Abstract
Objectives
The objective of this study was to characterize chromosomal mutations associated with resistance to tetracycline in Streptococcus pneumoniae.
Methods
Chronological appearance of mutations in two S. pneumoniae R6 mutants (R6M1TC-5 and R6M2TC-4) selected for resistance to tetracycline was determined by next-generation sequencing. A role for the mutations identified was confirmed by reconstructing resistance to tetracycline in a S. pneumoniae R6 WT background. RNA sequencing was performed on R6M1TC-5 and R6M2TC-4 and the relative expression of genes was reported according to R6. Differentially expressed genes were classified according to their ontology.
Results
WGS of R6M1TC-5 and R6M2TC-4 revealed mutations in the gene rpsJ coding for the ribosomal protein S10 and in the promoter region and coding sequences of the ABC genes patA and patB. These cells were cross-resistant to ciprofloxacin. Resistance reconstruction confirmed a role in resistance for the mutations in rpsJ and patA. Overexpression of the ABC transporter PatA/PatB or mutations in the coding sequence of patA contributed to resistance to tetracycline, ciprofloxacin and ethidium bromide, and was associated with a decreased accumulation of [3H]tetracycline. Comparative transcriptome profiling of the resistant mutants further revealed that, in addition to the overexpression of patA and patB, several genes of the thiamine biosynthesis and salvage pathway were increased in the two mutants, but also in clinical isolates resistant to tetracycline. This overexpression most likely contributes to the tetracycline resistance phenotype.
Conclusions
The combination of genomic and transcriptomic analysis coupled to functional studies has allowed the discovery of novel tetracycline resistance mutations in S. pneumoniae.
Keywords: S. pneumoniae, ABC transporters, rpsJ, thiamine, genomic, RNA-seq
Introduction
Streptococcus pneumoniae is a Gram-positive bacterium responsible for diseases such as otitis media, meningitis, community-acquired pneumonia and bacteraemia1 and for which resistance to antibiotics has become a worldwide concern. The prevalence of resistance varies between countries, but is globally high for β-lactams, macrolides, chloramphenicol and tetracycline, mainly due to the spread of MDR clones.2 Since the first report in 1967 of penicillin and tetracycline non-susceptible isolates,3–7 resistance has increased to a point where 24%, 35%, 42% and even >80% of disease-causing pneumococci are now resistant to tetracycline in the USA, France, Spain and some regions of Asia, respectively.8–10 Doxycycline, a tetracycline mostly used for the treatment of intracellular bacterial infections, has been proposed for the treatment of community-acquired pneumonia.11 However, cross-resistance between the tetracycline family compounds is observable in pneumococci,12,13 except for the new glycylcycline tigecycline.
Tetracycline inhibits protein synthesis by blocking the attachment of charged aminoacyl-tRNA to the A site on the ribosome, which prevents the introduction of new amino acids to the nascent peptide chain.14 Resistance to tetracycline in bacteria occurs through enzymatic inactivation,15 by active efflux or by ribosome protection from the acquisition of tet genes.16,17 In pneumococci, the acquisition of the gene tet(M) is the most common mechanism of resistance to tetracycline,6,18,19 while resistance mediated by tet(O) has only been reported sporadically.20 Both genes are located on Tn916-like mobile genetic elements and encode ribosomal protection proteins that have homology to elongation factors G.21 The GTPase activity of tet(M) and tet(O) appears to be important for the displacement of tetracycline from the ribosome.19 Resistance to tetracycline is now so frequent in S. pneumoniae that it is seldom used for treating this bacterium.
Besides mobile elements, chromosomal mutations have also been shown to participate in resistance to tetracycline. In Helicobacter pylori, the triple base pair mutation AGA926-928→TTC in both copies of the 16S rRNA gene caused high-level resistance to tetracycline, whereas strains with low-level resistance harboured only single or double base pair mutations.22,23 In Neisseria gonorrhoeae, point mutations in the rpsJ gene coding for ribosomal protein S10, the gene mtrR coding for a transcriptional regulator or the gene penB coding for a porin have also been implicated in resistance to tetracycline.24
Overexpression of multidrug efflux pumps of the ABC superfamily can contribute to resistance to structurally unrelated molecules, including tetracycline in Gram-negative bacteria and fluoroquinolones in S. pneumoniae.25–27 Fluoroquinolones still have an excellent activity against S. pneumoniae, but resistance in in vitro-selected or clinical strains was indeed shown to involve the overexpression of the multidrug ABC transporter PatA/PatB in addition to mutations in gyrase and topoisomerase genes.28–32 Here, we studied resistance to tetracycline in the absence of tet(M) in S. pneumoniae by DNA and RNA sequencing and found several novel mutations and gene overexpression, including one ribosomal protein, the ABC proteins PatA/PatB and thiamine biosynthetic enzymes that are associated for the first time with tetracycline resistance in S. pneumoniae.
Methods
Bacteria culture and strains
Strains used in this study are listed in Table S1 (available as Supplementary data at JAC Online). Pneumococci were grown in brain heart infusion broth (BHI, Difco) or on tryptic soy agar containing 5% defibrinated sheep's blood. Cultures were incubated for 16–24 h in a 5% CO2 atmosphere at 35°C. Tetracycline-resistant mutants of S. pneumoniae were obtained by successive passages of S. pneumoniae R6 WT on plates containing increasing concentrations of tetracycline, as previously described.32–35 Briefly, the selection for tetracycline resistance was conducted on Szybalski plates containing concentration gradients of tetracycline. Mutants were selected by scrapping colonies growing at the front of the highest concentration of tetracycline at each stage. This pool of colonies was then streaked onto plates containing either the same concentration of antibiotic or a gradient of increased antibiotic concentrations. End-point mutants corresponded to colonies growing on plates with the highest concentration of antibiotic and for which an additional increase in tetracycline concentration failed to yield resistant colonies. A total of five and four selection cycles were required to obtain the endpoint S. pneumoniae R6M1TC-5 and R6M2TC-4 mutants, respectively.
Next-generation DNA sequencing
Genomic DNAs were extracted using the Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer's instructions. The genomes of R6M1TC-5 and R6M2TC-4 were sequenced using a 454 Life Sciences GS-FLX system (Roche). Genome sequencing, assemblies and comparative analyses were performed at the McGill University Genome Québec Innovation Center. R6M1TC-5 and R6M2TC-4 generated aggregated genome sizes of 2 016 699 and 2 017 241 bp, respectively, at a mean 21× coverage. WGS was also performed on clones of strains R6M1TC-1, R6M1TC-2, R6M1TC-3, R6M1TC-4, R6M2TC-1, R6M2TC-2 and R6M2TC-3 using an Illumina MiSeq system (Centre de Recherche du CHU de Québec) and a 250 nt paired-end reads protocol. This generated genome assemblies covering at least 99% of the S. pneumoniae R6 genome at a mean 120× coverage. Sequence reads from each strain were filtered based on the quality score using Trimmomatic36 and aligned to the genome of S. pneumoniae R6 using the software bwa (bwa aln, version 0.5.9) with default parameters.37 The maximum number of mismatches was four, the seed length was 32 and two mismatches were allowed within the seed. The detection of SNPs was performed using samtools (version 0.1.18), bcftools (distributed with samtools) and vcfutils.pl (distributed with samtools),38 with a minimum of three reads to call a potential variation prior to further analysis. Mutations deduced from massively parallel sequencing were confirmed by PCR amplification and conventional DNA sequencing.
Sequencing data accession numbers
All sequencing data described in this paper have been deposited at the EBI SRA database under the study accession number PRJEB6539 and the sample accession numbers ERS480579 (R6M1TC-1), ERS480580 (R6M1TC-2), ERS480581 (R6M1TC-3a), ERS480582 (R6M1TC-3b), ERS480583 (R6M1TC-4a), ERS480584 (R6M1TC-4b), ERS480567 (R6M1TC-5), ERS480585 (R6M2TC-1a), ERS480586 (R6M2TC-1b), ERS480587 (R6M2TC-2a), ERS480588 (R6M2TC-2b), ERS480589 (R6M2TC-3a), ERS480590 (R6M2TC-3b) and ERS480568 (R6M2TC-4).
Genetic transformation and inactivation
Long PCR fragments (∼5 kb) containing the mutations of interest were amplified using primers listed in Table S2 and transformed in S. pneumoniae R6 as described previously.32,34 For introducing mutations that were less amenable to selection, short (500 bp) PCR fragments amplified from ribosomal protein S12 (rpsL) or dihydrofolate reductase (dhfr) gene variants conferring resistance to streptomycin and trimethoprim, respectively, were co-transformed and used as surrogate selection markers as previously described.32 Fragments containing the mutations from the entire patA-patB operon from R6M1TC-5 and R6M2TC-4 were produced by overlap PCR from two distinct fragments to increase the length of the PCR product.
Gene inactivation was performed using the plasmid pFF6 (kanamycin®) or pFF3 (chloramphenicol®).32 PCR fragments covering the middle region of the genes patA, patB, spr0632, spr0634, spr0637 and spr0638 were amplified using the primers listed in Table S2 and cloned into EamII05I-digested pFF6 or pFF3 to produce plasmids pFF6patA, pFF6patB, pFF6spr0632(R6), pFF3spr0632(CCRI22087), pFF6spr0634, pFF6spr0637 and pFF6spr0638. These were transformed into S. pneumoniae as previously described.34,39
MIC determination
MICs of tetracycline (Sigma), ciprofloxacin (Sigma) and ethidium bromide (Fluka) in the absence and the presence of 20 mg/L reserpine (Sigma) were determined by microdilution. All MIC measurements were performed with three independent biological replicates, each replicate being further assessed in technical duplicates.
RNA sequencing
Total RNA was isolated from S. pneumoniae R6M1TC-5, R6M2TC-4 and R6 WT grown to mid-log phase in BHI using the Qiagen RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNAs were quantified using 2100 BioAnalyzer RNA6000 Nano chips (Agilent) and 1 μg of total RNA was treated with Ribo-Zero™ rRNA Removal Kits (Bacteria) (Epicentre). RNA-seq libraries were produced from 50 ng of rRNA-depleted samples using the ScriptSeq™ v2 RNA-Seq Library Preparation Kit (Epicentre). The libraries were analysed using 2100 BioAnalyser High Sensitivity DNA chips and quantified by PicoGreen. The libraries were pooled, diluted to 8 pM and sequenced on an Illumina MiSeq system using a 250 bp paired-ends reads protocol.
Reads were mapped to the S. pneumoniae R6 reference genome as described above. Transcripts were assembled from the alignment files using the Cufflinks pipeline.40 Differential gene expression was computed with CuffDiff, and genes with a P value ≤0.015 were considered for further analysis.
Quantitative real-time PCR (qRT–PCR)
Total RNAs were extracted as described above and treated with DNase I (Ambion) to avoid any DNA contamination. The quality and integrity of the RNAs was assessed using a 2100 BioAnalyzer and RNA6000 Nano chips (Agilent). cDNAs were generated from 250 ng of total RNAs using the Superscript II reverse transcriptase (Invitrogen) and random hexamers according to the manufacturer's instructions. qRT–PCR assays were carried out with a Bio-Rad Cycler using SYBR Green I (Molecular Probes). A final volume of 10 μL was used for each reaction containing specific primers (Table S2) and iQ SYBR Green Supermix (Bio-Rad). All qRT–PCR data were normalized according to the amplification signals of 16S rRNA.
Gene ontology (GO)
Differentially expressed genes were classified according to their ontology using the Blast2GO software.41 If not mentioned, the default setups were used during the analysis. S. pneumoniae R6 ORFs were first blasted against the NCBI genome database and GO mapping performed on the blasted genomes to assign cellular component, molecular function and biological process ontology levels. Mapped genes where then screened using KEGG. GO enrichment analysis (Fisher's exact two-tailed t-test) was then performed for genes identified as overexpressed or down-regulated (P ≤ 0.015) in R6M1TC-5 and R6M2TC-4 by the CuffLinks pipeline, and also on the S. pneumoniae R6 genome as a control. GO terms with a value P ≤ 0.015 were considered significantly enriched in the group tested. Results are expressed as the proportion of sequences associated with a particular GO term in the tested group.
Tetracycline accumulation
Three independent S. pneumoniae cultures were independently grown to exponential phase (OD600 = 0.2–0.3) in BHI. Cells were centrifuged and washed once with 0.1 M phosphate buffer, pH 7. Cells were then concentrated 40× (∼109 cells/mL) in 0.1 M phosphate buffer containing 1 mM MgSO4, pH 7, and incubated at 37°C in the presence 1% glucose for 15 min. After pre-incubation with glucose, 0.05 μM [7-3H(N)]tetracycline (17.9 Ci/mmol, Moravek Biochemicals and Radiochemicals) was added to a final concentration of 5 μM of tetracycline. [3H]tetracycline was used within 1 week of arrival to avoid possible degradation.42 When required, 20 mg/L of the efflux pump inhibitor reserpine was added 15 min after the addition of [3H]tetracycline. Aliquots (0.1 mL) were withdrawn at specific intervals and accumulation was stopped by diluting with 1 mL of ice-cold 0.1 M phosphate buffer, pH 7, containing 100 mM LiCl. Samples were centrifuged immediately at 4°C for 2 min and washed once with 0.1 M ice-cold phosphate buffer/100 mM LiCl, pH 7. Pelleted cells were resuspended in 0.1 mL of 1× PBS and 5 mL of Ecolite was added before scintillation counts were measured using a LS6000TA Scintillation counter (Beckman). Results were normalized to bacterial count obtained prior to the addition of tetracycline at 0 and 37°C, since no growth was observed during the course of the experiment after the addition of the drug.
Results
Tetracycline-resistant mutants
Two S. pneumoniae mutants named R6M1TC-5 and R6M2TC-4 were independently selected for resistance to tetracycline from S. pneumoniae R6 WT (MIC of tetracycline = 0.125 mg/L) by serial passages in the presence of increasing tetracycline concentrations until they reached an MIC of 8 mg/L (Table 1). This is considered resistant according to recently revised breakpoints for tetracycline.43 Five and four passages on tetracycline-supplemented agar plates were required for the selection of R6M1TC-5 and R6M2TC-4, respectively. The R6M1TC-5 and R6M2TC-4 mutants were also cross-resistant to the fluoroquinolone ciprofloxacin and to the intercalating dye ethidium bromide (Table 1). A low-level cross-resistance was also observed in R6M1TC-5 and R6M2TC-4 to the new glycylcycline tigecycline. Interestingly, the efflux pump inhibitor reserpine decreased the ciprofloxacin and ethidium bromide MICs for both mutants to WT levels and also substantially increased their sensitivity to tetracycline (Table 1). Reserpine had no effect on the small cross-resistance to tigecycline (Table 1). While confirming a role for drug efflux in the multidrug resistance phenotype of S. pneumoniae R6M1TC-5 and R6M2TC-4, the reserpine-insensitive residual tetracycline resistance suggests that additional mechanisms are also likely to contribute to resistance in these mutants.
Table 1.
MICs for tetracycline-resistant strains used in this study
| MICs (mg/L) |
||||||||
|---|---|---|---|---|---|---|---|---|
| TET | TET + R | CIP | CIP + R | EtBr | EtBr + R | TGC | TGC + R | |
| R6 | 0.125 | 0.125 | 0.5 | 0.5 | 2 | 0.125 | 0.031 | 0.031 |
| R6M1TC-1 | 2 | 1 | 1 | 0.5 | 8 | 0.25 | 0.063 | 0.063 |
| R6M1TC-2 | 2 | 1 | 2 | 0.5 | 8 | 0.25 | 0.063 | 0.063 |
| R6M1TC-3 | 4 | 1 | 4 | 0.5 | 8 | 0.25 | 0.063 | 0.063 |
| R6M1TC-4 | 4 | 1 | 4 | 0.5 | 16 | 0.25 | 0.063 | 0.063 |
| R6M1TC-5 | 8 | 2 | 4 | 0.5 | 16 | 0.25 | 0.25 | 0.25 |
| R6M2TC-1 | 2 | 1 | 1 | 0.5 | 2 | 0.25 | 0.063 | 0.063 |
| R6M2TC-2 | 2 | 1 | 1 | 0.5 | 2 | 0.25 | 0.063 | 0.063 |
| R6M2TC-3 | 4 | 1 | 4 | 0.5 | 16 | 0.25 | 0.125 | 0.125 |
| R6M2TC-4 | 8 | 1 | 8 | 0.5 | 16 | 0.25 | 0.125 | 0.125 |
TET, tetracycline; R, reserpine (20 mg/L); CIP, ciprofloxacin; Etbr, ethidium bromide; TGC, tigecycline.
MICs are the results of three independent biological replicates.
WGS of tetracycline-resistant mutants
The genomes of each selection step leading to both R6M1TC-5 and R6M2TC-4 mutants were sequenced to understand the genomics of tetracycline resistance and the order of appearance of mutations linked to resistance. The genomes of S. pneumoniae R6M1TC-5 and R6M2TC-4 both contained nine mutations, which were distributed over seven and eight genomic loci, respectively (Table 2). Two of the mutated loci were common to both mutants, i.e. the gene spr0187 coding for ribosomal protein S10 and the gene spr1887 coding for the multidrug ABC transporter PatA, although mutations occurred at different positions in each mutant (Table 2). For the gene spr0187 (also known as rpsJ), the R6M1TC-5 mutant harboured a C157T transition (leading to a R53C mutation) in addition to a small deletion covering positions 175–180 (Table 2). Interestingly, intermediate mutants R6M1TC-1 to R6M1TC-4 presented instead a single G178T transversion in spr0187 (Table 2). The increasing tetracycline pressure thus appears to have selected for several distinct events in R6M1TC, the first one being selected early at position 178 and the others occurring late at positions 157 and 175–180 of rpsJ (which encompasses the initial G178T mutation) (Table 2). In contrast, a unique A169G transition leading to a K57E substitution was observed in spr0187 in R6M2TC-4, which was selected early at the first selection step (Table 2). For the patA locus, both mutants harboured mutations upstream of the start codon (possibly in the promoter region) and within the coding region (Table 2). Interestingly, in both cases the mutations in the promoter region of patA preceded those within the gene (Table 2). The number of patA mutations also correlated with the level of resistance to tetracycline (Table 2).
Table 2.
Chronological appearance of mutations in tetracycline-resistant mutants
| Locus IDa | Functiona | Mutationsb–d |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R6M1TC |
R6M2TC |
|||||||||
| 1e | 2f | 3g | 4h | 5 | 1i | 2j | 3k | 4 | ||
| spr0187 (rpsJ) | 30S ribosomal protein S10 |
G178T D60Y |
G178T D60Y |
G178T D60Y |
G178T D60Y |
C157T R53C Δ175–180 |
A169G K57E |
A169G K57E |
A169G K57E |
A169G K57E |
| spr0219 | phosphoglycerate mutase | WT | WT | WT | WT | T-52C | WT | WT | WT | WT |
| spr0291 | phosphotransferase system IIA component | WT | WT | WT | WT | WT | C-101G | C-101G | C-101G |
C-101G T-30C |
| spr0691 | biotin synthase | WT | WT | WT | WT | WT |
G356T A119D |
G356T A119D |
G356T A119D |
G356T A119D |
| spr0822 | agmatine deiminase | WT | WT | WT | WT | WT | WT | WT | WT | A95C Stop position 32 |
| spr0825 | hypothetical protein | G-61A | G-61A | G-61A | G-61A | G-61A | WT | WT | WT | WT |
| spr0960 | positive transcriptional regulator MutR | WT | WT | WT | WT | A-151C | WT | WT | WT | WT |
| spr1262 | transcriptional regulator Spx | WT | WT | T-11C | T-11C | T-11C | WT | WT | WT | WT |
| spr1377 | cystathionine γ-synthase | WT | WT | WT | WT | WT | WT | WT | WT | T290 CA97V |
| spr1777 | DNA-directed RNA polymerase subunit β | WT | WT | WT | WT | C1442T Syn |
WT | WT | WT | WT |
| spr1885 (patB) | ABC transporter ATP-binding protein/permease | WT | WT | WT | WT | WT | WT |
G794A G265E |
G794A G265E |
G794A G265E |
| spr1887 (patA) | ABC transporter ATP-binding protein/permease | G-35A |
G-35A C883A R295S |
G-35A G510A M170I C883A R295S |
G-35A G510A M170I C883A R295S |
G-35A G510A M170I C883A R295S |
WT | WT | G-48A |
G-48A G860A G287E |
| spr1991 | glycerol kinase | WT | WT | WT | WT | WT | WT | WT | WT | ΔG position 75 |
aID, identity. Nomenclature according to the genome annotation of S. pneumoniae R6.
bWhen mutations are within coding regions, the change in amino acids is also indicated in italics.
cIn non-coding sequence, the number preceded by a hyphen indicates the position upstream of the ATG.
dMutations in bold were further studied to determine their potential role in tetracycline resistance and mutations underlined are involved in drug resistance (tetracycline, ciprofloxacin, ethidium bromide).
eOne clone was sequenced. Unique mutations: spr1138 (C443A).
fOne clone was sequenced. Unique mutations: spr0428 (G250A); spr0857 (G1298T).
gTwo clones were sequenced. Unique mutations: spr0313 (A195G); spr1032 (G1216T); spr1367 (C511A); spr1865 (G224A); spr2035 (G-72A).
hTwo clones were sequenced. Unique mutations: spr0294 (T111C).
iTwo clones were sequenced. Unique mutations: spr0661 (G896T); spr0797: A to T 458 bp downstream of the gene.
jTwo clones were sequenced. Unique mutations: spr0661 (G896T/A); spr1250 (T999C).
kTwo clones were sequenced. Unique mutations: spr0232 (G191A).
Reconstruction of tetracycline resistance
Recurrent mutations are the most likely candidates to contribute to resistance and the role of the rpsJ and patA mutations was thus further assessed by resistance reconstruction. This was done by transforming S. pneumoniae R6 WT with variant alleles amplified from RM1TC-5 or R6M2TC-4 either alone or along with a PCR fragment covering the rpsL+ allele of S. pneumoniae CP1296 and conferring resistance to streptomycin that was used as a surrogate marker for the selection of transformants (see the Methods section). The rpsL+ PCR fragment had no impact on tetracycline susceptibility levels when transformed alone (Table 3). Each reconstructed strain was then tested for resistance to tetracycline, ciprofloxacin and ethidium bromide in the presence and absence of reserpine. The introduction of the rpsJ alleles from R6M1TC-1 (harbouring a G178T mutation) or R6M2TC-4 (harbouring a A169G mutation) into S. pneumoniae R6 WT increased the tetracycline MIC 4-fold for the S. pneumoniae R6rpsJ(R6M1TC-1) and S. pneumoniae R6rpsJ(R6M2TC) transformants (Table 3). Interestingly, the rpsJ allele from R6M1TC-5 was more potent at conferring resistance to tetracycline than the one from R6M1TC-1 [compare R6rpsJ(R6M1TC-1) and R6rpsJ(R6M1TC-5) in Table 3], which is consistent with their chronological order of appearance during the selection of the R6M1TC series of mutants (Table 3). To exclude the possibility of second-site mutations introduced during the transformation, we reintroduced the WT rpsJ allele in the S. pneumoniae R6rpsJ(R6M1TC-1) and R6rpsJ(R6M1TC-5) transformants and these cells became more susceptible to tetracycline [see R6rpsJ(R6M1TC-1)-rev(R6WT)-rpsL+ and R6rpsJ(R6M1TC-5)-rev(R6WT)-rpsL+ in Table 3].
Table 3.
Functional analysis of mutations detected in S. pneumoniae R6M1TC-5 and R6M2TC-4
| Straina | Mutated alleles |
MIC (mg/L)b |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rpsJ |
patA |
patB | TET | TET + R | CIP | CIP + R | Etbr | Etbr + R | ||
| promotor | CDS | |||||||||
| R6 | WT | WT | WT | WT | 0.125 | 0.125 | 0.5 | 0.5 | 2 | 0.25 |
| R6rpsL+ | WT | WT | WT | WT | 0.125 | 0.125 | 0.5 | 0.5 | 2 | 0.25 |
| R6M1TC-5 | R6M1TC-5 | R6M1TC-5 | R6M1TC-5 | WT | 8 | 2 | 4 | 0.5 | 16 | 0.25 |
| R6M2TC-4 | R6M2TC-4 | R6M2TC-4 | R6M2TC-4 | R6M2TC-4 | 8 | 1 | 8 | 0.5 | 16 | 0.25 |
| R6rpsJ(R6M1TC-1) | R6M1TC-1 | WT | WT | WT | 0.5 | 0.5 | 0.5 | 0.5 | 2 | 0.25 |
| R6rpsJ(R6M1TC-1)-rpsL+ | R6M1TC-1 | WT | WT | WT | 0.5 | ND | 0.5 | ND | ND | ND |
| R6rpsJ(R6M1TC-1)-rev(R6WT)-rpsL+ | WT | WT | WT | WT | 0.125 | ND | 0.5 | ND | ND | ND |
| R6rpsJ(R6M1TC-5) | R6MITC-5 | WT | WT | WT | 1 | 1 | 0.5 | 0.5 | 2 | 0.25 |
| R6rpsJ(R6M1TC-5)-rpsL+ | R6MITC-5 | WT | WT | WT | 1 | ND | 0.5 | ND | ND | ND |
| R6rpsJ(R6M1TC-5)-rev(R6WT)-rpsL+ | WT | WT | WT | WT | 0.125 | ND | 0.5 | ND | ND | ND |
| R6patA(R6M1TC) | WT | R6M1TC-5 | R6M1TC-5 | WT | 0.5 | 0.125 | 2 | 0.5 | 16 | 0.25 |
| R6p_patA(R6M1TC)-rpsL+ | WT | R6M1TC-5 | WT | WT | 0.25 | 0.125 | 1 | 0.5 | 8 | 0.25 |
| R6CDSpatA(R6M1TC)-rpsL+ | WT | WT | R6M1TC-5 | WT | 0.25 | 0.125 | 1 | 0.5 | 8 | 0.25 |
| R6CDSpatA(G510A)(R6M1TC)-rpsL+ | WT | WT | R6M1TC(G510A) | WT | 0.25 | 0.125 | 1 | 0.5 | 4 | 0.25 |
| R6CDSpatA(C883A)(R6M1TC)-rpsL+ | WT | WT | R6M1TC(C883A) | WT | 0.125 | 0.125 | 1 | 0.5 | 8 | 0.25 |
| R6rpsJ(R6M1TC)-patA(R6M1TC) | R6M1TC-5 | R6M1TC | R6M1TC-5 | WT | 2 | 0.5 | 2 | 0.5 | 16 | 0.25 |
| R6rpsJ(R6M2TC) | R6M2TC-4 | WT | WT | WT | 0.5 | 0.5 | 0.5 | 0.5 | 2 | 0.25 |
| R6patA-patB(R6M2TC) | WT | R6M2TC-4 | R6M2TC-4 | R6M2TC-4 | 0.25 | 0.125 | 1 | 0.5 | 16 | 0.25 |
| R6patA-patB(R6M2TC)-rpsL+ | WT | R6M2TC-4 | R6M2TC-4 | R6M2TC-4 | 0.25 | ND | 1 | ND | ND | ND |
| R6patA-patB(R6M2TC)-rev(patA)-rpsL+ | WT | WT | WT | R6M2TC-4 | 0.125 | ND | 0.5 | ND | ND | ND |
| R6p_patA(R6M2TC)-rpsL+ | WT | R6M2TC-4 | WT | WT | 0.25 | 0.125 | 1 | 0.5 | 8 | 0.25 |
| R6CDSpatA(R6M2TC)-rpsL+ | WT | WT | R6M2TC-4 | WT | 0.25 | 0.125 | 1 | 0.5 | 4 | 0.25 |
| R6CDSpatB(R6M2TC)-rpsL+ | WT | WT | WT | R6M2TC-4 | 0.125 | 0.125 | 0.5 | 0.5 | 2 | 0.25 |
| R6CDSpatA-patB(R6M2TC)-dhfr+ | WT | WT | R6M2TC-4 | R6M2TC-4 | 0.25 | 0.125 | 1 | 0.5 | 8 | 0.25 |
| R6rpsJ(R6M2TC)-patA(R6M2TC) | R6M2TC-4 | R6M2TC-4 | R6M2TC-4 | WT | 0.5 | 0.25 | 2 | 0.5 | 16 | 0.25 |
| R6rpsJ(R6M2TC)-patA(R6M2TC)-patB(R6M2TC) | R6M2TC-4 | R6M2TC-4 | R6M2TC-4 | R6M2TC-4 | 0.5 | 0.25 | 2 | 0.5 | 16 | 0.25 |
TET, tetracycline; R, reserpine (20 mg/L); CIP, ciprofloxacin; Etbr, ethidium bromide; ND, not done; CDS, coding sequence.
arpsL+ and dhfr+ indicate an allele conferring resistance to streptomycin and trimethoprim, respectively, that was co-transformed along with the PCR fragment of interest for selection purposes.
bMICs are the results of three independent biological replicates. MICs in bold are significantly different from the parent strain without the mutation.
The patA allele from R6M1TC-5 also accounted for a 4-fold increase in resistance to tetracycline in the S. pneumoniae R6patA(R6M1TC) transformant (Table 3), of which half is attributable to the G-35A mutation in the promoter region [R6p_patA(R6M1TC)-rpsL+ in Table 3] and the other half to the G510A mutation within the gene [compare R6CDSpatA(R6M1TC)-rpsL+, R6CDSpatA(G510A)(R6M1TC)-rpsL+ and R6CDSpatA(C883A)(R6M1TC)-rpsL+ in Table 3]. The patA allele from R6M2TC-4 was less effective at increasing tetracycline resistance, however, as it only doubled the tetracycline MIC for the S. pneumoniae R6patA(R6M2TC) transformant (Table 3). As expected, tetracycline resistance conferred by the patA mutations, but not by the rpsJ alleles, was reversible by reserpine (Table 3). Resistance reconstruction experiments also implicated mutations upstream and within patA in resistance to ciprofloxacin and ethidium bromide for both mutants (Table 3). As a final proof of the role of patA in resistance, we introduced the WT version of patA in S. pneumoniae R6patA-patB(R6M2TC), which became more susceptible to tetracycline [see R6patA-patB(R6M2TC)-rev(patA)-rpsL+ in Table 3].
Gene inactivation experiments further confirmed the role of the variant patA alleles in the resistance of R6M1TC-5 and R6M2TC-4 (Table 4). Inactivation of the gene coding for the ABC transporter PatB also sensitized both mutants to tetracycline and ciprofloxacin, which is consistent with the finding that PatA and PatB act as heterodimers to extrude toxic compounds in S. pneumoniae.44 In contrast to patA, however, no role in resistance could be attributed to the mutation selected in R6M2TC-4 in the coding region of patB (spr1885) by transformation into a tetracycline-susceptible S. pneumoniae background (Table 3).
Table 4.
Relative expression and functional inactivation of genes
| Strain | Genea | Mean fold expression compared with R6b | Gene inactivateda,c | MIC of tetracycline (mg/L)d | MIC of ciprofloxacin (mg/L)d |
|---|---|---|---|---|---|
| R6 | — | — | none | 0.125 | 0.5 |
| patA | — | patA | 0.125 | 0.5 | |
| patB | — | patB | 0.125 | 0.5 | |
| spr0632 | — | spr0632 | 0.125 | 0.5 | |
| tenA | — | tenA | 0.125 | 0.5 | |
| thiD | — | thiD | 0.125 | 0.5 | |
| thiE | — | thiE | 0.125 | 0.5 | |
| R6M1TC-5 | — | — | none | 8 | 4 |
| patA | 6.031 ± 3.647 | patA | 2 | 1 | |
| patB | — | patB | 4 | 1 | |
| spr0632 | 4.334 ± 1.048 | spr0632 | 4 | 4 | |
| tenA | 5.597 ± 0.997 | tenA | 8 | 4 | |
| thiD | 5.165 ± 1.374 | thiD | 8 | 4 | |
| thiE | 3.952 ± 0.880 | thiE | 8 | 4 | |
| spr1021 | 1.685 ± 0.247 | spr1021 | — | — | |
| R6p_patA(R6M1TC)-rpsL+ | — | — | none | 0.25 | — |
| patA | 7.782 ± 1.157 | — | — | — | |
| R6M2TC-4 | — | — | none | 8 | 8 |
| patA | 15.268 ± 5.841 | patA | 2 | 2 | |
| patB | — | patB | 2 | 2 | |
| spr0632 | 8.325 ± 2.587 | spr0632 | 2 | 4 | |
| tenA | 6.616 ± 1.513 | tenA | 2 | 8 | |
| thiD | 6.955 ± 0.541 | thiD | 4 | 8 | |
| thiE | 4.381 ± 0.570 | thiE | — | 8 | |
| spr1021 | 1.548 ± 0.199 | spr1021 | — | — | |
| R6p_patA(R6M1TC)-rpsL+ | — | — | none | 0.25 | — |
| patA | 11.426 ± 3.034 | — | — | — |
aS. pneumoniae R6 gene accessions: patA, spr1887; patB, spr1885; tenA, spr0634; thiD, spr0638; thiE, spr0637.
bMean fold expression was measured using qRT–PCR. Gene expressions were normalized on the 16S rRNA expression. The gene spr1021 was used as a control.
cInactivation of patA and patB was performed as a control.
dThe MICs are the results of three independent biological replicates. MICs in bold are significantly different from the parent strain without the inactivated gene.
Other strain-specific mutations were observed in the mutants (Table 2). These were often, but not always, detected in the most resistant mutants, but we could not, by gene transformation along with the rpsL+ allele, link any of these strain-specific mutations with tetracycline resistance (result not shown).
Decreased accumulation of tetracycline in S. pneumoniae-resistant mutants
Mutations upstream of patA have previously been linked to drug resistance in S. pneumoniae by inducing the overexpression of the transporter,35,45 although never for tetracycline. qRT–PCR also confirmed that patA was overexpressed in the S. pneumoniae R6M1TC-5 and R6M2TC-4 mutants, and introducing the G-35A and G-48A mutations in a S. pneumoniae R6 WT background confirmed that these were indeed responsible for patA overexpression (Table 4). As mutations in the coding regions of patA were also shown to contribute to tetracycline (Table 3) and ciprofloxacin (Table 3 and by Lupien et al.32) resistance, tetracycline accumulation experiments were conducted to directly address to role of both types of mutations (i.e. promoter versus coding region) on the transport capacity of the PatA/PatB heterodimer. S. pneumoniae R6M1TC-5 and R6M2TC-4 accumulated less [3H]tetracycline than the parental S. pneumoniae R6 WT (P ≤ 0.05) (Figure 1a). This is directly linked to the presence of patA variants, as S. pneumoniae R6patA(R6M1TC) and R6patA-patB(R6M2TC) harbouring promoter and coding region mutations derived from R6M1TC-5 and R6M2TC-4, respectively, displayed the same reduced accumulation of [3H]tetracycline as the original mutants (Figure 1b and c). Interestingly, while mutations in the promoter of patA and in its coding region conferred resistance to tetracycline when introduced on their own in S. pneumoniae R6 WT (Table 3), only the mutations in the promoter region were responsible for the decreased accumulation of tetracycline in the mutants (P ≤ 0.05) (Figure 1b and c). While mutations derived from the coding regions of patA/B in R6M2TC apparently enabled a small decrease in [3H]tetracycline accumulation compared with S. pneumoniae R6 WT (Figure 1c), the difference was not significant, but this might be due to the sensitivity of the transport experiment. The addition of reserpine, an inhibitor of efflux pumps,29,46 restored the accumulation of tetracycline in the S. pneumoniae R6patA(R6M1TC) and R6patA-patB(R6M2TC) transformants (Figure 1d).
Figure 1.
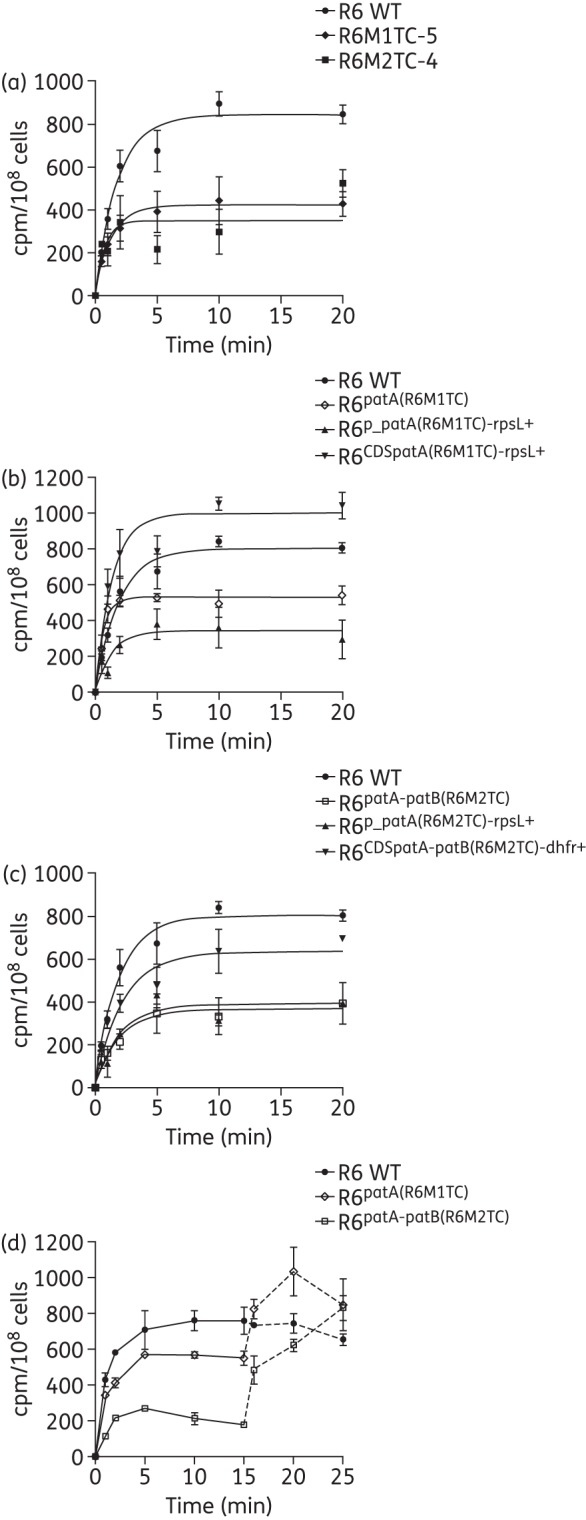
Accumulation of tetracycline in S. pneumoniae. (a) Accumulation of labelled tetracycline was measured in S. pneumoniae R6 WT, R6M1TC-5 and R6M2TC-4 at baseline and at 5 min intervals for a total of 20 min. (b) The role of mutations selected at the patA locus in R6M1TC-5 in accumulation of tetracycline was assessed by comparing the accumulation of labelled tetracycline in R6 WT, R6patA(R6M1TC), R6p_patA(R6M1TC)-rpsL+ and R6CDSpatA(R6M1TC)-rpsL+ for up to 20 min. (c) The role of mutations selected at the patA locus in R6M2TC-4 in accumulation of tetracycline was assessed by comparing the accumulation of labelled tetracycline in R6 WT, R6patA-patB(R6M2TC), R6p_patA(R6M2TC)-rpsL+ and R6CDSpatA-patB(R6M2TC)-dhfr+ for up to 20 min. Results represent the means of three independent replicates. A t-test was performed on normalized data at 10 and 20 min and the significance threshold set to P ≤ 0.05. (d) Accumulation of tetracycline in the presence of reserpine in R6 WT, R6patA(R6M1TC) and R6patA-patB(R6M2TC). Reserpine (20 mg/L) was added at 15 min.
RNA expression profiling in tetracycline-resistant mutants
The rpsJ and patA mutations could only partly explain the levels of tetracycline resistance of S. pneumoniae R6M1TC-5 and R6M2TC-4 (Table 3) and none of the other mutations identified in the genome of the mutants (Table 2) conferred resistance to tetracycline when introduced into S. pneumoniae R6 WT (not shown). We therefore hypothesized that resistance to tetracycline could also involve differences in gene expression (as in the case of patA/patB) and performed comparative gene expression profiling between the R6M1TC-5 and R6M2TC-4 mutants and their susceptible S. pneumoniae R6 WT parent by RNA-seq. Overall, 43 and 60 genes were found to be significantly overexpressed (P ≤ 0.015) in the R6M1TC-5 and R6M2TC-4 mutants compared with R6 WT, respectively (Figure 2 and Table S3). Moreover, 31 and 26 genes had their expression significantly decreased (P ≤ 0.015) in R6M1TC-5 and R6M2TC-4 compared with R6 WT, respectively (Figure 2 and Table S3). Overall, 13 overexpressed and 14 down-regulated genes were common to both mutants (Table S3).
Figure 2.
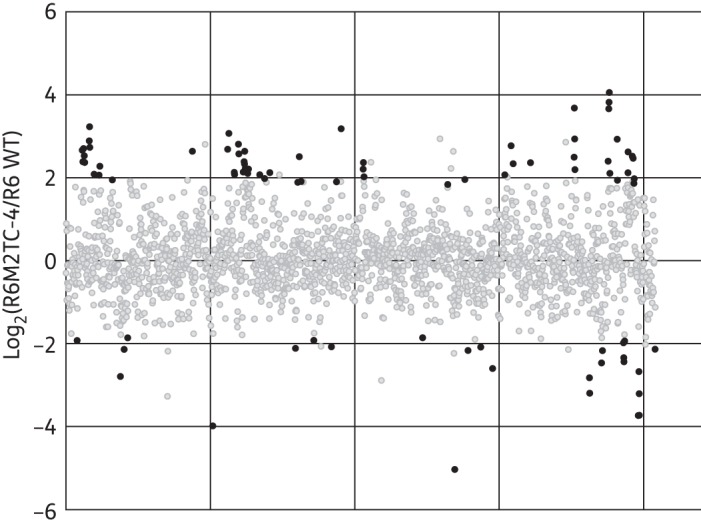
Gene expression alterations in S. pneumoniae tetracycline-resistant mutants. Gene expression was compared between S. pneumoniae R6 WT and R6M1TC-5 or R6M2TC-4 by RNA-seq. Results for R6M2TC-4 are shown. The dots represent the 2043 genes from the S. pneumoniae R6 genome sorted in numerical order on the abscissa. Black dots represent overexpressed and down-regulated genes (P ≤ 0.01) and grey dots represent not significantly altered genes.
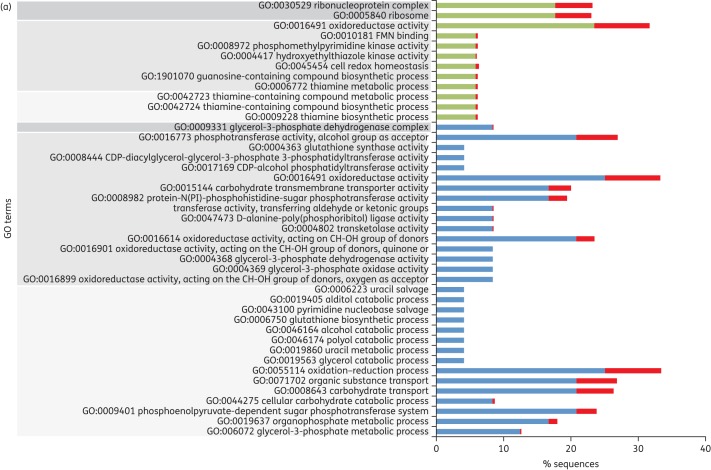
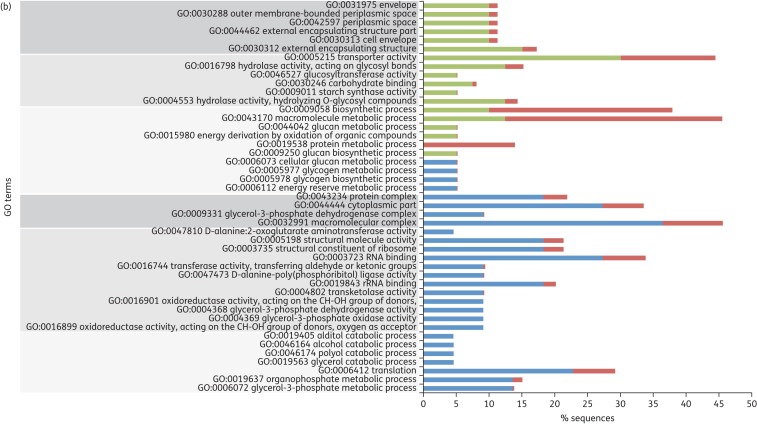
Clustering differentially expressed genes according to their ontology revealed that genes overexpressed in S. pneumoniae R6M1TC-5 were enriched for biological processes involving thiamine metabolism (Figure 3). For S. pneumoniae R6M2TC-4, genes identified as overexpressed by RNA-seq were enriched mainly for processes implicated in carbohydrate metabolism and transport functions (Figure 3) and were not directly linked to the metabolism of thiamine. Nonetheless, a thorough examination of the RNA-seq data revealed that the expression of most genes related to thiamine metabolism overexpressed in R6M1TC-5 were also increased in R6M2TC-4 when the cut-off P value for filtering differentially expressed genes was loosened to P ≤ 0.05. qRT–PCR later confirmed that genes spr0632, spr0634, spr0637 and spr0638, all involved in thiamine metabolism (see Figure S1), were indeed overexpressed in both mutants (Table 4). The expression of the spr0632 gene was further tested by qRT–PCR in 10 clinical isolates of S. pneumoniae (five susceptible and five resistant). Interestingly, an increased expression of spr0632 was observed in two resistant clinical isolates and in none of the susceptible ones (Table 5). Down-regulated genes also clustered in GO terms shared by the two tetracycline-resistant mutants (biological processes implicated in alcohol/polyol/glycerol or organophosphate metabolism; Figure 3).
Figure 3.
GO classification of genes whose expression is significantly altered in S. pneumoniae tetracycline-resistant mutants. Genes differentially expressed (P ≤ 0.015) in (a) R6M1TC-5 and (b) R6M2TC-4 compared with S. pneumoniae R6 WT were clustered according to their ontology. GO terms surrounded by dark grey, medium grey and light grey correspond to cellular component-, molecular function- and biological process-associated GO terms, respectively. The percentage of sequences associated with a specific GO term is shown for overexpressed genes (green), down-regulated genes (blue) and overall S. pneumoniae R6 genes (red). This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Table 5.
Expression of spr0632 in S. pneumoniae clinical isolates susceptible and resistant to tetracycline
| Strain | Specimen sourcea | Genotypeb |
Mean fold expression of spr0632c | Tetracycline MIC for WT/Δspr0632 (mg/L)d | ||
|---|---|---|---|---|---|---|
| tet(M) | tet(O) | rpsJ | ||||
| R6 | ATCC BAA-2555 | − | − | WT | 1 | 0.125/0.125 |
| CCRI 14730 | broncho-alveolar lavage | − | + | WT | 0.770 ± 0.097 | 32/NA |
| CCRI 14774 | throat | + | − | WT | 4.937 ± 1.510 | 2/NA |
| CCRI 15711 | sinus | + | − | WT | 0.858 ± 0.442 | 2/NA |
| CCRI 18414 | nose | − | + | WT | 0.950 ± 0.467 | 16/16 |
| CCRI 22087 | CSF | + | − | WT | 2.913 ± 0.772 | 32/16 |
| CCRI 55073 | respiratory tract | − | − | WT | 0.795 ± 0.360 | 0.125/NA |
| CCRI 60827 | respiratory tract | − | − | WT | 0.671 ± 0.315 | 0.125/NA |
| CCRI 1974 | NA | − | − | WT | 0.525 ± 0.060 | 0.125/NA |
| CCRI 9008 | sputum | NA | NA | NA | 1.104 ± 0.619 | 0.125/NA |
| CCRI 14598 | blood | NA | NA | NA | 0.581 ± 0.322 | 0.125/NA |
NA, not available.
aAll clinical isolates are from Canada, except for CCRI 1974, which is of unknown origin.
bPrimers used to amplified tet(M), tet(O) and rpsJ are listed in Table S1.
cSignificant increased expression is shown in bold.
dSensitization to tetracycline upon spr0632 inactivation is indicated in bold.
In addition to its central role in intermediary metabolism, vitamin B1 (i.e. thiamine) has also been reported to act as an important stress-response molecule against oxidative or osmotic stress in bacteria,47,48 plants49,50 and fungi.51 Interestingly, a triphosphorylated derivative of vitamin B1, namely thiamine triphosphate, was shown to act as an alarmone, initiating a reaction cascade involved in the adaptation of bacteria to stringent conditions such as amino acid starvation.52 We hypothesized that the overexpression of genes linked to thiamine metabolism in R6M1TC-5 and R6M2TC-4 could also have a role in favouring resistance to tetracycline. The gene spr0632 is overexpressed in R6M1TC-5 and R6M2TC-4 and codes for the ATPase subunit of an ABC transporter putatively involved in the transport of hydroxymethyl pyrimidine (HMP; one of the building blocks of thiamine) from the extracellular milieu (Figure S1). The inactivation of spr0632 by insertion–duplication mutagenesis decreased the level of tetracycline resistance of R6M1TC-5 and R6M2TC-4 by 2- and 4-fold, respectively (Table 4). A 2-fold sensitization to tetracycline was also observed upon inactivation of spr0632 in the clinical isolate CCRI 22087 initially overexpressing the gene, but not in the clinical isolate CCRI 18414 not overexpressing spr0632 (Table 5) [we were incapable of transforming strain CCRI 14774, which also had overexpression of spr0632 (Table 5)]. This is consistent with the lack of tetracycline sensitization upon inactivation of spr0632 in S. pneumoniae R6 WT or clinical isolates not overexpressing the gene (Tables 4 and 5) and suggests that overexpression is required for its contribution in resistance. In the case of R6M2TC-4, the inactivation of spr0632 also slightly sensitized to ciprofloxacin (Table 4). The inactivation of spr0634 and spr0638, coding for a thiaminase and for a phosphomethylpyrimidine kinase in the thiamine biosynthesis pathway (Figure S1), also affected the level of tetracycline susceptibility, but this time specifically in R6M2TC-4 (Table 4), which suggests that overexpression of some genes of the HMP salvage pathway is associated with tetracycline resistance.
Discussion
The selection of the S. pneumoniae tetracycline-resistant mutants R6M1TC-5 and R6M2TC-4 from S. pneumoniae R6 WT [which lacks tet(M) or tet(O)] by incremental increases in tetracycline concentrations revealed the possibility of S. pneumoniae becoming resistant to tetracycline through genomic alterations. For both mutants, the first selected event occurred at the level of the gene rpsJ coding for ribosomal protein S10. Based on the crystal structure of the 30S ribosomal subunit from Thermus thermophilus cross-linked with tetracycline,53 the mutations in the R6M1TC-5 and R6M2TC-4 mutants are all located in the vertex loop of ribosomal protein S10. This loop is composed of 50–60 amino acids and is located near the primary site of action of tetracycline, i.e. the aminoacyl-tRNA site.17 As previously proposed, amino acid changes located in the vicinity of this site could decrease the affinity for the antibiotic by altering the rRNA structure near the tetracycline-binding site.53 Interestingly, the amino acid from ribosomal protein S10 the nearest to the primary site in T. thermophilus (lysine-55) was found to be within 8–9 Å of bound tetracycline24,53 and the equivalent residue is mutated in R6M2TC-4 (K57E). Substitutions at homologous residues were also previously observed and implicated in tetracycline resistance in high-level resistant N. gonorrhoeae,24 confirming the importance of endogenous mutations near the ribosomal aminoacyl-tRNA site for tetracycline resistance in the absence of tet genes.
Multidrug efflux pumps are important contributors of resistance to antimicrobials and the S. pneumoniae multidrug ABC transporter PatA/B had already been implicated in resistance to ethidium bromide, berberine, novobiocin, acriflavine, erythromycin, fluoroquinolones, chloramphenicol and linezolid.35,54 A phenotypic microarray screening for 240 compounds against a panel of S. pneumoniae mutants inactivated for putative efflux pumps also identified PatA/PatB as a possible contributor to oxytetracycline resistance, but this was not further validated by an additional quantitative method like microdilution MIC.55 Here, we provide such evidence with S. pneumoniae transformants for which the introduction of a mutation upstream of patA facilitates the transcription of the PatA/PatB genes to foster resistance to tetracycline by decreasing its accumulation, most likely by increased efflux. While several other bacterial ABC transporters have been shown to confer resistance to structurally dissimilar compounds, including tetracycline and fluoroquinolones,25–27,56 this is the first known time that a reduced accumulation of tetracycline mediated by such transporter has been directly measured. The mutation in the promoter region of patA most likely increases transcription rates by loosening a terminator-like structure upstream of the gene. It was recently proposed that mutations upstream of patA could lead to its overexpression by altering the strength of a terminator-like stem-loop structure in the 5′ untranslated region (UTR) of patA.45,57 Also, constitutive overexpression of patAB in fluoroquinolone-resistant isolates may be caused by mutations in a Rho-independent transcriptional terminator structure located upstream of patA gene.57 An analysis of the DNA sequence upstream of patA using RibEx58 indeed suggested the presence of three stem-loop structures (a transcriptional terminator, an anti-terminator and an anti-anti-terminator structure) in S. pneumoniae R6 (Figure S2). Interestingly, the strength (ΔG) of the transcriptional terminator stem-loop structure is decreased by the presence of the mutations detected upstream of patA in R6M1TC-5 and R6M2TC-4 mutants (Figure S2), suggesting that these alterations are responsible for the increase in patA transcription (Table 4). Similarly, small deletions impeding the formation of a transcriptional attenuator upstream of tet(M) have been shown to increase its transcription rate in S. pneumoniae clinical isolates unsusceptible to tetracycline.59 In S. pneumoniae, tetracycline resistance conferred by ribosomal protection proteins such as Tet(M) would lead to an excess of unbound tetracycline, and overexpression of the chromosomally encoded PatA/PatB efflux pump may be exploited by the pneumococcus to efflux the excess tetracycline. It would thus be interesting to assess whether patA/patB overexpression also occurs in S. pneumoniae clinical isolates resistant to tetracycline due to the presence of mobile elements of the Tn916 family.
Mutations in the coding region of patA also contributed to resistance to tetracycline and ciprofloxacin (Table 2), possibly by altering the specificity of the transporter. A TMHMM analysis of the PatA sequence revealed that every mutation observed in R6M1TC-5 and R6M2TC-4 was located in transmembrane domains (TMs). A comparison with the crystal structure of the bacterial ABC exporter SAV1866 from Staphylococcus aureus60 also supported the location of mutation M170I within TM4 and the location of mutations G287E and R295S within TM6. Residues from transmembrane helices TM1, TM4–TM6 and TM10–TM12 in the homologous human MDR1 have been shown to interact with the substrates of the transporter.61 Unfortunately, no well-defined substrate binding sites have been identified for bacterial ABC exporters62,63 to hypothesize more precisely about the role of these mutations in the activity of the transporter. Nonetheless, it is intriguing that, for both R6M1TC-5 and R6M2TC-4 mutants, a single mutation in the coding region of patA enhanced resistance against structurally different molecules. Mutations affecting substrate specificity have also been reported previously for other multidrug efflux pumps.64,65 These mutations can possibly alter the tertiary structure of the transporter and thereby modulate the substrate profile or provide additional substrate binding sites relevant for the recognition of a given substrate. In the case of R6M1TC, one possibility would be that the M170I substitution in PatA increases its specificity for tetracycline and excludes it from rRNA by favouring its trapping at the membrane in the transporter before its extrusion out of the cell.
The expression profile of R6M1TC-5 and R6M2TC-4 has highlighted that gene expression alterations other than the overexpression of patA/patB can foster resistance to tetracycline (Figure 2 and Table S3). Comparative expression profiling by RNA-seq and qRT–PCR indeed revealed that key genes involved in the salvage of HMP in the thiamine biosynthesis pathway were overexpressed in the R6M1TC-5 and R6M2TC-4 mutants (Tables 4, Table S3 and Figure S1). This appears to be coherent with resistance to tetracycline since the inactivation of spr0632, which is responsible for the uptake of HMP from the extracellular milieu, increased tetracycline susceptibility in both mutants. Interestingly, increased expression of thiamine metabolism gene was also observed in clinical isolates resistant to tetracycline. An increased expression of spr0632 could be detected in two tetracycline-resistant pneumococcal isolates, but in none of the susceptible isolates tested (Table 5). While the number of isolates tested is small, these results indicate that the expression of thiamine metabolism genes can be increased in clinical isolates and further work will undoubtedly help refine our understanding of the clinical significance of thiamine metabolism and resistance to tetracycline. The inactivation of the spr0632 gene in one tetracycline-resistant clinical isolate reduced its resistance to tetracycline. The activity of several enzymes of carbohydrate metabolism, such as pyruvate dehydrogenase, 2-oxoglutarate dehydrogenase and transketolase, requires thiamine pyrophosphate as a co-factor.66 Resistance to tetracycline may require increased energetic needs, which would translate into a higher thiamine demand from these enzymes. Alternatively, thiamine derivatives have been shown to act as signals involved in the adaptation of bacteria to stressful conditions52,67 and it will be interesting to conduct metabolomics studies with extracts derived from mutants R6M1TC-5 and R6M2TC-4 to fully understand the changes happening in the thiamine biosynthesis pathway.
Overall, the combined use of WGS, RNA-seq and functional analyses revealed that endogenous tetracycline resistance mechanisms can be selected in S. pneumoniae in the absence of tet(M) and tet(O). One early mutation is the S10 ribosomal protein, but the clinical relevance of antibiotic efflux mediated by the ABC transporter PatA/PatB in S. pneumoniae was reiterated by the demonstration that the overexpression of this transporter can foster resistance to tetracycline in addition to resistance to fluoroquinolones30–32,44 and linezolid.35 A novel link between thiamine metabolism and resistance to tetracycline was also revealed.
Funding
This work was supported by a Canadian Institutes of Health Research grant to M. O.
Transparency declarations
None to declare.
Author contributions
A. L., P. L. and M. O. conceived and designed the experiments. A. L. and H. G. performed the experiments. A. L. and P. L. conducted the bioinformatics analyses of the data. A. L. and P. L. wrote the paper (with contributions from all of the authors). M. G. B. provided the clinical S. pneumoniae isolates. All authors have read and approved the manuscript for publication.
Supplementary data
Acknowledgements
We thank the McGill University Genome Quebec Innovation Centre for performing the sequencing of R6M1TC-5 and R6M2TC-4 mutants. A. L. received a studentship from programme de bourses de leadership et développement durable de l'Université Laval, and M. O. holds the Canada Research Chair in Antimicrobial Resistance.
References
- 1.Levine OS, O'Brien KL, Knoll M, et al. Pneumococcal vaccination in developing countries. Lancet 2006; 367: 1880–2. [DOI] [PubMed] [Google Scholar]
- 2.McGee L, McDougal L, Zhou J, et al. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol 2001; 39: 2565–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansman D. BMM. A resistant pneumococcus. Lancet 1967; 290: 264–5. [Google Scholar]
- 4.Parra EL, Ramos V, Sanabria O, et al. Serotype and genotype distribution among invasive Streptococcus pneumoniae isolates in Colombia, 2005–2010. PLoS One 2014; 9: e84993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou L, Ma X, Gao W, et al. Molecular characteristics of erythromycin-resistant Streptococcus pneumoniae from pediatric patients younger than five years in Beijing, 2010. BMC Microbiol 2012; 12: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montanari MP, Cochetti I, Mingoia M, et al. Phenotypic and molecular characterization of tetracycline- and erythromycin-resistant strains of Streptococcus pneumoniae. Antimicrob Agents Chemother 2003; 47: 2236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sa-Leao R, Tomasz A, Sanches IS, et al. Carriage of internationally spread clones of Streptococcus pneumoniae with unusual drug resistance patterns in children attending day care centers in Lisbon, Portugal. J Infect Dis 2000; 182: 1153–60. [DOI] [PubMed] [Google Scholar]
- 8.Jones RN, Sader HS, Moet GJ, et al. Declining antimicrobial susceptibility of Streptococcus pneumoniae in the United States: report from the SENTRY Antimicrobial Surveillance Program (1998–2009). Diagn Microbiol Infect Dis 2010; 68: 334–6. [DOI] [PubMed] [Google Scholar]
- 9.Wyres KL, van Tonder A, Lambertsen LM, et al. Evidence of antimicrobial resistance-conferring genetic elements among pneumococci isolated prior to 1974. BMC Genomics 2013; 14: 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim SH, Song JH, Chung DR, et al. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob Agents Chemother 2012; 56: 1418–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson JR. Doxycycline for treatment of community-acquired pneumonia. Clin Infect Dis 2002; 35: 632; author reply-3. [DOI] [PubMed] [Google Scholar]
- 12.Izdebski R, Sadowy E, Fiett J, et al. Clonal diversity and resistance mechanisms in tetracycline-nonsusceptible Streptococcus pneumoniae isolates in Poland. Antimicrob Agents Chemother 2007; 51: 1155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lederman ER, Gleeson TD, Driscoll T, et al. Doxycycline sensitivity of S. pneumoniae isolates. Clin Infect Dis 2003; 36: 1091. [DOI] [PubMed] [Google Scholar]
- 14.Connell SR, Tracz DM, Nierhaus KH, et al. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob Agents Chemother 2003; 47: 3675–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W, Moore IF, Koteva KP, et al. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J Biol Chem 2004; 279: 52346–52. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen F, Starosta AL, Arenz S, et al. Tetracycline antibiotics and resistance mechanisms. Biol Chem 2014; 395: 559–75. [DOI] [PubMed] [Google Scholar]
- 17.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 2001; 65: 232–60; second page, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doherty N, Trzcinski K, Pickerill P, et al. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob Agents Chemother 2000; 44: 2979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widdowson CA, Klugman KP. The molecular mechanisms of tetracycline resistance in the pneumococcus. Microb Drug Resist 1998; 4: 79–84. [DOI] [PubMed] [Google Scholar]
- 20.Widdowson CA, Klugman KP, Hanslo D. Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob Agents Chemother 1996; 40: 2891–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donhofer A, Franckenberg S, Wickles S, et al. Structural basis for TetM-mediated tetracycline resistance. Proc Natl Acad Sci USA 2012; 109: 16900–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu JY, Kim JJ, Reddy R, et al. Tetracycline-resistant clinical Helicobacter pylori isolates with and without mutations in 16S rRNA-encoding genes. Antimicrob Agents Chemother 2005; 49: 578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dailidiene D, Bertoli MT, Miciuleviciene J, et al. Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob Agents Chemother 2002; 46: 3940–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu M, Nandi S, Davies C, et al. High-level chromosomally mediated tetracycline resistance in Neisseria gonorrhoeae results from a point mutation in the rpsJ gene encoding ribosomal protein S10 in combination with the mtrR and penB resistance determinants. Antimicrob Agents Chemother 2005; 49: 4327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huda N, Lee EW, Chen J, et al. Molecular cloning and characterization of an ABC multidrug efflux pump, VcaM, in Non-O1 Vibrio cholerae. Antimicrob Agents Chemother 2003; 47: 2413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Hamad A, Upton M, Burnie J. Molecular cloning and characterization of SmrA, a novel ABC multidrug efflux pump from Stenotrophomonas maltophilia. J Antimicrob Chemother 2009; 64: 731–4. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo T, Chen J, Minato Y, et al. SmdAB, a heterodimeric ABC-Type multidrug efflux pump, in Serratia marcescens. J Bacteriol 2008; 190: 648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avrain L, Garvey M, Mesaros N, et al. Selection of quinolone resistance in Streptococcus pneumoniae exposed in vitro to subinhibitory drug concentrations. J Antimicrob Chemother 2007; 60: 965–72. [DOI] [PubMed] [Google Scholar]
- 29.Garvey MI, Piddock LJ. The efflux pump inhibitor reserpine selects multidrug-resistant Streptococcus pneumoniae strains that overexpress the ABC transporters PatA and PatB. Antimicrob Agents Chemother 2008; 52: 1677–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Garch F, Lismond A, Piddock LJ, et al. Fluoroquinolones induce the expression of patA and patB, which encode ABC efflux pumps in Streptococcus pneumoniae. J Antimicrob Chemother 2010; 65: 2076–82. [DOI] [PubMed] [Google Scholar]
- 31.Garvey MI, Baylay AJ, Wong RL, et al. Overexpression of patA and patB, which encode ABC transporters, is associated with fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 2011; 55: 190–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupien A, Billal DS, Fani F, et al. Genomic characterization of ciprofloxacin resistance in a laboratory-derived mutant and a clinical isolate of Streptococcus pneumoniae. Antimicrob Agents Chemother 2013; 57: 4911–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fani F, Brotherton MC, Leprohon P, et al. Genomic analysis and reconstruction of cefotaxime resistance in Streptococcus pneumoniae. J Antimicrob Chemother 2013; 68: 1718–27. [DOI] [PubMed] [Google Scholar]
- 34.Fani F, Leprohon P, Legare D, et al. Whole genome sequencing of penicillin-resistant Streptococcus pneumoniae reveals mutations in penicillin-binding proteins and in a putative iron permease. Genome Biol 2011; 12: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng J, Lupien A, Gingras H, et al. Genome sequencing of linezolid-resistant Streptococcus pneumoniae mutants reveals novel mechanisms of resistance. Genome Res 2009; 19: 1214–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Billal DS, Feng J, Leprohon P, et al. Whole genome analysis of linezolid resistance in Streptococcus pneumoniae reveals resistance and compensatory mutations. BMC Genomics 2011; 12: 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conesa A, Gotz S, Garcia-Gomez JM, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005; 21: 3674–6. [DOI] [PubMed] [Google Scholar]
- 42.McMurry L, Levy SB. Two transport systems for tetracycline in sensitive Escherichia coli: critical role for an initial rapid uptake system insensitive to energy inhibitors. Antimicrob Agents Chemother 1978; 14: 201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dallas SD, McGee L, Limbago B, et al. Development of doxycycline MIC and disk diffusion interpretive breakpoints and revision of tetracycline breakpoints for Streptococcus pneumoniae. J Clin Microbiol 2013; 51: 1798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boncoeur E, Durmort C, Bernay B, et al. PatA and PatB form a functional heterodimeric ABC multidrug efflux transporter responsible for the resistance of Streptococcus pneumoniae to fluoroquinolones. Biochemistry 2012; 51: 7755–65. [DOI] [PubMed] [Google Scholar]
- 45.Croucher NJ, Mitchell AM, Gould KA, et al. Dominant role of nucleotide substitution in the diversification of serotype 3 pneumococci over decades and during a single infection. PLoS Genet 2013; 9: e1003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitz FJ, Fluit AC, Luckefahr M, et al. The effect of reserpine, an inhibitor of multidrug efflux pumps, on the in-vitro activities of ciprofloxacin, sparfloxacin and moxifloxacin against clinical isolates of Staphylococcus aureus. J Antimicrob Chemother 1998; 42: 807–10. [DOI] [PubMed] [Google Scholar]
- 47.Jung IL, Kim IG. Thiamine protects against paraquat-induced damage: scavenging activity of reactive oxygen species. Environ Toxicol Pharmacol 2003; 15: 19–26. [DOI] [PubMed] [Google Scholar]
- 48.Fukui K, Wakamatsu T, Agari Y, et al. Inactivation of the DNA repair genes mutS, mutL or the anti-recombination gene mutS2 leads to activation of vitamin B1 biosynthesis genes. PLoS One 2011; 6: e19053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tunc-Ozdemir M, Miller G, Song L, et al. Thiamin confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol 2009; 151: 421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rapala-Kozik M, Wolak N, Kujda M, et al. The upregulation of thiamine (vitamin B1) biosynthesis in Arabidopsis thaliana seedlings under salt and osmotic stress conditions is mediated by abscisic acid at the early stages of this stress response. BMC Plant Biol 2012; 12: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowalska E, Kujda M, Wolak N, et al. Altered expression and activities of enzymes involved in thiamine diphosphate biosynthesis in Saccharomyces cerevisiae under oxidative and osmotic stress. FEMS Yeast Res 2012; 12: 534–46. [DOI] [PubMed] [Google Scholar]
- 52.Lakaye B, Wirtzfeld B, Wins P, et al. Thiamine triphosphate, a new signal required for optimal growth of Escherichia coli during amino acid starvation. J Biol Chem 2004; 279: 17142–7. [DOI] [PubMed] [Google Scholar]
- 53.Brodersen DE, Clemons WM, Jr, Carter AP, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 2000; 103: 1143–54. [DOI] [PubMed] [Google Scholar]
- 54.Robertson GT, Doyle TB, Lynch AS. Use of an efflux-deficient Streptococcus pneumoniae strain panel to identify ABC-class multidrug transporters involved in intrinsic resistance to antimicrobial agents. Antimicrob Agents Chemother 2005; 49: 4781–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tocci N, Iannelli F, Bidossi A, et al. Functional analysis of pneumococcal drug efflux pumps associates the MATE DinF transporter with quinolone susceptibility. Antimicrob Agents Chemother 2013; 57: 248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandez-Moreno MA, Carbo L, Cuesta T, et al. A silent ABC transporter isolated from Streptomyces rochei F20 induces multidrug resistance. J Bacteriol 1998; 180: 4017–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baylay AJ, Piddock LJV. Clinically relevant fluoroquinolone resistance due to constitutive overexpression of the PatAB ABC transporter in Streptococcus pneumoniae is conferred by disruption of a transcriptional attenuator. J Antimicrob Chemother 2015; 70: 670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abreu-Goodger C, Merino E. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res 2005; 33: W690–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grohs P, Trieu-Cuot P, Podglajen I, et al. Molecular basis for different levels of tet(M) expression in Streptococcus pneumoniae clinical isolates. Antimicrob Agents Chemother 2012; 56: 5040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature 2006; 443: 180–5. [DOI] [PubMed] [Google Scholar]
- 61.Loo TW, Clarke DM. Recent progress in understanding the mechanism of P-glycoprotein-mediated drug efflux. J Membr Biol 2005; 206: 173–85. [DOI] [PubMed] [Google Scholar]
- 62.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol 2004; 11: 918–26. [DOI] [PubMed] [Google Scholar]
- 63.Higgins CF. Multiple molecular mechanisms for multidrug resistance transporters. Nature 2007; 446: 749–57. [DOI] [PubMed] [Google Scholar]
- 64.Bohnert JA, Schuster S, Fahnrich E, et al. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF). J Antimicrob Chemother 2007; 59: 1216–22. [DOI] [PubMed] [Google Scholar]
- 65.Vettoretti L, Plesiat P, Muller C, et al. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother 2009; 53: 1987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du Q, Wang H, Xie J. Thiamin (vitamin B1) biosynthesis and regulation: a rich source of antimicrobial drug targets? Int J Biol Sci 2011; 7: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bettendorff L, Wirtzfeld B, Makarchikov AF, et al. Discovery of a natural thiamine adenine nucleotide. Nat Chem Biol 2007; 3: 211–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.