Abstract

The Notch signaling pathway is critical in development, neuronal maintenance, and hematopoiesis. An obligate step in the activation of this pathway is cleavage of its transmembrane (TM) domain by γ-secretase. While the soluble domains have been extensively studied, little has been done to characterize its TM and flanking juxtamembrane (JM) segments. Here, we present the results of nuclear magnetic resonance (NMR) studies of the human Notch1 TM/JM domain. The TM domain is largely α-helical. While the flanking JM segments do not adopt regular secondary structure, they interact with the membrane surface, suggesting membrane interactions may play a role in modulating its cleavage by γ-secretase and subsequent NOTCH signaling function.
The Notch signaling pathway is essential to development, neuronal maintenance, and hematopoiesis. Notch signaling also controls neurogenesis, synaptic plasticity, axonal and dendritic growth, and neuronal death.1−9 In this pathway, the Notch receptor (Figure 1A) is cleaved in its luminal domain by a furin-like convertase in the Golgi.9 This cleavage event leaves the protein as a heterodimer with the extracellular domain (ECD) linked to the combined TM and intracellular domains.10,11 The protein is then transported to the plasma membrane. Trans binding of a membrane-bound protein ligand from a neighboring cell to the extracellular EGF domain of the Notch protein12 triggers trans-endocytosis of the Notch-bound ligand.13 This induces a force on the NOTCH ECD that mechanically extends the negative regulatory region of the protein, including residues 1449–1731 (LNR and HD domains).14 This exposes the previously buried S2 cut site (Figure 1B) to the ADAM 10/17 metalloprotease,15−17 which cleaves the protein to release its ECD in the committed step of the signaling pathway. While the exact order of the subsequent processing, trafficking, and cleavage steps is still being investigated, Notch is endocytosed and cleaved in its TMD by γ-secretase,18 releasing a small extracellular peptide (Nβ)19 and the large Notch intracellular domain (NICD).20−23 After translocation to the nucleus, the NICD forms a transcriptional activator complex with CSL24 and mastermind (MAML in mammals)25 that targets a number of different genes.26,27 The signaling cascade is terminated when the NICD C-terminal PEST domain is phosphorylated by CDK8 and targeted for polyubiquitination and proteosomal degradation.28−30 Researchers have explored the structure of the water-soluble domains of Notch protein-associated proteins.14,31−37 However, the NOTCH TM and flanking juxtamembrane (TM/JM) domains have not been examined.
Figure 1.
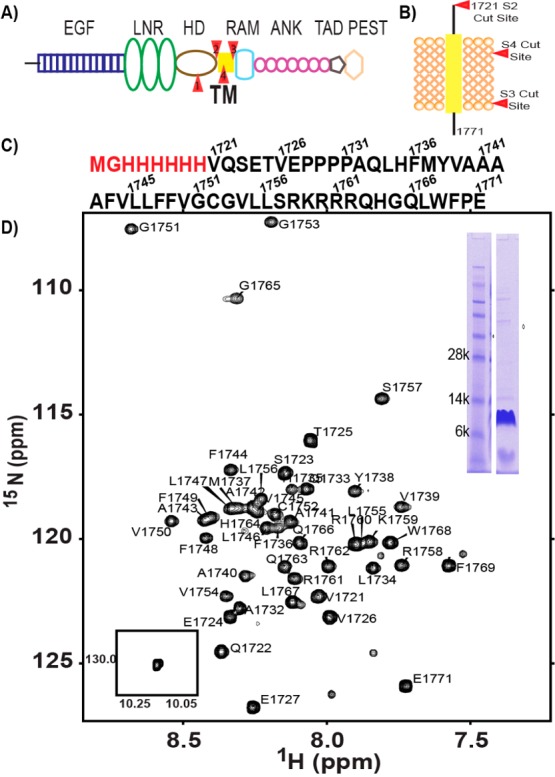
(A) Domain organization of full length Notch1. Sites of proteolysis are colored red. (B) Notch1 TM/JM segment with proteolysis sites colored red. S3 and S4 are γ-secretase cut sites. (C) Sequence of the Notch1 TM/JM segment. (D) Assigned 900 MHz 15N TROSY-HSQC spectrum of the Notch1 TM segment in 15% DMPC/DH6PC bicelles (q = 0.33) at pH 5.5 and 318 K. Backbone amide 1H–15N peaks have been assigned for all of the non-proline residues. The NMR sample included 2 mM DTT, 10% D2O, and 1 mM EDTA. A sodium dodecyl sulfate–polyacrylamide gel of the NMR sample and the single indole side chain 1H–15N peak are shown in the insets.
As noted, cleavage of this domain is an essential step in the Notch signaling pathway, and the prevention of this event can cause significant dysregulation and disease.38,39 Moreover, toxicity caused by inhibition of γ-secretase cleavage of the NOTCH TMD has stymied efforts to prevent or treat Alzheimer’s disease by inhibiting γ-secretase cleavage of the amyloid precursor protein.40 In this paper, we present the purification and preliminary structural characterization of the combined Notch TM/JM domains.
An N-terminally His6-tagged construct of human Notch 1 residues 1721 (S2 cleavage site) to 1771 was selected for study (Figure 1C) on the basis that it includes both N- and C-terminal juxtamembrane regions, that it is expressed well in Escherichia coli, and that it yields a well-resolved NMR spectrum in bicelle model membranes (Figure 1D). As detailed in the Supporting Information, whole E. coli cell lysates harboring this recombinant Notch1 TM/JM domain were mixed with the harsh zwitterionic detergent Empigen to solubilize all membrane proteins, followed by addition of Ni(II) metal ion affinity resin and elution of all non-His6-tagged proteins. The resin with the pure bound Notch1 TM/JM domain was subsequently re-equilibrated with lyso-phospholipid micelles and then DMPC/DH6PC bicelles to refold the protein, followed by elution of the pure Notch TM/JM domain in bicelles
Following screening (Figure S1 of the Supporting Information), optimal NMR sample conditions were determined to be 0.5 mM TM/JM in 15% DMPC/DH6PC bicelles (q = 0.33) at pH 5.5, where the TM/JM-to-(DH6PC+DMPC) molar ratio is approximately 1:600. We then conducted NMR experiments at 900 MHz using standard TROSY-based three-dimensional experiments (HNCA, HNCO, and HNCACB) in conjunction with both uniform and amino acid-selective labeling (see the Supporting Information) to accomplish nearly complete backbone and Cβ resonance assignment for the TM/JM domain (Figure 1D; BioMagResBank entry 26565). The secondary structure was then determined using both chemical shift index and TALOS-N analyses of the chemical shifts.41−44 It appears that most of the TM domain is encompassed by an α-helix that extends from residue 1732 to 1761, with a point of uncertainty being the secondary structure of the tetraproline motif preceding this segment. The flanking JM segments appear to have little regular secondary structure (see Figure S2 of the Supporting Information).
We also collected NMR R1, R2, and HN nuclear Overhauser effect (NOE) data, which provide insight into the flexibility of the Notch1 TM/JM domain (Figure 2). Low R1R2 and NOE values indicate that sites in the N-terminal JM segment of the protein are very flexible, while generally high values indicate that the transmembrane domain is well-ordered. The C-terminal JM domain appears to be somewhat more ordered than the N-terminus. The elevated R2 value for Val1754 is suggestive of local intermediate time scale motions at this site, which is interesting given that γ-secretase cleaves Notch between sites 1753 and 1754.19,44
Figure 2.
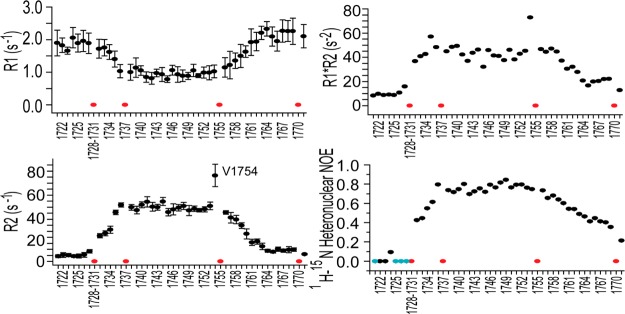
15N NMR relaxation measurements recorded on a 900 MHz magnet reveal the global dynamics of the Notch1 TM/JM segment in 15% DMPC/DH6PC bicelles (q = 0.33) at pH 5.5 and 318 K. R1 is the longitudinal relaxation rate, and R2 is the transverse relaxation rate. Error bars give the uncertainty associated with the fits of the relaxation decays to yield the reported values. Cyan circles represent peaks with negative values; red circles represent either proline sites or instances in which extensive peak overlap prevented the determination of a reliable value. Additional discussion of the surprsingly high R1R2 plateau for the TM domain is found in the Supporting Information.
We next examined the topology of the TM/JM domain in its bicelle environment. A water-soluble paramagnetic probe, Gd(III)-DTPA, a lipid-soluble probe, 16-DSA, and a weakly apolar probe, 3-cyano-PROXYL (that prefers the water–-membrane interface), were added to bicellar Notch TM/JM samples, and the paramagnet-induced intensity changes in the TROSY resonances were quantitated. From these data (Figure 3), it is clear that the TM domain ends at the cluster of basic residues starting at R1758. The topology near the start of the transmembrane helix at position 1732 seems to be more complex. The 3-cyano-PROXYL probe (Figure 3) and NMR CLEANEX-PM water/amide hydrogen exchange data (Figure S3 of the Supporting Information) indicate that the first turn of the TM helix at residues 1732–1736 is located at the water–bilayer interface. Moreover, a spike in the hydrogen exchange rates at residues 1735 and 1736 (Figure S3) suggests local instability of the TM helix. While the tetraproline segment located at sites 1728–1731 was not observed in our NMR spectra, the fact that the amide sites flanking this segment are hydrogen exchange-resistant and inaccessible to all three paramagnetic probes might be interpreted to suggest some tertiary structure involving the tetraproline motif and the preceding segment of residues 1724–1727. However, it is difficult to reconcile ordered tertiary structure with the high dynamics observed for the segment of residues 1724–1727 (Figure 2). A more plausible explanation is that the Gd(III)-DTPA data for this segment are anomalous for a not-yet-determined reason. If these data are disregarded, all other measurements are consistent with a model in which the N-terminus is disordered and located in the aqueous phase but is anchored to the membrane surface by the tetraproline motif, which sits in the water–bilayer interface, oriented 90° with respect to the transmembrane domain.
Figure 3.

Membrane topology probed for the 15N Notch1 TM/JM segment in 15% DMPC/DH6PC bicelles (q = 0.33) at pH 5.5 and 318 K. A plot of the ratio of site-specific peak intensities from samples containing a paramagnet vs those from a diamagnetic reference sample reveals the probe accessibility of each residue. The Gd(III)-DTPA accessibility is colored blue and the 16-DSA accessibility red. The bottom panel represents a similar plot using 3-cyano-PROXYL. Sites with negative bars represent either proline residues or instances in which extensive peak overlap prevented reliable analysis.
The short C-terminal JM segment from R1758 to Q1766 is seen to be solvent-exposed. However, this segment is followed by residues that are significantly broadened by 16-DSA and 3-cyano-PROXYL but not by Gd(III)-DTPA, indicating that the end of the C-terminal JM segment actually dips back into the membrane as a consequence of the Leu-Trp-Phe sequence located near the C-terminus. This model is strongly supported by the hydrogen exchange data (Figure S3 of the Supporting Information).
Even at the very rough level of resolution provided by the data of this work, it appears that the NOTCH TM/JM domain is structurally very different from the corresponding domain of the APP,45,46 also a γ-secretase substrate. APP has a kinked TM domain and a C-terminal JM domain that interacts with the membrane surface only after a disordered and water-exposed 40-residue segment connecting its TM domain to a distal C-terminal amphipathic helix. These features differ from those of the apparently unbroken helix of the NOTCH TM domain followed by a short (9 residue) flexible cytosolic linker to the membrane-interacting Leu-Trp-Phe segment. While both APP and Notch have N-terminal JM segments that interact with the membrane surface, APP has a surface-bound helix followed by a soluble connecting loop to the TMD, whereas the Notch TM helix appears to be preceded by an interfacial tetraproline segment, with no connecting water-exposed loop. While a more complete compare-and-contrast analysis of the structures of APP and Notch1 TM/JM domains will await completion of the Notch1 TM/JM structure, this work indicates that there are significant differences in their structures. These differences might be exploited in strategies to inhibit cleavage of APP by γ-secretase while still permitting normal (healthy) processing of NOTCH.
Acknowledgments
We thank Dr. Rob McFeeters for his insight into the assignment process and Dr. Markus Voehler for NMR assistance.
Supporting Information Available
Methods, supporting results and discussion, and three supporting figures. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b00456.
This work was supported by National Institutes of Health (NIH) Grants RO1 GM106672 and U54 GM094608. The NMR instrumentation used in this work was supported by NIH Grant S10 RR026677 and National Science Foundation Grant DBI-0922862.
The authors declare no competing financial interest.
Supplementary Material
References
- Huppert S. S.; Le A.; Schroeter E. H.; Mumm J. S.; Saxena M. T.; Milner L. A.; Kopan R. (2000) Nature 405, 966–970. [DOI] [PubMed] [Google Scholar]
- Redmond L.; Oh S. R.; Hicks C.; Weinmaster G.; Ghosh A. (2000) Nat. Neurosci. 3, 30–40. [DOI] [PubMed] [Google Scholar]
- Presente A.; Andres A.; Nye J. S. (2001) NeuroReport 12, 3321–3325. [DOI] [PubMed] [Google Scholar]
- Lowell S.; Benchoua A.; Heavey B.; Smith A. G. (2006) PLoS Biol. 4, e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigas A.; D’Altri T.; Espinosa L. (2012) Curr. Top. Microbiol. Immunol. 360, 1–18. [DOI] [PubMed] [Google Scholar]
- Kopan R.; Ilagan M. X. (2009) Cell 137, 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. J. (2006) Nat. Rev. Mol. Cell Biol. 7, 678–689. [DOI] [PubMed] [Google Scholar]
- Ables J. L.; Breunig J. J.; Eisch A. J.; Rakic P. (2011) Nat. Rev. Neurosci. 12, 269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logeat F.; Bessia C.; Brou C.; LeBail O.; Jarriault S.; Seidah N. G.; Israel A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8108–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaumueller C. M.; Qi H.; Zagouras P.; Artavanis-Tsakonas S. (1997) Cell 90, 281–291. [DOI] [PubMed] [Google Scholar]
- Rand M. D.; Grimm L. M.; Artavanis-Tsakonas S.; Patriub V.; Blacklow S. C.; Sklar J.; Aster J. C. (2000) Mol. Cell. Biol. 20, 1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordle J.; Redfieldz C.; Stacey M.; van der Merwe P. A.; Willis A. C.; Champion B. R.; Hambleton S.; Handford P. A. (2008) J. Biol. Chem. 283, 11785–11793. [DOI] [PubMed] [Google Scholar]
- Nichols J. T.; Miyamoto A.; Olsen S. L.; D’Souza B.; Yao C.; Weinmaster G. (2007) J. Cell Biol. 176, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon W. R.; Vardar-Ulu D.; Histen G.; Sanchez-Irizarry C.; Aster J. C.; Blacklow S. C. (2007) Nat. Struct. Mol. Biol. 14, 295–300. [DOI] [PubMed] [Google Scholar]
- Brou C.; Logeat F.; Gupta N.; Bessia C.; LeBail O.; Doedens J. R.; Cumano A.; Roux P.; Black R. A.; Israel A. (2000) Mol. Cell 5, 207–216. [DOI] [PubMed] [Google Scholar]
- Lieber T.; Kidd S.; Young M. W. (2002) Genes Dev. 16, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm J. S.; Schroeter E. H.; Saxena M. T.; Griesemer A.; Tian X.; Pan D. J.; Ray W. J.; Kopan R. (2000) Mol. Cell 5, 197–206. [DOI] [PubMed] [Google Scholar]
- Vooijs M.; Schroeter E. H.; Pan Y.; Blandford M.; Kopan R. (2004) J. Biol. Chem. 279, 50864–50873. [DOI] [PubMed] [Google Scholar]
- Chandu D.; Huppert S. S.; Kopan R. (2006) J. Neurochem. 96, 228–235. [DOI] [PubMed] [Google Scholar]
- Okochi M.; Fukumori A.; Jiang J.; Itoh N.; Kimura R.; Steiner H.; Haass C.; Tagami S.; Takeda M. (2006) J. Biol. Chem. 281, 7890–7898. [DOI] [PubMed] [Google Scholar]
- Okochi M.; Steiner H.; Fukumori A.; Tanii H.; Tomita T.; Tanaka T.; Iwatsubo T.; Kudo T.; Takeda M.; Haass C. (2002) EMBO J. 21, 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenkowski P.; Ye W.; Wang R.; Wolfe M. S.; Selkoe D. J. (2008) J. Biol. Chem. 283, 22529–22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M. S.; Kopan R. (2004) Science 305, 1119–1123. [DOI] [PubMed] [Google Scholar]
- Kovall R. A. (2008) Oncogene 27, 5099–5109. [DOI] [PubMed] [Google Scholar]
- Wu L.; Aster J. C.; Blacklow S. C.; Lake R.; Artavanis-Tsakonas S.; Griffin J. D. (2000) Nat. Genet. 26, 484–489. [DOI] [PubMed] [Google Scholar]
- Iso T.; Kedes L.; Hamamori Y. (2003) J. Cell. Physiol. 194, 237–255. [DOI] [PubMed] [Google Scholar]
- Kageyama R.; Ohtsuka T. (1999) Cell Res. 9, 179–188. [DOI] [PubMed] [Google Scholar]
- Fryer C. J.; Lamar E.; Turbachova I.; Kintner C.; Jones K. A. (2002) Genes Dev. 16, 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C. J.; White J. B.; Jones K. A. (2004) Mol. Cell 16, 509–520. [DOI] [PubMed] [Google Scholar]
- Tsunematsu R.; Nakayama K.; Oike Y.; Nishiyama M.; Ishida N.; Hatakeyama S.; Bessho Y.; Kageyama R.; Suda T.; Nakayama K. I. (2004) J. Biol. Chem. 279, 9417–9423. [DOI] [PubMed] [Google Scholar]
- Vardar D.; North C. L.; Sanchez-Irizarry C.; Aster J. C.; Blacklow S. C. (2003) Biochemistry 42, 7061–7067. [DOI] [PubMed] [Google Scholar]
- Gordon W. R.; Roy M.; Vardar-Ulu D.; Garfinkel M.; Mansour M. R.; Aster J. C.; Blacklow S. C. (2009) Blood 113, 4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehebauer M. T.; Chirgadze D. Y.; Hayward P.; Martinez Arias A.; Blundell T. L. (2005) Biochem. J. 392, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubman O. Y.; Kopan R.; Waksman G.; Korolev S. (2005) Protein Sci. 14, 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel M. E.; Leahy D. J.; Barrick D. (2005) Structure 13, 1599–1611. [DOI] [PubMed] [Google Scholar]
- Nam Y.; Sliz P.; Song L.; Aster J. C.; Blacklow S. C. (2006) Cell 124, 973–983. [DOI] [PubMed] [Google Scholar]
- Gordon W. R.; Arnett K. L.; Blacklow S. C. (2008) J. Cell Sci. 121, 3109–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio O.; van Gijn M. E.; Bezdek A. C.; Pellegrinet L.; van Es J. H.; Zimber-Strobl U.; Strobl L. J.; Honjo T.; Clevers H.; Radtke F. (2008) EMBO Rep. 9, 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong G. T.; Manfra D.; Poulet F. M.; Zhang Q.; Josien H.; Bara T.; Engstrom L.; Pinzon-Ortiz M.; Fine J. S.; Lee H. J.; Zhang L.; Higgins G. A.; Parker E. M. (2004) J. Biol. Chem. 279, 12876–12882. [DOI] [PubMed] [Google Scholar]
- De Strooper B.; Chavez Gutierrez L. (2015) Annu. Rev. Pharmacol. Toxicol. 55, 419–437. [DOI] [PubMed] [Google Scholar]
- Shen Y.; Bax A. (2013) J. Biomol. NMR 56, 227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Sykes B. D.; Richards F. M. (1992) Biochemistry 31, 1647–1651. [DOI] [PubMed] [Google Scholar]
- Wishart D. S.; Sykes B. D. (1994) Methods Enzymol. 239, 363–392. [DOI] [PubMed] [Google Scholar]
- Wishart D. S. (2011) Prog. Nucl. Magn. Reson. Spectrosc. 58, 62–87. [DOI] [PubMed] [Google Scholar]
- Barrett P. J.; Song Y.; Van Horn W. D.; Hustedt E. J.; Schafer J. M.; Hadziselimovic A.; Beel A. J.; Sanders C. R. (2012) Science 336, 1168–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel A. J.; Mobley C. K.; Kim H. J.; Tian F.; Hadziselimovic A.; Jap B.; Prestegard J. H.; Sanders C. R. (2008) Biochemistry 47, 9428–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


