Abstract
Sphingomyelin (SM) and cholesterol (Chol) are considered essential for the formation of lipid rafts; however, the types of molecular interactions involved in this process, such as intermolecular hydrogen bonding, are not well understood. Since, unlike other phospholipids, SM is characterized by the presence of an amide group, it is essential to determine the orientation of the amide and its order in the lipid bilayers to understand the nature of the hydrogen bonds in lipid rafts. For this study, 1′-13C-2-15N-labeled and 2′-13C-2-15N-labeled SMs were prepared, and the rotational-axis direction and order parameters of the SM amide in bilayers were determined based on 13C and 15N chemical-shift anisotropies and intramolecular 13C-15N dipole coupling constants. Results revealed that the amide orientation was minimally affected by Chol, whereas the order was enhanced significantly in its presence. Thus, Chol likely promotes the formation of an intermolecular hydrogen-bond network involving the SM amide without significantly changing its orientation, providing a higher order to the SM amide. To our knowledge, this study offers new insight into the significance of the SM amide orientation with regard to molecular recognition in lipid rafts, and therefore provides a deeper understanding of the mechanism of their formation.
Introduction
Since the lipid rafts hypothesis was first proposed in the late 1990s (1), the formation of microdomains in biomembranes has been investigated because of its implications for membrane processes such as signal transduction, protein sorting, and microbial infection (2–6). Lipid rafts are thought to be associated with a liquid-ordered phase, which is characterized by tight packing but relatively high lateral lipid mobility. In contrast, unsaturated phosphatidylcholines (PCs) are loosely packed and develop a liquid-disordered phase that coexists with the liquid-ordered domain under certain conditions. In general, cholesterol (Chol) and sphingolipids such as sphingomyelin (SM) are considered essential for the formation of lipid rafts (7–9).
The structure of SM is characterized by the presence of amide and hydroxy groups, which presumably are responsible for the formation of collateral intra- and intermolecular hydrogen bonds in membranes, making SM a preferable raft constituent compared with PC. Numerous theoretical and experimental studies have been conducted in an attempt to reveal the nature of the interactions involving the amide and hydroxy groups of SMs. For example, in lipid bilayers the SM amide group has been reported to interact with water, the hydroxy and amide groups of other SM molecules, and the hydroxy group of Chol, and possible hydrogen bonding with Chol has drawn the most attention as a key factor in raft formation (10–14). However, the detailed nature of the molecular interaction, including hydrogen bonds in the SM bilayers, has not been fully elucidated due to limited experimental data on SM conformation and dynamics.
Recently, Yamaguchi et al. (15) determined the partial conformation of an SM molecule in a membrane environment using bicelles as a raft model, revealing that its conformation is relatively stable and capable of forming intermolecular hydrogen bonds between amide groups. In addition, the comprehensive and unambiguous order profiles of the SM alkyl chains were recently determined using site-specifically deuterated SMs (16,17). The results demonstrated a higher thermal stability of the upper portions of the acyl chains, which further suggests intermolecular hydrogen-bond formation between SM amides. To gain further insights into the hydrogen bonding involving the SM amides, detailed information about the dynamic and spatial properties of the amide moiety in a bilayer membrane is required.
In previous studies (18,19), we reported the rotation-axis direction and order parameter of 6-F-Chol in PC and SM bilayers by analyzing 19F chemical shift anisotropy (CSA) and intramolecular 13C-19F dipole coupling constants in conjunction with 2H NMR data. This method can be applied to analyze the orientation of the SM amide plane in lipid bilayers. Hence, we prepared two types of 13C/15N doubly labeled SMs, 1′-13C-2-15N-SM and 2′-13C-2-15N-SM (Fig. 1), for this study and analyzed 13C and 15N CSAs, as well as intramolecular 13C-15N dipole coupling constants, to determine the rotational-axis direction and order parameter of the SM amides in bilayer membranes with and without Chol. In this study, we used 50 mol % Chol to ensure the formation of the ordered phase as well as to make the condition identical to that used in our previous study on the order profile of SM alkyl chains (16,17).
Figure 1.

Structure of 13C/15N doubly labeled SMs. To see this figure in color, go online.
Materials and Methods
Details regarding the synthetic procedures used for the 13C/15N doubly labeled SMs and calculations of CSA tensors are described in the Supporting Material.
Sample preparation and solid-state NMR measurements
For static CSA measurements of SM amides, powdered 1′-13C-SM or 2-15N-SM (10.8 mg, 14.8 μmol), prepared in the same manner as the 13C/15N doubly labeled SMs, was transferred into a 5-mm magic-angle spinning (MAS) rotor. For preparation of membrane samples, labeled SM (8.0 mg, 10.9 μmol) with or without Chol (4.2 mg, 10.9 μmol) was dissolved in MeOH-CHCl3 and the solvent was removed under vacuum for at least 12 h. The dried membrane film was hydrated with 1 mL of water. After a few minutes of sonication (Branson 1510 ultrasonic cleaner (Danbury, CT)) and vortexing (Voltex-2Genie (Scientific Industries, Bohemia, NY)) at 55°C, the suspension was freeze-thawed three times to make multilamellar vesicles. Then the vesicle solution was lyophilized and rehydrated with 8 μL (for SM membranes) or 12.2 μL (for SM-Chol membranes) using deuterated water. The suspension was vortexed and freeze-thawed several times, and transferred into a 3.2-mm glass tube (Wilmad, Vineland, NJ). The glass tube was sealed with epoxy glue and inserted into a 5-mm MAS rotor.
All of the solid-state NMR spectra were recorded on a 300-MHz CMX300 spectrometer (Chemagnetics; Varian, Palo Alto, CA). The 13C and 15N cross-polarization (CP)-MAS spectra were recorded with a CP contact time of 3 ms and a recycle delay of 2.5 s at 45°C under various MAS frequencies. Two-pulse phase modulation (TPPM) 1H decoupling (20) was applied during acquisition with a decoupling power of 65 kHz.
13C-15N rotational-echo double-resonance NMR (REDOR) spectra were acquired with a 5-mm triple-resonance MAS probe for 1H, 13C, and 15N (Varian) using xy-8 phase cycling for 15N irradiations (21). The MAS frequency was 5000 Hz and the rotor temperature was maintained at 45°C. The 1H 90° pulse width was 3.3 μs and the 180° pulse widths for 13C and 15N were 8.5 and 8.3 μs, respectively. The contact time for CP was set to 3 ms. The recycle delay was 2.5 s, the sweep width was 30 kHz, and the number of scans was ∼16,000. TPPM 1H decoupling (20) was applied during acquisition with a decoupling power of 65 kHz.
Results and Discussion
Preparation of 13C and 15N doubly labeled SMs
An N-stearoyl SM was selected for this study because it is the most abundant SM constituent in bovine brain (22). We prepared two types of 13C and 15N doubly labeled SMs (Fig. 1): 1′-13C-2-15N-SM for 13C and 15N CSA and intramolecular dipole coupling measurements, and 2′-13C-2-15N-SM for intramolecular dipole coupling measurements. These labeled SMs were synthesized by coupling 13C-labeled stearic acids with 2-15N-lyso-SM prepared using a modification of a previously reported procedure (Fig. 2) (15,23). Briefly, starting from 15N-l-serine, the formation of a Weireb amide followed by a substitution reaction with a vinyl Grignard reagent provided a vinyl ketone. The ketone was reduced to a secondary alcohol with LiAlH(Ot-Bu)3 and the resulting allyl alcohol was subjected to olefin metathesis using a Grubbs catalyst. Introduction of a choline group and deprotection of a Boc group gave TBS-protected 2-15N-lyso-SM. Finally, coupling with the p-nitrophenyl ester of 1′-13C-stearic acid or 2′-13C-stearic acid (for details regarding the preparation of these esters, see the Supporting Material) furnished the appropriately labeled SMs (Fig. 2). For the static CSA measurements of powdered samples, we also prepared singly labeled SMs (1′-13C-SM and 2-15N-SM) in the same manner.
Figure 2.
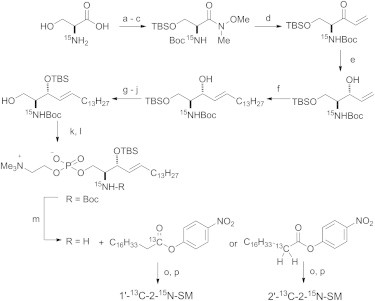
Synthesis of 13C/15N doubly labeled SMs. (a–p) Reagents and conditions: (a) (Boc)2O, NaOH, H2O, dioxane, rt, 2 h; (b) NH(Me)OMe∙HCl, NMM, EDC, CH2Cl2, −15°C, 4 h; (c) TBSCl, imidazole, DMF, rt, 1 h (86% for three steps); (d) vinyl MgBr, THF, rt, 1.5 h (86%); (e) LiAlH(OtBu)3, EtOH, −78°C, 2 h (93%); (f) 1-pentadecene, Grubbs II catalyst, p-quinone, CH2Cl2, 55°C, 2 h (64%); (g) TBAF, THF, rt, 35 min (93%); (h) PivCl, pyridine, −30°C, 2 h (89%); (i) TBSOTf, 2,6-lutidine, CH2Cl2, 0°C, 30 min; (j) DBU, MeOH, rt, 16 h (40% for two steps); (k) 2-chloro-1,3,2-dioxaphosphoran-2-oxide, Et3N, DMAP, benzene, 0°C, 1.5 h; (l) Me3N, CH3CN, 85°C, 46 h (90% for two steps); (m) TFA, CH2Cl2, 0°C, 1 h; (o) Et3N, DMAP, THF, rt, 21 h; and (p) TBAF, THF, rt, 28 h (53% for three steps).
13C and 15N CSA of SM amides
With the labeled SMs available, we obtained static solid-state NMR spectra of powdered 1′-13C-SM and 2-15N-SM to determine the original CSA tensor values for the amide 13C and 15N of SM, which have not been documented previously. Spectra were recorded with slow MAS under 1H decoupling, and spinning side band patterns were simulated with SIMPSON 3.0 software (24) (Fig. 3). Table 1 lists the principal CSA values, which are typical of amide bonds (25).
Figure 3.

(a and b) Amide regions of CP-MAS spectra (black lines) of powdered 1′-13C-SM (a) and 2-15N-SM (b) with spectral simulation using SIMPSON (red lines). Spectra were recorded at 45°C under MAS of (a) 1.5 kHz or (b) 0.7 kHz. To see this Figure in color, go online.
Table 1.
Principal CSA values (ppm) for powdered SM amides
| δ11 | δ22 | δ33 | δiso | |
|---|---|---|---|---|
| 13C of SM amide | 251 () | 185 () | 93 () | 176 |
| 15N of SM amide | 210 () | 67 () | 54 () | 110 |
The possible errors were within ±3 ppm.
Next, to obtain the CSA principal-axis system with respect to the amide frame, we calculated the CSA tensors using the gauge including atomic orbital (GIAO) method (26). The truncated structure shown in Fig. S1 was used to input data for the calculations. Before performing the CSA tensor calculations, we optimized the structure using Becke’s three-parameter hybrid functional (27) and the Lee, Yang, and Parr correlation functional (B3LYP) (28) method using a 6-311G(d) basis set. For CSA tensor calculations using the GIAO method, the density-functional theory (DFT) method was adopted with a 6-311G++(2d,2p) basis. Fig. 4 shows the calculated CSA principal-axis systems (for more details, see Tables S1–S5), in which δ33 of 13C and δ22 of 15N are perpendicular to the amide plane and the remaining principal axes are on the plane. These tensor directions are consistent with those typical of amide bonds (25).
Figure 4.

Top view of the amide group of SM from calculated 13C and 15N CSA principal-axis systems. The axes of and are perpendicular to the amide plane. Experimental principal values in ppm (Table 1) are shown in parentheses. To see this figure in color, go online.
Similar measurements were performed in hydrated membrane systems with and without equimolar Chol. Since pure stearoyl SM membrane forms gel phase below the phase transition temperature (44°C) (29), which provides broadened solid-state NMR spectra, it is necessary to measure the spectra above 44°C. In addition, in our previous report (17), we found that there is no critical difference in the 2H NMR spectra of deuterated SM bilayers between 45°C and 50°C in either the presence or absence of Chol; thus, the NMR spectra were recorded at 45°C for SM and SM-Chol membranes. In membrane environments, lipid molecules undergo fast axial rotation, giving rise to an axially symmetric signal pattern that is defined by the two tensor values δ┴ and δ//. Fig. 5 shows the 13C CP-MAS spectra of 1′-13C-2-15N-SM in hydrated bilayers with SIMPSON simulation spectra, which resulted in estimates for the δ┴ and δ// values (Table 2), in conjunction with the anisotropic parameter, Δδ = δ// − δ┴. The 15N CSA values of the SM amide group in membranes were obtained in a similar manner (Table 2; Fig. S2).
Figure 5.
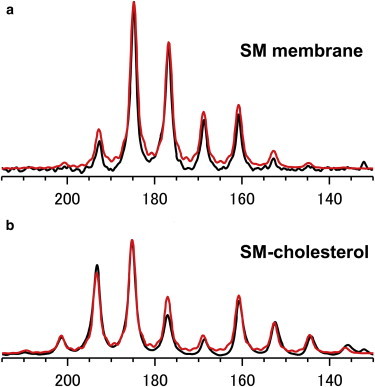
13C CP-MAS spectra of hydrated 1′-13C-2-15N-SM with spectral simulations using SIMPSON (red lines). (a and b) 1′-13C-2-15N-SM (a) and 1′-13C-2-15N-SM/Chol (b) at a 1:1 molar ratio were hydrated with 50 wt% D2O, and both spectra were recorded at 45°C under MAS of 600 Hz. To see this figure in color, go online.
Table 2.
13C and 15N CSA principal values (ppm) of SM amide in hydrated membranes at 45°C
|
13C |
δiso |
15N |
δiso | |||||
|---|---|---|---|---|---|---|---|---|
| δ// | δ┴ | ΔδCa | δ// | δ┴ | ΔδNa | |||
| SM | 151.0 | 188.5 | −37.5 | 176 | 86 | 122 | −36 | 110 |
| SM/Chol (1:1) | 139.0 | 194.5 | −55.5 | 176 | 82 | 124 | −42 | 110 |
Anisotropic parameter Δδ = δ// − δ┴. The possible errors were within ±3 ppm.
Magnitude of the C-N intramolecular dipole coupling of SM amides
Next, to determine the rotational-axis direction and the molecular order parameter of SM amide in bilayers, we measured the intramolecular C-N dipole couplings of 1′-13C-2-15N-SM and 2′-13C-2-15N-SM using the C-N REDOR method (30). Fig. 6 a shows the REDOR spectra of the 1′-13C-2-15N-SM/Chol membrane with a dephasing time of 4 ms at 45°C, which demonstrates a significant reduction in the 13C signal due to the intramolecular REDOR dephasing effect from 15N. Fig. 6 b plots the experimental REDOR dephasing values (ΔS/S0) for 1′-13C-2-15N-SM in bilayers. A comparison with theoretical dephasing curves revealed that the intramolecular dipole coupling values between C1′ and N were 265 and 150 Hz for the SM-Chol and SM membranes, respectively. Note that intermolecular C-N dipole coupling was negligible at these dephasing times because intermolecular REDOR between 1′-13C-SM and 2′-15N-SM was marginal, at most ΔS/S0 = 2%, even at much longer dephasing times (≥40 ms; unpublished results).
Figure 6.

Intramolecular REDOR experiments for 1′-13C-2-15N-SM. (a) 13C-15N REDOR spectra for the SM-Chol membrane (1′-13C-2-15N-SM/Chol = 1:1 in 50 wt% D2O) with a dephasing time of 4 ms at 45°C. The S0 (black line) and S (red line) spectra were obtained with and without 15N-irradiation, respectively. (b) REDOR dephasing effects for (●) SM-Chol membranes and (▪) SM-only membranes at various dephasing times. ΔS was (S0 − S). The best-fit theoretical curves were obtained with D = 265 Hz (red line) and 150 Hz (blue line) for SM-Chol and SM membranes, respectively. The possible errors were within ±20 Hz. To see this figure in color, go online.
Similarly, the intramolecular REDOR between C2′ and amide N was measured using 2′-13C-2-15N-SM bilayers. As shown in Fig. 7, the intramolecular dipole coupling for SM-Chol was 13 Hz as determined from theoretical curve fitting, whereas the SM membrane did not produce any significant REDOR dephasing effect even at a dephasing time of 160 ms. This suggests that dipole coupling is almost nonexistent, probably because the average angle between the rotation axis and the C2′-N bond is close to the magic angle.
Figure 7.

Intramolecular REDOR plots for 2′-13C-2-15N-SM in membranes in the (●) presence and (▪) absence of Chol. ΔS was (S0 − S). Intramolecular REDOR dephasing was not observed for the SM membrane. The best-fit theoretical curve for SM-Chol with D = 13 Hz is shown. The possible errors were within ±2 Hz. To see this figure in color, go online.
Calculations of the rotation-axis angles and order parameter for SM amides
The rotation axis is defined by the polar coordinates α (azimuthal angle) and β (polar angle) connecting the rotation axis with the 13C CSA tensor axes (Fig. 8). To account for the wobbling of the rotation axis, the order parameter (S), which varies from 1 to 0, was introduced. When S = 1, no molecular wobbling with respect to the rotation axis is present, and when S = 0, every orientation is allowed (such as in solution) (31). Although Saupe order tensor analysis is more exact when the wobbling motion of the rotation axis is anisotropic (32), approximating the motion using the single order parameter S, which assumes a fast isotropic wobbling, also is acceptable (31).
Figure 8.

The rotation-axis direction is defined by the polar coordinates α (azimuthal angle) and β (polar angle) with respect to the 13C CSA tensor axes. Angle θ is the angle between the rotation axis and the C1′-N bond. The oxygen atom on C1′ was omitted for clarity. To see this figure in color, go online.
Using α, β, and S values, the anisotropic parameter of the amide 13C () can be calculated as (18,33)
Similarly, as a function of α, β, and S, is given by
where terms containing (α − 129.4°) appear because the direction overlaps due to a −129.4° rotation around the normal of the amide plane (Fig. 4).
The magnitude of dipolar coupling is known to depend on molecular motion as well as internuclear distance. Therefore, for a fixed internuclear distance such as C1′-N and C2′-N in the doubly labeled SMs, attenuation of the magnitude of coupling should be due only to motional averaging. When the C1′-N bond vector is rotating around the axis, the C1′-N dipolar coupling constant can be provided by the following equation (31):
where θ is the angle between the C1′-N vector and the rotation axis (Fig. 8) and denotes the dipolar coupling constant for the static C1′-N interaction, which was calculated to be 1303 Hz based on the C1′-N internuclear distance of 1.33 Å. Angle θ is related to α and β by cosθ = sinβ cos(138.4° − α) because the angle between the C1′-N vector and is 138.4° (Fig. 4).
Similarly, the C2′-N dipolar coupling constant is provided by
where ϕ is the angle between the C2′-N vector and the rotation axis, which has the relation cosϕ = sinβ cos(103.8° − α), because the angle between the C2′-N vector and δC 22 is 103.8° (Fig. 4). The value for was calculated to be 219 Hz based on the C2′-N internuclear distance of 2.41 Å.
The best-fit combinations of α, β, and S were obtained by minimizing the root mean-square deviation (RMSD) between the experimental and calculated dipolar coupling constants D (DC1′-N and DC2′-N) and anisotropic parameters Δδ (ΔδC and ΔδN):
Each value in this equation was normalized by its rigid-limit value (Dmax or Δδmax) to equalize the contributions from dipolar couplings and chemical shifts (34).
Fig. 9 shows the 2D RMSD plots for SM and SM-Chol membranes at S values of 0.67 and 0.84, respectively, which provide the minimum RMSD values. From the figure, the combinations of α, β, and S were elucidated as listed in Table 3, although β has two possible values for each membrane. Although Fig. 9 seems to indicate another minimum points at α = ∼140° and β = ∼45° and 135°, we can rule out these combinations because those RMSD values are 0.10 for SM and 0.16 for SM-Chol membranes, and thus are much higher than those listed in Table 3. It is also clear from the contour gradient in Fig. 9 that α angles in Table 3 have larger uncertainty than β angles. These results indicate that the rotational-axis directions are very similar regardless of the presence or absence of Chol, whereas the order parameter S is enhanced significantly by Chol. The value of S is related to wobbling in the rotation axis, and a value of 0.84 leads to a cone semi-angle of 14° for oscillations of the rotation axis when a Gaussian distribution of the wobbling angle is assumed (35,36). In contrast, an S value of 0.67 indicates an angle of 21°.
Figure 9.

(a and b) 2D RMSD contour plots for (a) SM at S = 0.67 and (b) SM-Chol at S = 0.84; β has two possible values. To see this figure in color, go online.
Table 3.
Order parameter S and (α, β) values that provide a minimum RMSD
| RMSD | S | α | βa | |
|---|---|---|---|---|
| SM | 0.03 | 0.67 ± 0.05 | 103° ± 10° | 34° ± 3° (146 ± 3°) |
| SM-Chol | 0.045 | 0.84 ± 0.05 | 99° ± 10° | 31° ± 3° (149 ± 3°) |
The uncertainties take into account the propagation of experimental errors.
Although the two possible β values give two potential rotational-axis directions, the values in the parentheses are less probable when the conformation of SM is considered (see text).
Of note are the small RMSD values in both membranes (Table 3), which validate the current assumption that the wobbling motion of the amide rotation axis is fast and isotropic. As described above, if the wobbling motion of the rotation axis is anisotropic, Saupe order tensor analysis using Szz and Sxx−Syy values should be adopted (32), and therefore the approximation using a single order parameter S (i.e., the Sxx−Syy value is ignored) would provide much larger RMSD values.
Because of the two possible β values (Fig. 9; Table 3), each membrane has two possible rotational-axis directions. Fig. 10 a shows the two possible orientations of the amide plane of the SM-Chol membrane according to the values listed in Table 3. A recent report revealed that SM forms bicelles with dihexanoylphosphocholine (15), and its partial conformation in bicelles was determined, indicating that the conformation around the amide is relatively stable based on coupling constants typical of anti-rotamers. Based on the conformation of SM, the orientation corresponding to a β of 31° (Fig. 10 b) is more reasonable because the long axis of an SM molecule appears parallel to the rotation axis, whereas the rotation axis is not directed along the SM long axis when β is 149° (Fig. 10 c). Therefore, the rotational axes of SM amide are α = 103°, β = 34°, and S = 0.67 for SM bilayers, and α = 99°, β = 31°, and S = 0.84 for SM-Chol bilayers.
Figure 10.
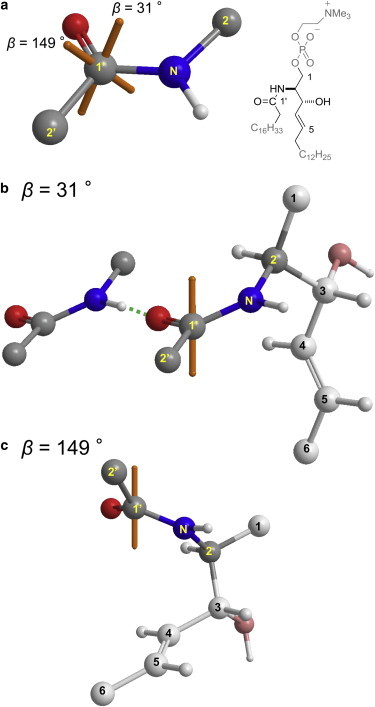
(a–c) Two rotation-axis directions for an SM-Chol membrane (a) with respect to the amide plane and partial orientations of an SM molecule for (b) β = 31° or (c) 149°, based on the conformation of SM deduced from bicelle experiments (15). The orange axes indicate the rotation axes. Considering the direction of an acyl chain, orientation b is more likely than c. Structure b also shows one possible intermolecular hydrogen bond involving the SM amide group. To see this figure in color, go online.
Implications for amide orientation analysis
Here, we first discuss the validity of the above analysis, in which structural heterogeneity of the amide orientation is assigned entirely to wobble. Judging from the typical static CSA values of SM in the powder state (Table 1), it is reasonable to consider that the amide plane is rigid. On that basis, we assumed that the amide plane takes a single major orientation with wobbling. Of course, there are two other possibilities: 1) stable plural orientations of the amide plane exist with respect to the rotation axis and 2) the plural orientations exchange rapidly. The latter case can be regarded as wobbling of the amide plane, and therefore our assumption holds true. The former case would provide larger RMSD values because the calculation of dipolar and anisotropy values in the RMSD equation is based on the assumption that the SM amide takes a single orientation. The small RMSD values (Table 3) suggest that the dynamics of the amide plane can be approximated as a single major orientation with wobbling, although the residual RMSDs might indicate the presence of minor orientations.
SM molecules feature two potential hydrogen-bond donors in the bilayer interfacial region, i.e., amide and hydroxy groups, whereas PCs contain none of these. Consequently, the formation of interlipid hydrogen bonds is often proposed as a mechanism of raft formation (10–14,37–39). Interestingly, this study demonstrates that the SM amide orientation is not affected significantly by the presence or absence of Chol. There are two possible explanations for this: first, the direct hydrogen bonding between the SM amide and Chol may not be as strong as previously proposed. This seems in line with our previous observations (16,17) that Chol is situated more deeply in SM membranes than in saturated PC membranes; more specifically, the rigid alicyclic parts of Chol are located in the middle region of alkyl chains of SM. The resultant depth difference between the SM amide and the Chol OH group would induce an orientational change of the SM amide plane upon formation of the hydrogen bond. A recent simulation study also suggested minimal formation of hydrogen bonds between the SM amide and Chol (40). Second, it is also possible to form a water-bridged hydrogen bond between the SM amide and Chol without significantly altering the amide orientation. On the basis of our data, we are unable to distinguish between these two possibilities; however, detailed simulation studies based on our data may help to resolve this issue.
In contrast, Chol significantly enhanced the order parameter S of SM amide from 0.67 to 0.84, indicating that Chol restricts the rotation axis wobbling of the SM amide. A possible explanation for this result is that the interlipid distances were shortened due to the ordering effect of Chol on the SM alkyl chains, consequently promoting the intermolecular hydrogen-bond networks involving the SM amide. In fact, the orientation and conformation around the SM amide, as shown in Fig. 10 b, seems suitable for forming intermolecular hydrogen bonds with neighboring SM amides. In other words, Chol is likely to shift the equilibrium toward formation of the amide-associated hydrogen bonds through its ordering effect on the SM alkyl chains. Collectively, the upper regions of SM-Chol membranes appear to be stabilized by the hydrogen bonds involving the SM amide, and the middle regions are ordered by the rigid alicyclic skeleton of Chol, both of which synergistically contribute to form the ordered phase of SM-Chol membranes.
Conclusions
In this study we revealed the orientation and order parameter of the amide moiety of SM in the presence and absence of Chol by using C-N dipole couplings and chemical-shift anisotropies, and successfully preparing 13C and 15N doubly labeled SMs. We demonstrated that the amide orientation was minimally affected by Chol, whereas the order was enhanced significantly in the presence of Chol. These findings can be explained by the notion that the ordering effect of Chol on the SM alkyl chains tightens membrane packing, shortens the interlipid distances, and promotes the intermolecular hydrogen bonds involving the SM amide. In fact, the orientation of the SM amide (Fig. 10 b), as deduced from the results presented here as well as a previously determined conformation of SM (15), seems suitable for forming the intermolecular hydrogen bonds between SM amides, although it is also possible to form intermolecular hydrogen bonds with the OH group of Chol. The hydrogen bonds thus formed restrict the wobbling of the SM amide and enhance its order. Considering the dynamic nature of bilayer membranes, the SM amide is more likely to rapidly undergo hydrogen-bond formation and dissociation than to form stable intermolecular hydrogen bonds. Therefore, it is more reasonable to assume that the stabilization effect of Chol on the hydrophobic regions of SM shifts the dynamic equilibrium toward intermolecular hydrogen-bond formation involving the SM amide. In this context, this study provides new (to our knowledge) insights into the significance of the SM amide orientation with regard to molecular recognition in lipid rafts, and therefore enhances our understanding of lipid rafts and the mechanism of their formation. Finally, it is expected that the results of this study, in conjunction with previously reported comprehensive deuterium order profiles of SM alkyl chains (16,17,41–43), will provide basic information for molecular-dynamics studies of SM membranes.
Author Contributions
N.M. designed research, T.Y. and Y.M. performed research, and all authors analyzed data. N.M. and M.M. wrote the article.
Acknowledgments
The authors thank Dr. Naoya Inazumi (Osaka University), Prof. Tohru Oishi (Kyushu University), and Dr. Yuichi Umegawa (Osaka University) for their help with organic synthesis and solid-state NMR measurements.
This work was supported by Grants-in-Aid for Scientific Research (A) (No. 25242073), (B) (No. 20310132), and (S) (No. 18101010) from MEXT, Japan, and by a SUNBOR grant from the Suntory Institute for Bioorganic Research, Japan.
Editor: Klaus Gawrisch.
Footnotes
Nobuaki Matsumori’s present address is Department of Chemistry, Graduate School of Science, Kyushu University, Higashi-ku, Fukuoka 812-8581, Japan.
Supporting Material
References
- 1.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Anderson R.G.W., Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. doi: 10.1126/science.1068886. [DOI] [PubMed] [Google Scholar]
- 3.Binder W.H., Barragan V., Menger F.M. Domains and rafts in lipid membranes. Angew. Chem. Int. Ed. Engl. 2003;42:5802–5827. doi: 10.1002/anie.200300586. [DOI] [PubMed] [Google Scholar]
- 4.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 5.Ikonen E. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 2001;13:470–477. doi: 10.1016/s0955-0674(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 6.Brown D.A., London E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed S.N., Brown D.A., London E. On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes. Biochemistry. 1997;36:10944–10953. doi: 10.1021/bi971167g. [DOI] [PubMed] [Google Scholar]
- 8.Baumgart T., Hess S.T., Webb W.W. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 9.Silvius J.R., del Giudice D., Lafleur M. Cholesterol at different bilayer concentrations can promote or antagonize lateral segregation of phospholipids of differing acyl chain length. Biochemistry. 1996;35:15198–15208. doi: 10.1021/bi9615506. [DOI] [PubMed] [Google Scholar]
- 10.Simons K., Vaz W.L.C. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 11.Li X.-M., Momsen M.M., Brown R.E. Cholesterol decreases the interfacial elasticity and detergent solubility of sphingomyelins. Biochemistry. 2001;40:5954–5963. doi: 10.1021/bi002791n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bittman R., Kasireddy C.R., Slotte J.P. Interaction of cholesterol with sphingomyelin in monolayers and vesicles. Biochemistry. 1994;33:11776–11781. doi: 10.1021/bi00205a013. [DOI] [PubMed] [Google Scholar]
- 13.Sankaram M.B., Thompson T.E. Interaction of cholesterol with various glycerophospholipids and sphingomyelin. Biochemistry. 1990;29:10670–10675. doi: 10.1021/bi00499a014. [DOI] [PubMed] [Google Scholar]
- 14.Zidar J., Merzel F., Janežic D. Liquid-ordered phase formation in cholesterol/sphingomyelin bilayers: all-atom molecular dynamics simulations. J. Phys. Chem. B. 2009;113:15795–15802. doi: 10.1021/jp907138h. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi T., Suzuki T., Murata M. NMR-based conformational analysis of sphingomyelin in bicelles. Bioorg. Med. Chem. 2012;20:270–278. doi: 10.1016/j.bmc.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Matsumori N., Yasuda T., Murata M. Comprehensive molecular motion capture for sphingomyelin by site-specific deuterium labeling. Biochemistry. 2012;51:8363–8370. doi: 10.1021/bi3009399. [DOI] [PubMed] [Google Scholar]
- 17.Yasuda T., Kinoshita M., Matsumori N. Detailed comparison of deuterium quadrupole profiles between sphingomyelin and phosphatidylcholine bilayers. Biophys. J. 2014;106:631–638. doi: 10.1016/j.bpj.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumori N., Kasai Y., Nomura K. Orientation of fluorinated cholesterol in lipid bilayers analyzed by 19F tensor calculation and solid-state NMR. J. Am. Chem. Soc. 2008;130:4757–4766. doi: 10.1021/ja077580l. [DOI] [PubMed] [Google Scholar]
- 19.Matsumori N., Okazaki H., Murata M. Fluorinated cholesterol retains domain-forming activity in sphingomyelin bilayers. Chem. Phys. Lipids. 2011;164:401–408. doi: 10.1016/j.chemphyslip.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Bennett A.E., Rienstra C.M., Griffin R.G. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 21.Gullion T., Baker D.B., Conradi M.S. New, compensated Carr-Purcell sequences. J. Magn. Reson. 1990;89:479–484. [Google Scholar]
- 22.Jungalwala F.B., Hayssen V., McCluer R.H. Separation of molecular species of sphingomyelin by reversed-phase high-performance liquid chromatography. J. Lipid Res. 1979;20:579–587. [PubMed] [Google Scholar]
- 23.Yamamoto T., Hasegawa H., Katsumura S. Versatile synthetic method for sphingolipids and functionalized sphingosine derivatives via olefin cross metathesis. Org. Lett. 2006;8:5569–5572. doi: 10.1021/ol062258l. [DOI] [PubMed] [Google Scholar]
- 24.Bak M., Rasmussen J.T., Nielsen N.C. SIMPSON: a general simulation program for solid-state NMR spectroscopy. J. Magn. Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 25.Saitô H., Ando I., Ramamoorthy A. Chemical shift tensor—the heart of NMR: insights into biological aspects of proteins. Prog. Nucl. Magn. Reson. Spectrosc. 2010;57:181–228. doi: 10.1016/j.pnmrs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ditchfield R. Self-consistent perturbation theory of diamagnetism 1. A gauge-invariant LCAO method for NMR chemical shifts. Mol. Phys. 1974;27:789–807. [Google Scholar]
- 27.Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 28.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 29.Jaikishan S., Björkbom A., Slotte J.P. Sphingomyelin analogs with branched N-acyl chains: the position of branching dramatically affects acyl chain order and sterol interactions in bilayer membranes. Biochim. Biophys. Acta. 2010;1798:1987–1994. doi: 10.1016/j.bbamem.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Gullion T., Schaefer J. Rotational-echo double-resonance NMR. J. Magn. Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Ulrich A.S. Solid state F-19 NMR methods for studying biomembranes. Prog. Nucl. Magn. Reson. Spectrosc. 2005;46:1–21. [Google Scholar]
- 32.Sternberg U., Witter R., Ulrich A.S. All-atom molecular dynamics simulations using orientational constraints from anisotropic NMR samples. J. Biomol. NMR. 2007;38:23–39. doi: 10.1007/s10858-007-9142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bechinger B., Sizun C. Alignment and structural analysis of membrane polypeptides by 15N and 31P solid-state NMR spectroscopy. Concepts Magn. Reson. A. 2003;18A:130–145. [Google Scholar]
- 34.Tang M., Waring A.J., Hong M. Orientation of a β-hairpin antimicrobial peptide in lipid bilayers from two-dimensional dipolar chemical-shift correlation NMR. Biophys. J. 2006;90:3616–3624. doi: 10.1529/biophysj.105.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaikh S.R., Cherezov V., Wassall S.R. Molecular organization of cholesterol in unsaturated phosphatidylethanolamines: X-ray diffraction and solid state 2H NMR reveal differences with phosphatidylcholines. J. Am. Chem. Soc. 2006;128:5375–5383. doi: 10.1021/ja057949b. [DOI] [PubMed] [Google Scholar]
- 36.Oldfield E., Meadows M., Jacobs R. Spectroscopic studies of specifically deuterium labeled membrane systems. Nuclear magnetic resonance investigation of the effects of cholesterol in model systems. Biochemistry. 1978;17:2727–2740. doi: 10.1021/bi00607a006. [DOI] [PubMed] [Google Scholar]
- 37.Khelashvili G.A., Scott H.L. Combined Monte Carlo and molecular dynamics simulation of hydrated 18:0 sphingomyelin-cholesterol lipid bilayers. J. Chem. Phys. 2004;120:9841–9847. doi: 10.1063/1.1724814. [DOI] [PubMed] [Google Scholar]
- 38.Veiga M.P., Arrondo J.L.R., Marsh D. Interaction of cholesterol with sphingomyelin in mixed membranes containing phosphatidylcholine, studied by spin-label ESR and IR spectroscopies. A possible stabilization of gel-phase sphingolipid domains by cholesterol. Biochemistry. 2001;40:2614–2622. doi: 10.1021/bi0019803. [DOI] [PubMed] [Google Scholar]
- 39.Epand R.M. Cholesterol in bilayers of sphingomyelin or dihydrosphingomyelin at concentrations found in ocular lens membranes. Biophys. J. 2003;84:3102–3110. doi: 10.1016/S0006-3495(03)70035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z., Bhide S.Y., Berkowitz M.L. Molecular dynamics simulations of bilayers containing mixtures of sphingomyelin with cholesterol and phosphatidylcholine with cholesterol. J. Phys. Chem. B. 2007;111:12888–12897. doi: 10.1021/jp074037i. [DOI] [PubMed] [Google Scholar]
- 41.Bunge A., Müller P., Huster D. Characterization of the ternary mixture of sphingomyelin, POPC, and cholesterol: support for an inhomogeneous lipid distribution at high temperatures. Biophys. J. 2008;94:2680–2690. doi: 10.1529/biophysj.107.112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartels T., Lankalapalli R.S., Brown M.F. Raftlike mixtures of sphingomyelin and cholesterol investigated by solid-state 2H NMR spectroscopy. J. Am. Chem. Soc. 2008;130:14521–14532. doi: 10.1021/ja801789t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veatch S.L., Soubias O., Gawrisch K. Critical fluctuations in domain-forming lipid mixtures. Proc. Natl. Acad. Sci. USA. 2007;104:17650–17655. doi: 10.1073/pnas.0703513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


