from the heating thermograms and the obtained differences were normalized to 1 mol of DNA to obtain the partial molar heat capacity of DNA as a function of temperature. Data were analyzed in the same way as described in Bončina et al. (24). DSC experiments were performed using the Nano DSC instrument (TA Instruments, New Castle, DE).
Results and Discussion
Thermodynamic analysis of binding-induced structural transitions
We investigated thermodynamics of Tel22 structural transitions in solution at various temperatures, concentrations of K+ or Na+ ions, and in the presence or absence of bisquinolinium ligands Phen-DC3 and 360A-Br (Fig. S1); these ligands are known for their high affinity for G-quartets, most likely due to strong π-π stacking interactions (25–28). The analysis of calorimetric (DSC and ITC) and spectroscopic (CD and FL) data obtained in solutions with K+ (see Fig. 2; Figs. S4 and S6) or Na+ ions (Figs. S3, S5, and S7) suggests that the observed unfolding and binding processes may be described by the model mechanism that involves five macroscopic states (Fig. 1). Reversibility of folding/unfolding of Tel22 in the absence of ligands and in the presence of K+ or Na+ ions (U ↔ I ↔ Q; Fig. S3) was verified by DSC while the reversibility of ligand binding (Q + L ↔ Q′L + L ↔ Q″L2) was verified by gel electrophoresis (Fig. S11). For a given model mechanism (Fig. 1), one can write a general equation for the spectroscopic and calorimetric properties of the ligand-DNA solution, measured in the presence of K+ or Na+ ions (Eq. 1), from which the corresponding contributions of the buffer are subtracted. Such property, X, can be presented as a linear combination of molar ratios, αj = cj/cDNA of all species, j, predicted by the model mechanism (j = L, U, I, Q, Q′L, and Q″L2),
| (1) |
In Eq. 1, Xj represents the property of the solute j at a given pressure, and it includes the temperature T, the salt type (KCl and NaCl), the salt concentration, and the total DNA concentration cDNA. Because according to the model and , where r = cL,tot/cDNA = nL,tot/nDNA (cL,tot is the total ligand concentration), one obtains
| (2) |
where the changes , , , and refer to each step in the model mechanism presented in Fig. 1. From Eq. 2 are derived various model functions (see Eqs. S3–S6 and S9 in the Supporting Material) expressed in terms of a set of adjustable parameters that describe the CD (X = [Θ] = Θ/(cDNAl) = normalized ellipticity at given λ), FL (X = [F] = IF/(cDNAl) = normalized emitted fluorescence measured at given λ), and DSC and ITC (X = H = enthalpy of solution per mol of DNA) experiments. Each step in the suggested mechanism is described in terms of the corresponding changes of three standard thermodynamic parameters that are independent of the Na+ or K+ concentration. Two of them, and , depend on temperature and are thus determined at the reference temperature T0 = 298.15 K, while the third parameter, , is assumed to be temperature-independent. These three parameters define the standard free energy and enthalpy of folding or binding, and , at any value of T through the Gibbs-Helmholtz relation and the Kirchhoff’s law .
Figure 2.
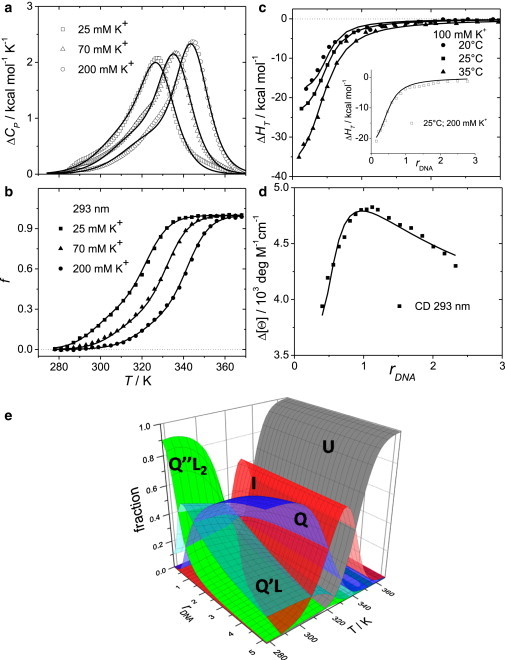
Model analysis of experimental data. Best-fit global model functions (lines) show good agreement with experimental data. Symbols represent unfolding data obtained by DSC and CD spectroscopy (a and b), and ligand (Phen-DC3) binding data obtained by ITC and CD spectroscopy (c and d) that is measured as a function of temperature, K+ ion concentration, and DNA/ligand molar ratio rDNA = cDNA/cL,tot (see the Supporting Material for details). (e) The corresponding fractions of Tel22 species are presented as a function of the DNA/ligand molar ratio and temperature in the presence of ligand Phen-DC3 in 100 mM K+ solution predicted by global thermodynamic analysis of data in terms of the proposed mechanism (Fig. 1). To see this figure in color, go online.
Figure 1.
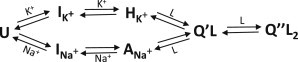
Mechanism of Tel22 structural transitions induced by binding of K+ or Na+ ions and bisquinolinium ligand (L = Phen-DC3 or 360A-Br) consistent with all experimental data.
Each step is usually described in terms of the apparent (note that X+ stands for Na+ or K+), which depends on the salt concentration. Its relation with the true thermodynamic is given by
| (3) |
where parameter Δni represents the number of ions released or uptaken in the transition step i and is assumed to be independent of T (29). Note that equilibrium molar concentration of unbound X+, [X+], appearing in Eq. 3, is normalized to 1 M concentration in the reference (standard) state. Four thermodynamic parameters (, , , and Δni for each step i in the suggested mechanism) define each equilibrium constant, , appearing in the proposed model presented in Fig. 1. In other words, 16 parameters specify the populations of species U, I, Q, Q′L, and Q″L2 in the solution at any value of T, ligand, salt, and DNA concentration , and consequently also the corresponding CD, FL, ITC, and DSC model functions (right-hand side of Eqs. S3–S6 and S9). Global fitting of the model functions to the experimental CD, FL, ITC, and DSC data was based on the nonlinear Levenberg-Marquardt χ2 regression procedure. The first step in the global fitting procedure was a semiglobal model analysis of DSC and CD folding/unfolding data of the ligand-free Tel22 (U ↔ I ↔ Q equilibrium) in the presence of K+ (Fig. 2, a and b) or Na+ (Fig. S3) ions in which eight adjustable parameters were used (Table S4 in the Supporting Material). In the second step, these parameters were used in the description of the binding isotherms (Figs. 2, c and d, and S4–S7) as fixed values. Moreover, due to the high correlation between the adjustable parameters describing the Q′L + L ↔ Q″L2 step, we were forced to reduce the number of the adjustable parameters. In this light, we assumed that the heat capacity and the number of ions released is the same for Q + L ↔ Q′L and Q′L + L ↔ Q″L2 step (Table S2). This assumption may be supported by the observed structural features of a number of (aromatic) ligand-quadruplex complexes in which the interacting surfaces of the first and the second aromatic ligand molecules, bound at the opposite ends of the G-quadruplex, are similar (30,31). Similar surface areas buried upon binding of the ligand in each step should result in similar changes in heat capacities, and, due to similar electrostatic interactions, the accompanying number of released ions should be approximately the same. Taken together, nine adjustable parameters were used in the fitting procedure in the case of Phen-DC3 (ITC and CD data; Table S5) and 11 in the case of 360A-Br (ITC, CD, and FL data).
In the absence of ligand, the folding/unfolding was described as a three-state process involving U, I, and Q (24). In K+ or Na+ solutions, I can be considered to be a mixture of so-called G-triplex conformations (IK+ or INa+) (19,32), which, according to the measured CD spectra, exhibit structural properties similar to HK+ or ANa+ (Fig. 3). Each stage of folding is in the presence of K+ [U → IK+ → HK+] and Na+ [U → INa+ → ANa+], characterized by an extensive enthalpy-entropy compensation (33) (Fig. 4 and Table S1) and a large negative change in the heat capacity accompanying the first transition step [U → I, ΔC°P ≈ −400 cal mol−1 K−1]. These thermodynamic parameters are comparable with those reported for the thrombin binding aptamer folding/unfolding transition (34). Q is more thermodynamically stable (ΔG°U→Q is lower) in solutions with K+ than with Na+ ions, which is a general characteristic of the G-quadruplex stability (35,36).
Figure 3.
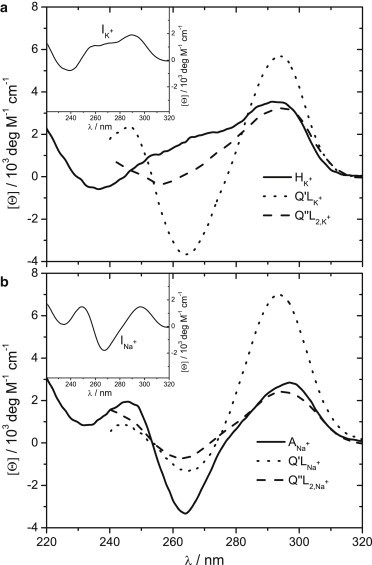
Structural features monitored by CD spectroscopy. Spectra corresponding to hybrid (HK+) and antiparallel (ANa+) structures, complexes with one (Q′L) and two bound ligand (L = Phen-DC3) molecules (Q″L2) and folding intermediates (IK+ and INa+, see inset) at 25°C in the presence of 100 mM K+ (a) or Na+ (b) ions. CD spectra of intermediates and complexes were estimated by deconvolution of the measured spectra based on the model-predicted populations of species (Figs. S4–S7) and spectrum of Q (ANa+ or HK+) form.
Figure 4.
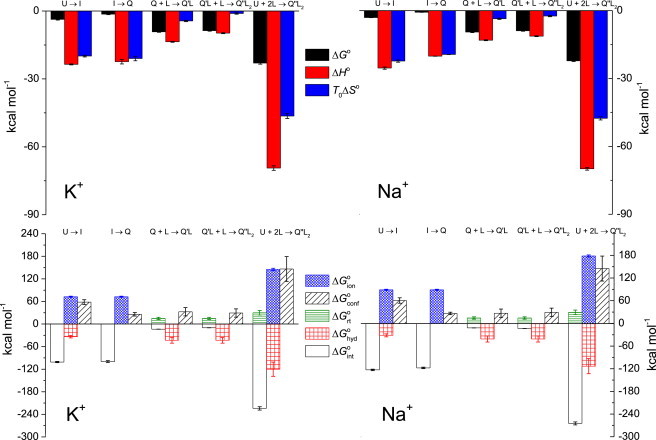
Thermodynamic profiles (top) and Gibbs free energy contributions (driving forces) for each step in the Tel22 binding-coupled folding mechanism in the presence of ligand (Phen-DC3) and 100 mM K+ or Na+ (bottom) at 25°C. The errors of ΔG° contributions (Eq. 4) were calculated by combining the errors of experimental quantities (ΔG°, ΔH°, and ΔC°P; global model analysis) and errors reported in the literature (ΔG°ion, ΔG°hyd, and ΔG°rt) (46,48). To see this figure in color, go online.
Binding of Phen-DC3 and 360A-Br to Tel22 is successfully described by the sequential binding model Q + L ↔ Q′L + L ↔ Q″L2 (Figs. 2 and S4–S7), which assumes that Tel22 unfolded and intermediate states contain no binding sites. All the ligand binding experiments were performed at conditions at which the model analysis of Tel22 melting curves (Figs. 2 and S3) predicts the presence of the folding intermediates I. The fact that the global analysis of the measured binding data is appropriate (good quality of fit, reasonable values of thermodynamic parameters) only when the model-predicted population of I is taken into account, supports the suggested linkage between the folding and binding processes (Figs. 1, 2, a and b, and S4–S7). It should be noted that this linkage could be better supported by thermal unfolding experiments performed with ligand-DNA complexes using DSC or CD (37). Our attempts to perform such experiments and the corresponding data analysis were not successful. Namely, the detection of influence of ligand binding on melting transitions, monitored by DSC and CD spectroscopy, requires ligand concentrations that are much higher than those used in our titration experiments. In other words, due to low solubility of ligands in 100 mM K+ or Na+ solutions, the melting experiments cannot be performed at reversible conditions. Moreover, an adequate thermodynamic analysis of titration experiments conducted at very low K+ or Na+ concentrations, at which the model analysis predicts the presence of U and I, cannot be performed due to the aggregation of ligand-G-quadruplex complexes.
Taken together, Fig. 1 represents the simplest model (Model 1) of ion- and ligand-binding-induced structural alterations in the presence of K+ and Na+ ions, consistent with all the experimental data, whose fitting gives reliable values of thermodynamic parameters. To provide evidence that slightly simpler models are a worse fit to the data, we present in Fig. S8 some characteristics of the best global fit of the U ↔ Q + L ↔ Q′L + L ↔ Q″L2 model (Model 2), which assumes that the state I is not populated, and the model that takes into account the U ↔ I ↔ Q equilibrium and assumes that L binds to two equivalent independent binding sites on Q (Model 3). As shown in Fig. S8, Model 2 cannot describe the DSC thermograms and ITC data measured at 35°C, while Model 3 fails to describe CD titration data. On the other hand, more-complex models involve too many adjustable parameters that are highly correlated and thus cannot be determined with sufficient accuracy. Our analysis emphasizes an important advantage of the global fitting over the traditional fitting of the model to limited datasets (29). For example, ITC data alone (measured at T < 30°C) can be successfully described by the simplified Model 3 (no I present in the solution), however, according to other available experimental data (DSC and CD titration), such analysis results in thermodynamic binding parameters that have no physical meaning.
Thermodynamics and structural features
CD spectra (Figs. 3 and S9) suggest for both ligands (Phen-DC3, 360A-Br) that their binding is accompanied by quadruplex conformational changes and that the resulting complexes (Q′L, Q″L2) have similar structures in solutions with K+ and Na+ ions. CD spectra of Q′L and Q″L2 complexes show characteristics of the ANa+ spectrum (15). Interestingly, difference CD spectra (Fig. S10) corresponding to the Q + L ↔ Q′L and Q′L + L ↔ Q″L2 binding events are almost mirror images, suggesting that binding of the first ligand molecule to one end of the quadruplex induces changes in CD spectrum that are opposite to those induced by binding of the second ligand molecule to the other end of the quadruplex.
The influence of the bound ligand on the conformation of the folded quadruplexes was examined also by gel electrophoresis experiments, which show that quadruplexes complexed with the ligand are electrophoretically faster than the ligand-free quadruplexes (Fig. S11). According to our model analysis, binding of dicationic ligand displaces only approximately one (nonspecifically) bound cation, which means that the net (negative) charge of the ligand-quadruplex complexes is lower than that of the ligand-free quadruplexes. Thus, if ligand binding to the quadruplex is a rigid-body association, the surface net charge of the ligand-quadruplex complex should be lower than that of the ligand-free quadruplex, and consequently the studied complexes should exhibit lower gel-mobility than the ligand-free quadruplexes (12,38). By contrast, our results (Fig. S11) show an opposite effect, i.e., an increase in mobility of the ligand-quadruplex complexes that may be, in principle, ascribed to increased hydrodynamical compactness and/or increased surface net charge of the ligand-quadruplex complexes. Because both of the two effects can arise from the increased compactness of the ligand-quadruplex complex (despite lower net, i.e., negative charge, its surface net charge can increase), we believe that the observed increased mobility of the complexes results, very likely, from the ligand-induced conformational changes of the quadruplexes. In addition, electrophoresis results suggest that, Q′ and Q″ structures differ from ANa+ and HK+ and also from the possible all parallel Tel22 quadruplex conformation that has been shown to be gel-electrophoretically slower than ANa+ and HK+ (39).
The observed thermodynamic characteristics of ligand binding to Tel22 in K+ and Na+ solutions are very similar. For both ligands, the Q binding affinity for the first ligand molecule is higher than for the second ligand (Table S2). Both steps are enthalpy-driven, accompanied by negative change in entropy and heat capacity (Fig. 4 and Table S2). This suggests that ligand binding is driven mainly by ligand-quadruplex π-π stacking (ΔH° < 0) and by displacement of water from the ligand-quadruplex binding interface (ΔC°P < 0). Moreover, for both ligands, the overall thermodynamics of binding-coupled folding (U + L → Q′L or U + 2L → Q″L2) is nearly the same in the K+ and Na+ environments (Fig. 4 and Table S2). This supports our suggestion that in solutions with either of the two ions, the ligand-bound structures are similar. In addition, HK+ + L → Q′LK+ and ANa+ + L → Q′LNa+ (and HK+ + 2L → Q″L2,K+ and ANa+ + 2L → Q″L2,Na+) events are also accompanied by similar energetic contributions (Fig. 4 and Table S2). Because according to the thermodynamic parameters that characterize unfolding of ANa+ and HK+ (Table S1), the ANa+ and HK+ conformations are energetically very similar, this observation is consistent with the suggested similarity of the ligand-quadruplex structures in the presence of K+ and Na+.
Driving forces of binding-induced structural alterations
Several relatively recent articles discuss the thermodynamic forces that may control folding of G-quadruplexes (40–42). We present here, to the best of our knowledge, the first attempt at quantitative dissection of ΔG° accompanying the folding and ligand binding-coupled structural transitions of ht-DNA to more fundamental contributions (Fig. 4 and Table S3). Following the additivity approach (43–45), ΔG° can be treated as a sum of the main contributions,
| (4) |
in which the first contribution, ΔG°solv, ascribed to the solvation effects (reorganization of water molecules surrounding Tel22, ions, and ligands) may be further expressed as ΔG°solv = ΔG°ion + ΔG°hyd. The ΔG°ion contribution reflects the dehydration of K+ or Na+ ions accompanying their coordinative binding within Tel22. Its enthalpic and entropic origin has been well characterized by Marcus (46) and results in the estimates of ΔG°ion,K+ = 72.6 kcal mol−1 and ΔG°ion,Na+ = 89.6 kcal mol−1. The ΔG°hyd contribution is ascribed to the desolvation/solvation of Tel22 and ligand molecules and may be interpreted mainly as a hydrophobic contribution to the overall ΔG° of folding and/or binding (43,44,47). At 25°C it may be estimated by an empirical relation as ΔG°hyd = ΔC°P·80(±10)K (48,49), where ΔC°P is the corresponding heat capacity change determined by the global model analysis of experimental data (Figs. 2 and S1) combined with the careful treatment of the corresponding baselines (DSC and ITC). The second main contribution, ΔG°int, reflects the specific intra- and intermolecular interactions (van der Waals (base-stacking), H-bonds, Coulombic interactions, and cation coordination) that stabilize a particular Tel22 conformation. It can be considered mainly as an enthalpic contribution and estimated as the difference between the measured enthalpy change, ΔH°, and the corresponding enthalpy of ion dehydration, ΔH°ion (46). Thus, ΔG°int ≈ ΔH° ‒ ΔH°ion. The third main contribution, ΔG°rt, is considered as an entropic contribution due to changes of rotational and translational freedom of ligand and Tel22 (lost upon ligand binding): ΔG°rt = ‒TΔS°rt. Similarly, the fourth main contribution ΔG°conf is interpreted as an entropic contribution accompanying the changes of conformational freedom of Tel22, ΔG°conf = ‒TΔS°conf. It may be estimated as ΔG°conf = ΔG° ‒ ΔG°solv ‒ ΔG°int ‒ ΔG°rt. At this point, we would like to mention that in our dissection of ΔG° the free energy contribution due to the release of the nonspecifically bound cations, which accompanies unfolding of Tel22 and ligand binding to it, has been neglected. Namely, according to the prediction of the polyelectrolyte theory (50), this electrostatic contribution is relatively small (its absolute value is smaller than the error of the least accurate contribution in Eq. 4).
We are well aware that, due to approximations involved in the additivity approach presented above, the interpretation of ΔG° contributions should be taken with great care. It should be emphasized that the main aim of using Eq. 4 is to show the importance of various types of interactions and conformational changes in the formation of folding intermediates and ligand-quadruplex complexes. To demonstrate this, the ΔG° contributions do not need to be specified with high precision (see below). In this light, the described dissection of energetics (Fig. 4) enables us to characterize the dominant driving forces involved in the Tel22 binding-induced structural transitions in the following way:
-
1)
U → I transition accompanied by specific binding (dehydration) of one cation appears to be driven by specific interactions and hydrophobic desolvation because [‒ (ΔG°int + ΔG°hyd) (ΔG°conf + ΔG°ion)]. It seems that even though base-stacking, H-bonding, and cation coordination are needed for the early stage of Tel22 secondary structure formation, intermediate states would not be significantly populated without being stabilized by hydrophobic desolvation (if ΔG°hyd ≈ 0 ⇒ ‒(ΔG°int + ΔG°hyd) (ΔG°conf + ΔG°ion) ⇒ ΔG°U→I > 0; I formation unfavorable). Although IK+ and INa+ are structurally different (Fig. 3), the observation that ΔG°hyd,Na+ ≈ ΔG°hyd,K+ suggests that hydrophobic desolvation are equally important for driving the first step of Tel22 folding in Na+ and K+ solutions.
-
2)
I → Q transition accompanied by specific binding (dehydration) of one cation appears to be driven entirely by specific interactions that overcome loss of conformational freedom and unfavorable ion dehydration [‒ΔG°int (ΔG°conf + ΔG°ion); ΔG°hyd ≈ 0]. The conformational entropy loss is approximately two times lower than in the case of the U → I step, which is in accordance with structural properties of I that are closer to Q than to U (Fig. 4). The observation that for the U → I and I → Q step, ΔG°int,Na+ < ΔG°int,K+, suggests that specific interactions are more favorable for stabilizing I and Q in the presence of Na+ ions, which can be ascribed to energetically more favorable coordination of Na+ compared to K+ ions.
-
3)
Q + L → Q′L and Q′L + L → Q″L2 steps resulting in the formation of Q′L and Q″L2 conformations that differ from the corresponding ligand-free structures Q are driven predominantly by removal of water from ligand-Tel22 interacting surface, which, together with specific (ligand-Tel22 stacking) interactions, overcome unfavorable loss of conformational, translational, and rotational freedom. Even if we overestimate the magnitude of ΔG°rt, by taking as an approximation ΔG°rt ≈ 15 kcal mol−1 (51,52), the ΔG°conf is still very high (Fig. 4). This means that the Q + L → Q′L and Q′L + L → Q″L2 steps are accompanied by a large Tel22 conformational entropy loss, which is in accordance with the observed ligand-induced conformational changes (Figs. 3 and S9).
Conclusion
The global model analysis of a wide variety of experimental data enabled us to describe the ht-DNA (Tel22) behavior in aqueous solutions at various conditions (temperature, salt, and ligand concentration; Fig. 2 e). It resulted in the proposed hierarchy of forces that drive the structural alterations of Tel22 in the absence and presence of G-quadruplex specific ligands, Phen-DC3 and 360A-Br. We believe that such an approach can generally be used in discussing the heterogeneity of both the ht-quadruplex structural polymorphism and its folding and binding pathways.
Author Contributions
J.L. and G.V. designed research; M.B., F.H., and B.I. performed research; M.-P.T.-F. and F.H. contributed new reagents; M.B., J.L., and S.H. analyzed data; and J.L., M.B., G.V., and S.H. wrote the article.
Acknowledgments
Financial support of the Slovenian Research Agency through grant No. P1-0201 and by COST action No. MP0802 is gratefully acknowledged. J.L. thanks the Excellent NMR - Future Innovation for Sustainable Technologies Centre of Excellence for purchasing the DSC apparatus, which was used to perform this study. M.-P.T.-F thanks Agence Nationale pour la Recherche (ANR), ANR-Gquad, for funding F.H. (postdoctoral fellowship).
Editor: Timothy Lohman.
Supporting Material
References
- 1.Biffi G., Tannahill D., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Sullivan R.J., Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqui-Jain A., Grand C.L., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zahler A.M., Williamson J.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 5.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 7.Ambrus A., Chen D., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan A.T., Kuryavyi V., Patel D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray R.D., Petraccone L., Chaires J.B. Characterization of a K+-induced conformational switch in a human telomeric DNA oligonucleotide using 2-aminopurine fluorescence. Biochemistry. 2010;49:179–194. doi: 10.1021/bi901357r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luu K.N., Phan A.T., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 12.Nicoludis J.M., Barrett S.P., Yatsunyk L.A. Interaction of human telomeric DNA with n-methyl mesoporphyrin IX. Nucleic Acids Res. 2012;40:5432–5447. doi: 10.1093/nar/gks152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman K.M., Reszka A.P., Thurston D.E. Biaryl polyamides as a new class of DNA quadruplex-binding ligands. Chem. Commun. (Camb.) 2009;27:4097–4099. doi: 10.1039/b902359c. [DOI] [PubMed] [Google Scholar]
- 14.Granzhan A., Ihmels H., Jäger K. Diazonia- and tetraazoniapolycyclic cations as motif for quadruplex-DNA ligands. Chem. Commun. (Camb.) 2009;10:1249–1251. doi: 10.1039/b812891j. [DOI] [PubMed] [Google Scholar]
- 15.Garner T.P., Williams H.E.L., Searle M.S. Selectivity of small molecule ligands for parallel and anti-parallel DNA G-quadruplex structures. Org. Biomol. Chem. 2009;7:4194–4200. doi: 10.1039/b910505k. [DOI] [PubMed] [Google Scholar]
- 16.Chen M., Song G., Qu X. Small-molecule selectively recognizes human telomeric G-quadruplex DNA and regulates its conformational switch. Biophys. J. 2009;97:2014–2023. doi: 10.1016/j.bpj.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu J.F., Chang T.C., Li H.W. Single-molecule TPM studies on the conversion of human telomeric DNA. Biophys. J. 2010;98:1608–1616. doi: 10.1016/j.bpj.2009.12.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Čeru S., Šket P., Plavec J. A new pathway of DNA G-quadruplex formation. Angew. Chem. Int. Ed. Engl. 2014;53:4881–4884. doi: 10.1002/anie.201400531. [DOI] [PubMed] [Google Scholar]
- 19.Koirala D., Mashimo T., Sugiyama H. Intramolecular folding in three tandem guanine repeats of human telomeric DNA. Chem. Commun. (Camb.) 2012;48:2006–2008. doi: 10.1039/c2cc16752b. [DOI] [PubMed] [Google Scholar]
- 20.Gray R.D., Buscaglia R., Chaires J.B. Populated intermediates in the thermal unfolding of the human telomeric quadruplex. J. Am. Chem. Soc. 2012;134:16834–16844. doi: 10.1021/ja307543z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stadlbauer P., Trantírek L., Sponer J. Triplex intermediates in folding of human telomeric quadruplexes probed by microsecond-scale molecular dynamics simulations. Biochimie. 2014;105:22–35. doi: 10.1016/j.biochi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Dettler J.M., Buscaglia R., Lewis E.A. DSC deconvolution of the structural complexity of c-MYC P1 promoter G-quadruplexes. Biophys. J. 2011;100:1517–1525. doi: 10.1016/j.bpj.2011.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantor C.R., Warshaw M.M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 24.Bončina M., Lah J., Vesnaver G. Energetic basis of human telomeric DNA folding into G-quadruplex structures. J. Am. Chem. Soc. 2012;134:9657–9663. doi: 10.1021/ja300605n. [DOI] [PubMed] [Google Scholar]
- 25.De Cian A., Delemos E., Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 26.Monchaud D., Teulade-Fichou M.P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 27.Sidibe A., Hamon F., Riou J.F. Effects of a halogenated G-quadruplex ligand from the pyridine dicarboxamide series on the terminal sequence of XpYp telomere in HT1080 cells. Biochimie. 2012;94:2559–2568. doi: 10.1016/j.biochi.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Chung W.J., Heddi B., Phan A.T. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC3. Angew. Chem. Int. Ed. Engl. 2014;53:999–1002. doi: 10.1002/anie.201308063. [DOI] [PubMed] [Google Scholar]
- 29.Drobnak I., Vesnaver G., Lah J. Model-based thermodynamic analysis of reversible unfolding processes. J. Phys. Chem. B. 2010;114:8713–8722. doi: 10.1021/jp100525m. [DOI] [PubMed] [Google Scholar]
- 30.Gavathiotis E., Heald R.A., Searle M.S. Drug recognition and stabilization of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J. Mol. Biol. 2003;334:25–36. doi: 10.1016/j.jmb.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Parkinson G.N., Cuenca F., Neidle S. Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes. J. Mol. Biol. 2008;381:1145–1156. doi: 10.1016/j.jmb.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 32.Mashimo T., Yagi H., Sugiyama H. Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J. Am. Chem. Soc. 2010;132:14910–14918. doi: 10.1021/ja105806u. [DOI] [PubMed] [Google Scholar]
- 33.Olsen C.M., Gmeiner W.H., Marky L.A. Unfolding of G-quadruplexes: energetic, and ion and water contributions of G-quartet stacking. J. Phys. Chem. B. 2006;110:6962–6969. doi: 10.1021/jp0574697. [DOI] [PubMed] [Google Scholar]
- 34.Limongelli V., De Tito S., Parrinello M. The G-triplex DNA. Angew. Chem. Int. Ed. Engl. 2013;52:2269–2273. doi: 10.1002/anie.201206522. [DOI] [PubMed] [Google Scholar]
- 35.Gray R.D., Li J., Chaires J.B. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 37.Brandts J.F., Lin L.-N. Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry. 1990;29:6927–6940. doi: 10.1021/bi00481a024. [DOI] [PubMed] [Google Scholar]
- 38.De Cian A., Mergny J.L. Quadruplex ligands may act as molecular chaperones for tetramolecular quadruplex formation. Nucleic Acids Res. 2007;35:2483–2493. doi: 10.1093/nar/gkm098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buscaglia R., Miller M.C., Chaires J.B. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013;41:7934–7946. doi: 10.1093/nar/gkt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lane A.N., Chaires J.B., Trent J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smirnov I.V., Shafer R.H. Electrostatics dominate quadruplex stability. Biopolymers. 2007;85:91–101. doi: 10.1002/bip.20609. [DOI] [PubMed] [Google Scholar]
- 42.Gray R.D., Chaires J.B. Linkage of cation binding and folding in human telomeric quadruplex DNA. Biophys. Chem. 2011;159:205–209. doi: 10.1016/j.bpc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haq I., Ladbury J.E., Chaires J.B. Specific binding of Hoechst 33258 to the d(CGCAAATTTGCG)2 duplex: calorimetric and spectroscopic studies. J. Mol. Biol. 1997;271:244–257. doi: 10.1006/jmbi.1997.1170. [DOI] [PubMed] [Google Scholar]
- 44.Lah J., Vesnaver G. Energetic diversity of DNA minor-groove recognition by small molecules displayed through some model ligand-DNA systems. J. Mol. Biol. 2004;342:73–89. doi: 10.1016/j.jmb.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Lah J., Drobnak I., Vesnaver G. What drives the binding of minor groove-directed ligands to DNA hairpins? Nucleic Acids Res. 2008;36:897–904. doi: 10.1093/nar/gkm1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcus Y.A. John Wiley; New York: 1986. Ion Solvation. [Google Scholar]
- 47.Majhi P.R., Qi J., Shafer R.H. Heat capacity changes associated with guanine quadruplex formation: an isothermal titration calorimetry study. Biopolymers. 2008;89:302–309. doi: 10.1002/bip.20918. [DOI] [PubMed] [Google Scholar]
- 48.Baldwin R.L. Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. USA. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spolar R.S., Record M.T., Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 50.Record M.T., Jr., Anderson C.F., Lohman T.M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 51.Finkelstein A.V., Janin J. The price of lost freedom: entropy of bimolecular complex formation. Protein Eng. 1989;3:1–3. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- 52.Dolenc J., Baron R., van Gunsteren W.F. Configurational entropy change of netropsin and distamycin upon DNA minor-groove binding. Biophys. J. 2006;91:1460–1470. doi: 10.1529/biophysj.105.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


