Abstract
The transition to selfing in Capsella rubella accompanies its recent divergence from the ancestral outcrossing C. grandiflora species about 100,000 years ago. Whether the change in mating system was accompanied by the evolution of additional reproductive barriers that enforced species divergence remained unknown. Here, we show that C. rubella and C. grandiflora are reproductively separated by an endosperm-based, non-reciprocal postzygotic hybridization barrier. While hybridizations of C. rubella maternal plants with C. grandiflora pollen donors resulted in complete seed abortion caused by endosperm cellularization failure, the reciprocal hybridization resulted in the formation of small seeds with precociously cellularized endosperm. Strikingly, the transcriptomic response of both hybridizations mimicked respectively the response of paternal and maternal excess hybridizations in Arabidopsis thaliana, suggesting unbalanced genome strength causes hybridization failure in both species. These results provide strong support for the theory that crosses between plants of different mating systems will be unbalanced, with the outcrosser behaving like a plant of increased ploidy, evoking a response that resembles an interploidy-type seed failure. Seed incompatilibity of C. rubella pollinated by C. grandiflora followed the Bateson-Dobzhansky-Muller model, involving negative genetic interaction of multiple paternal C. grandiflora loci with at least one maternal C. rubella locus. Given that both species only recently diverged, our data suggest that a fast evolving mechanism underlies the post-zygotic hybridization barrier(s) separating both species.
Author Summary
Changes in mating system are supposed to change levels of sexual conflict, causing reduced conflict in inbreeding compared to outbreeding species. The recently diverged species pair of the inbreeding Capsella rubella and the outbreeding C. grandiflora provides the opportunity to test this hypothesis. While hybridizations of C. rubella maternal plants with C. grandiflora pollen donors gave rise to seeds with phenotypic similarities to paternal excess hybridizations in Arabidopsis thaliana, the reciprocal hybridization had similarities to maternal excess hybridizations. These results lend support to the hypothesis that selfing reduces sexual conflict, causing the outcrossing parent to have an increased effective ploidy compared to the selfing parent, resulting in unbalanced genome ratios after hybridization and seed failure. Seed failure correlates with either precocious or delayed endosperm cellularization, in agreement with the endosperm being the battleground for sexual conflict in flowering plants. C. grandiflora and C. rubella have only recently diverged, suggesting that the genes building the hybridization barrier are rapidly evolving as a consequence of sexual conflict.
Introduction
The highly selfing species Capsella rubella separated about 100,000 years ago from the obligate outcrosser C. grandiflora [1,2]. While C. rubella is found throughout much of southern and western Europe, C. grandiflora is restricted primarily to the northwest of Greece [3,4]. The breakdown of self-incompatibility in Capsella was concurrent with species divergence [1,3,4], which was associated with a rapid loss of diversity in the newly founded C. rubella [5].
Changes in mating system have been proposed to change levels of sexual conflict, with parental conflict being less intense in self-pollinating plants than in outcrossers [6]. The sexual conflict theory implies that maternally and paternally inherited genes are not equal in relation to maternal investment in offspring, while paternally inherited genes promote maternal provisioning of the progeny, maternally inherited genes counteract this activity [7,8]. Based on this theory, if such genes evolved under different levels of selection pressure in two different species, hybridization should lead to unbalanced maternal investment to offspring, leading to the establishment of a postzygotic hybridization barrier.
Nonequivalence of maternal and paternal genomes can be explained by genomic imprinting, an epigenetic phenomenon causing differential expression of genes depending on their parent-of-origin [9]. In flowering plants, genomic imprinting plays a predominant role in the endosperm, a terminal nutritive tissue supporting embryo growth that is consumed by the embryo during seed development or after germination [10]. Most angiosperms follow a nuclear-type of endosperm development, where the endosperm initially develops as a syncytium and cellularization is triggered after a defined number of mitotic cycles [11,12]. Endosperm cellularization is a crucial developmental transition, which in case of failure triggers embryo arrest [13–15]. Studies on different plant species consistently reported interspecies hybrid seed lethality due to endosperm cellularization failure [7,16–21]. Similar endosperm defects have been observed in hybridizations of plants that differ in ploidy, suggesting a common mechanistic basis [13,21,22]. Specifically in Arabidopsis, hybridization between a maternal tetraploid plant and a paternal diploid plant results in precocious endosperm cellularization, while the reciprocal hybridization causes increased endosperm growth and delayed or failure of endosperm cellularization [13]. Interspecies postzygotic hybridization barriers that are established in the endosperm can often be bypassed by changing the ploidy of one parental species [23–25]. This suggests that these barriers have a quantitative basis and that endosperm-based hybridization barriers are a consequence of deregulated imprinted genes [26–28]. These theoretical considerations are supported by the imprinted gene ADMETOS, which is causally responsible for triploid seed abortion [29]. As parental conflict is predictably less intense in self-pollinating plants than in outcrossers [7], in hybridizations of plants with differing mating systems, outcrossing parents are expected to behave like plants of increased ploidy. Therefore, there should be symptoms of maternal excess when the outcrosser is the seed parent and symptoms of paternal excess in the reciprocal cross [6]. While endosperm phenotypes in response to interspecies crosses lent support for this hypothesis [6], molecular evidence has been lacking thus far. To close this knowledge gap, we have investigated the interspecies hybridization barrier between the closely related species pair C. rubella and C. grandiflora on a morphological, genetic, and molecular level. Our results provide strong support for the theory that crosses between plants of different mating systems will be unbalanced, with the outcrosser behaving like a plant of increased ploidy, evoking a response that resembles an interploidy-type seed failure.
Results
The postzygotic barrier between C. rubella and C. grandiflora non-reciprocally affects seed development
The recently diverged C. rubella and C. grandiflora species are reproductively separated by different mating systems [1,3,4]. However, whether there are additional reproductive barriers between both species has not yet been investigated. We therefore performed reciprocal crosses between C. grandiflora and C. rubella and analyzed the resulting seed set. Pollinations in both directions were successful and resulted in similar numbers of seeds per silique between reciprocal crosses and intra-species crosses, revealing that there were no pollen incompatibilities between C. rubella and C. grandiflora (S1 Table). However, hybrid seeds developed abnormally and aborted at different frequencies; while about 40% of seeds after crosses of C. grandiflora × C. rubella (female × male) were abnormal (Fig 1A and 1B), corresponding to a germination rate of about 60% (Fig 1C), in the reciprocal cross all seeds aborted (Fig 1A, 1B and 1C). Hybrid seedlings of the cross C. grandiflora × C. rubella were variable in size and germination time point (Fig 1D) but developed into fertile adult plants. Intra-species crosses resulted in seed abortion rates below 5% and germination rates above 80% (Fig 1A and 1C). The seeds of the parental species were similar in size and weight (Fig 1B, 1E and 1F; P > 0.1, t-test). Seeds of the cross C. rubella × C. grandiflora were significantly larger than C. rubella seeds but lighter compared to both parental seeds (Fig 1E and 1F; P < 0.05, t-test), while seeds of the reciprocal cross were significantly smaller and lighter (Fig 1E and 1F; P < 0.01, t-test).
Fig 1. Cross direction-dependent incompatibility affects development of Capsella rubella and C. grandiflora hybrid seeds.
Percentage (A) and phenotypes (B) of aborted and non-aborted seeds of C. rubella, C. grandiflora and reciprocal hybrids of both species. Scale bars reflect 1 mm. (C) Percentage of germinated seeds of indicated crosses. (D) Seedlings of indicated crosses 10 days after germination. Seed area (E) and seed weight (F) of indicated crosses. Error bars show standard deviation. Significance was determined by t test analysis. * P < 0.05, ** P < 0.01 ns, not significant. In all graphs numbers on top of the bars correspond to number of analyzed seeds.
Abortion of C. rubella × C. grandiflora hybrid seeds is a consequence of abnormal endosperm development
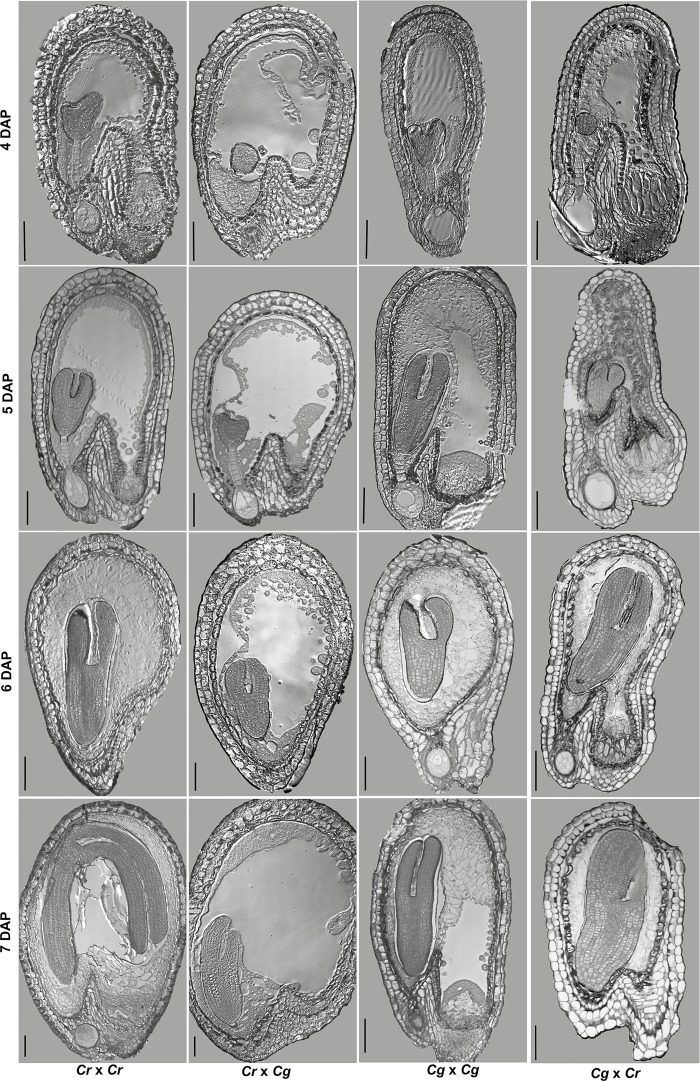
Detailed analysis of parental and hybrid seeds at defined time points (4–7 days after pollination, DAP) revealed differences in the timing of endosperm cellularization in hybrid seeds compared to parental seeds (Fig 2). In both parental species cellularization was completed at 6 DAP, with the embryo having reached the torpedo stage. In contrast, the endosperm of C. rubella × C. grandiflora hybrid seeds was not cellularized at 7 DAP and embryo development was substantially delayed (Fig 2). Conversely, in the reciprocal cross, cellularization happened precociously at 4 DAP and was completed at 5 DAP. The seed coat did not expand properly, leaving insufficient space for the embryo to bend.
Fig 2. Hybrid seed incompatibility between C. rubella and C. grandiflora correlates with endosperm cellularization defects.
Sections of C. rubella, C. grandiflora and reciprocal hybrid seeds at 4–7 days after pollination (DAP). Scale bar 100μm.
Delayed or complete block of endosperm cellularization has been correlated with embryo arrest [15]. We therefore tested whether abortion of hybrid C. rubella × C. grandiflora seeds is a consequence of abnormal endosperm development or rather caused by an embryo defect. To distinguish between both possibilities we isolated hybrid embryos at 13 DAP, when the endosperm was already completely collapsed, but the embryo was still viable (Fig 3A and 3B). Of 14 isolated embryos 9 developed normally and based on flower size the F1 plants were clearly recognized as hybrids (Fig 3C). C. grandiflora × C. rubella hybrids were easily obtained and resembled in flower size the reciprocal hybrid (Fig 3C). Together we conclude that abortion of C. rubella × C. grandiflora hybrid seeds is a consequence of abnormal endosperm development and most likely endosperm cellularization failure, which can be bypassed by embryo rescue.
Fig 3. C. rubella × C. grandiflora hybrid embryos are viable, revealing a major role of endosperm defects in hybrid seed incompatibility.
(A) C. rubella × C. grandiflora hybrid seeds at 13 days after pollination (DAP). (B) Section through seeds derived from crosses of C. rubella × C. rubella (left) and C. rubella × C. grandiflora (right) at 13 DAP. The sections reveal that hybrid embryos reach the torpedo stage at which development arrests. (C) Comparison of adult plants of all crosses (C. grandiflora × C. grandiflora (Cg), C. grandiflora × C. rubella (Cg × Cr), C. rubella × C. grandiflora (Cr × Cg), C. rubella × C. rubella (Cr)). Flower phenotypes of the respective genotypes are shown on top.
Endosperm cellularization failure is not coupled to proliferation abnormalities
In interploidy crosses of A. thaliana, increased paternal ploidy causes increased endosperm proliferation and cellularization failure [13], mimicking crosses of C. rubella × C. grandiflora. Conversely, in seeds with increased maternal ploidy, endosperm proliferation is decreased and cellularization occurs precociously [13]. We therefore tested whether the endosperm cellularization defects in reciprocal Capsella crosses were reflected by changes in endosperm proliferation. Crosses of C. rubella × C. grandiflora resulted in similar proliferation rates compared to proliferation rates in the maternal C. rubella species, with nuclei numbers doubling between 3–5 DAP (Figs 4 and S1). Nuclei numbers decreased at 5–7 DAP in C. rubella, correlating with progression of embryo development. Failure of endosperm cellularization and arrest of embryo development correlated with unchanged nuclei numbers in C. rubella × C. grandiflora hybrid seeds between 5 and 7 DAP (Fig 4).
Fig 4. Hybrid seed incompatibility is not a consequence of endosperm proliferation defects.
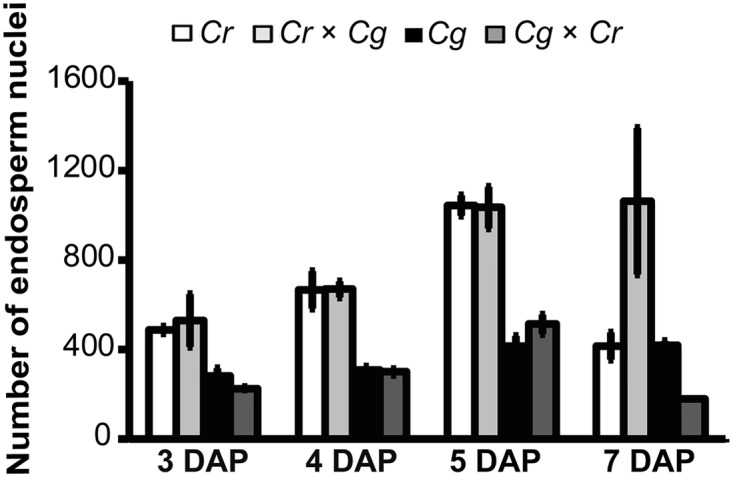
Endosperm nuclei numbers for each cross (C. grandiflora × C. grandiflora (Cg), C. grandiflora × C. rubella (Cg × Cr), C. rubella × C. grandiflora (Cr × Cg), C. rubella × C. rubella (Cr)) at indicated days after pollination (DAP). Three seeds per cross where counted. Error bars show standard error.
In the reciprocal cross, C. grandiflora × C. rubella nuclei proliferation followed that of the maternal C. grandiflora parent and nuclei numbers also doubled between 3–5 DAP. However, C. grandiflora as well as C. grandiflora x C. rubella hybrid seeds had only half the nuclei numbers compared to C. rubella and C. rubella x C. grandiflora seeds, revealing different endosperm proliferation rates between both parental species. At 7 DAP nuclei numbers in C. grandiflora x C. rubella seeds were only half that compared to C. grandiflora seeds, which is likely a consequence of increased endosperm consumption by the hybrid embryo. Together, we conclude that endosperm proliferation in the hybrids followed that of their maternal parental species, suggesting that the hybridization block caused by failure in endosperm cellularization is not a consequence of abnormal endosperm proliferation.
Interspecies and interploidy hybridizations cause similar molecular defects
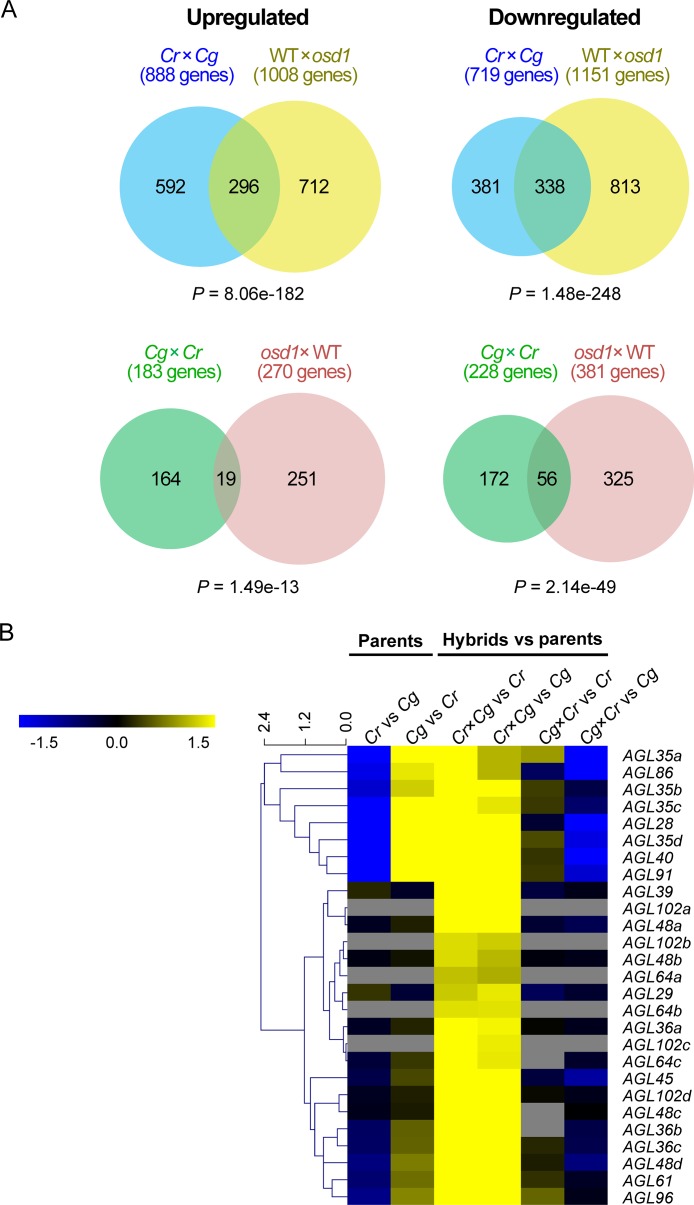
The observed non-reciprocal defects in Capsella hybrid seeds resembled defects in response to interploidy hybridizations in Arabidopsis [13]. To test whether the similarity at the phenotypic level was also reflected at the molecular level, we generated genome-wide expression data of C. rubella and C. grandiflora parental seeds and reciprocal hybrid seeds. We filtered for genes exhibiting transgressive expression towards both parents, thus having either increased or decreased expression levels compared to both parents. For each gene we identified the closest homolog in Arabidopsis and analyzed the expression of those genes in interploidy hybrid seeds generated using the omission of second division1 (osd1) mutant [30,31]. Loss of OSD1 causes the formation of unreduced male and female gametes at high frequency [32], allowing to mimic interploidy hybridizations. A large proportion of genes were similarly deregulated in C. rubella × C. grandiflora hybrid seeds and seeds of paternal excess hybridizations in Arabidopsis (Fig 5A, S2 Table), while substantially fewer genes overlapped with deregulated genes in maternal excess seeds (S2 Fig). Conversely, in the reciprocal cross a substantial number of downregulated genes overlapped with downregulated genes in maternal excess hybridizations in Arabidopsis (Fig 5A, S2 Table), whereas the overlap with deregulated genes in paternal excess seeds was less pronounced (S2 Fig). These data reveal that interspecies and interploidy hybridizations cause a similar molecular response; while C. rubella × C. grandiflora hybrid seeds mimic a paternal excess phenotype, C. grandiflora × C. rubella hybrid seeds mimic a maternal excess phenotype. Notably, genes related to microtubular activity were enriched among downregulated genes in both C. rubella × C. grandiflora and Arabidopsis paternal excess hybrid seeds, while upregulated genes were enriched for genes involved in cell wall modification and specifically in glycosyl hydrolyzing activity (S3 Table). Microtubules are required in the phragmoplast for the transport of vesicles to construct a new cell wall [33], while glycosyl hydrolyzing enzymes can degrade pectin, which is assumed to be a key step in the deconstruction of plant cell walls [34]. Therefore, reduced expression of genes that are potentially required to build the phragmopast together with increased expression of genes that degrade cell walls correlates with the observed cellularization failure in interploidy and interspecies hybrid seeds (S3 Table). Conversely, in the reciprocal cross C. grandiflora × C. rubella, pectinesterase encoding genes were significantly downregulated, suggesting that inhibition of pectin degradation promotes cellularization.
Fig 5. Molecular response to reciprocal hybridizations of C. rubella × C. grandiflora is similar to interploidy hybridizations in Arabidopsis.
(A) Genes up- and downregulated in C. rubella × C. grandiflora reciprocal hybrid seeds compared to both parents overlap with genes deregulated in Arabidopsis interploidy seeds. The Arabidopsis osd1 mutant produces unreduced gametes, mimicking an interploidy hybridization when crossed with wild-type (WT). P values reflecting significance of overlap were calculated using a hypergeometric test. (B) Heatmap of expression log2 fold changes of selected AGL genes between samples. Capsella AGLs with several homologs in Arabidopsis are marked by small letters.
Increased expression of type I MADS-box AGAMOUS-LIKE (AGL) genes has been functionally connected to interploidy and interspecies seed arrest [15,25,35–37]. To test whether the same concept applies to Capsella interspecies hybrids, we isolated type I AGLs in the C. rubella genome according to the Phytozome annotation (S4 Table). Consistently, we identified a large number of AGLs being upregulated in C. rubella × C. grandiflora seeds compared to both parents (Fig 5B, S4 Table) and confirmed these results for selected AGLs (homologs of AGL28, AGL36a, AGL61) in independent experiments (S3 Fig). AGL62 and PHE1 have been functionally connected to interspecies seed abortion [25,35], however, read numbers were too low to detect significant expression changes for either gene. Nevertheless, by qPCR analysis we could detect strongly increased expression levels of both genes in C. rubella × C. grandiflora hybrid seeds (S3 Fig). We also tested expression of ADM, which is causally responsible for triploid seed abortion in Arabidopsis [29]. However, increased ADM expression in C. rubella × C. grandiflora hybrid seeds was only detected quite late during seed development at 7 DAP (S3 Fig), making it unlikely that increased ADM expression is causally responsible for interspecies seed arrest.
Multiple genetic loci control incompatibility of C. rubella × C. grandiflora hybrid seeds
Seeds derived from selfed F1 plants aborted at a frequency of 5.7% (n = 7 plants, total number of seeds = 207), suggesting a multilocus genetic interaction. To estimate the number of loci involved in postzygotic hybrid incompatibility in the cross combination C. rubella × C. grandiflora we generated an F2 population of plants from selfed F1 individuals and backcrossed them as pollen donors to C. rubella plants (C. rubella × F2). We developed a genetic model assuming that one C. rubella maternal locus negatively interacts with two or three C. grandiflora paternally derived loci in a Bateson–Dobzhansky–Muller-type interaction, whereby divergent alleles at two or more loci negatively interact in hybrids to reduce fitness [38,39] (S4 Fig, S5 Table). Thus, based on this model hybrid seed abortion only occurs when two or three unlinked C. grandiflora paternal loci are inherited together. The frequency distribution of F2 plants (n = 250) triggering specific seed abortion rates in the backcrosses to C. rubella (Fig 6A) fitted the prediction of the model with three C. grandiflora paternal loci negatively interacting with one maternal C. rubella locus (Fig 6B), revealing a multilocus negative genetic interaction underlying hybrid incompatibility between C. rubella and C. grandiflora.
Fig 6. C. rubella × C. grandiflora hybrid seed incompatibility involves multiple genetic loci.
(A) Frequency of F2 plants derived from crosses of C. grandiflora × C. rubella producing defined rates of seed abortion when backcrossed to C. rubella maternal plants. (B) Theoretical distribution of F2 plants producing defined rates of seed abortion when assuming that one maternal locus from C. rubella negatively interacts with two or three paternal loci from C. grandiflora. Experimental data correspond to data shown in (A). Significance of the observed distributions with the predictions of the models has been tested by Chi-square and P>0.05 is marked by an asterisk. Ns, non significant. (C) LOD scores for paternal C. grandiflora QTLs affecting abortion of C. rubella × C. grandiflora hybrid seeds. The purple line (top) represents the LOD score of each marker and the purple line (bottom) represents the effects. The red line represents the significance threshold as estimated by 1,000 permutations.
To identify the paternal C. grandiflora genetic loci contributing to postzygotic hybrid incompatibility with a C. rubella seed parent, we determined seed abortion rates after crossing a RIL population of C. grandiflora / C. rubella [40] to C. rubella maternal plants (S5A Fig). The parental accessions of the RIL population gave rise to 100% seed abortion in the C. rubella × C. grandiflora cross and 50% seed abortion in the reciprocal cross (S5B Fig).
The majority of RILs did not trigger seed abortion (S5A Fig), suggesting that hybrid incompatibility genes are purged from this population. Nevertheless, we could identify two significant QTLs located on chromosomes 2 and 8 (Fig 6C) that showed a positive correlation between C. grandiflora alleles and seed abortion. Both QTLs explain about 30% of the phenotypic variance, supporting the idea that there are more than two genes involved in conferring hybrid seed abortion and that incompatibility genes likely have been lost in the RIL population.
Discussion
Recent divergence of C. rubella and C. grandiflora has been associated with the loss of self-incompatibility in C. rubella [1,2]. This drastic change in mating system most probably decreased gene flow between the two species, increasing genetic divergence and allowing additional reproductive barriers to be established. Our data lend support to this hypothesis by revealing a cross direction-dependent, non-reciprocal effect on hybrid seed development. Hybrid seeds resulting from C. rubella × C. grandiflora hybridizations were completely unviable; while the other cross direction resulted in smaller seeds that however were partially viable. Similar examples of unidirectional hybrid incompatibility were previously reported [6,41,42] and have been conceptionalized in the "weak inbreeder/strong outbreeder" (WISO) hypothesis [6]. Based on this hypothesis, in crosses between plants with differing mating systems outcrossing parents are expected to cause a stronger detrimental effect on seed development than selfing parents, as parental conflict is less intense in self-pollinating plants than in outcrossers [6]. Our data provide strong support for this hypothesis, suggesting that genes evolving under parental conflict may build the interspecies hybridization barrier between C. rubella and C. grandiflora. In angiosperms, the endosperm is the battleground for parental conflict [7], consistent with our data revealing that the endosperm is causally responsible for interspecies seed arrest. In C. rubella × C. grandiflora hybrid seeds, endosperm cellularization fails, which likely causes embryo arrest [15]. Supporting this idea, viable hybrids of C. rubella × C. grandiflora could easily be generated using in vitro embryo rescue. Conversely, arrest of C. grandiflora × C. rubella hybrid seeds is likely caused by precocious endosperm cellularization, in agreement with data showing that precocious endosperm cellularization causes decreased seed size and in extreme cases seed arrest [13,43]. Similar to data from rice interspecies hybrids revealing that endosperm cellularization and endosperm proliferation are uncoupled [21], we did not detect obvious endosperm proliferation defects that could explain differences in endosperm cellularization. Therefore, the observed correlation between precocious or delayed endosperm cellularization with respectively decreased or increased endosperm proliferation in response to interploidy hybridizations [13,44] seems unlikely to reflect a general mechanism coupling endosperm proliferation to cellularization. Our transcriptome data revealed that endosperm cellularization failure in response to interspecies and interploidy hybridizations correlated with increased expression of enzymes that hydrolyze glycosidic bonds between carbohydrates and could therefore degrade cell walls [45]. This suggests that the timing of endosperm cellularization is transcriptionally controlled and requires suppression of cell wall degrading enzymes.
Our data suggest that there are three or more C. grandiflora paternal loci that confer seed abortion in combination with a C. rubella seed parent. Similarly, the hybridization barrier between A. thaliana and A. arenosa is controlled by many genes participating in a complex genetic network [46], suggesting that postzygotic hybrid incompatibility that manifests in the endosperm is built by rather complex genetic interactions. Consistent with a scenario that there are many small effect genes contributing to hybrid seed abortion in Capsella, the identified QTLs explained only a low percentage of the phenotypic variation. Nevertheless, it is possible that major QTLs contributing to hybrid seed lethality have been purged from the RIL population and only small effect QTLs are maintained. The Bateson-Dobzhansky-Muller (BDM) model provides a theoretical framework explaining postzygotic reproductive isolation and substantial evidence in support of this model have accumulated over recent years [47–49]. This model stipulates that the interaction of independently evolving loci is innocuous in their native genomic context, however, the interaction between them is deleterious in hybrids [38,39]. Several studies in multiple plant species indicate that the genetic basis of reproductive isolation may involve relatively few loci [49–52]. This is contrasted by other studies supporting a scenario of multiple small incompatibilities accumulating over time [46,53–56]. Our study lends support to the latter, suggesting that multiple small incompatibilities can quickly evolve. A role of imprinted genes in building the interspecies hybridization barrier could provide an explanation for the fast evolution of hybridization barriers.
In conclusion, this study supports the “weak inbreeder/strong outcrosser” hypothesis [6] and suggests that a fast evolving, multi-locus mechanism underlies the post-zygotic barrier affecting C. rubella × C. grandiflora seed survival.
Material and Methods
Plant material and growth conditions
All seeds were surface sterilized using 5% Sodium Hypochlorite solution under the fume hood. After sterilization seeds were plated on MS media containing 1% sucrose. After stratification for two days in the dark at 4°C seedlings were grown in a growth room under long-day photoperiod (16 hrs light and 8 hrs darkness) at 22°C light and 20°C darkness temperature and a light intensity of 110 μE. All seedlings were transferred to pots and plants were grown in a growth chamber at 60% humidity and daily cycles of 16 hrs light at 21°C and 8 hrs darkness at 18°C.
For all crosses, designated female partners were emasculated, and the pistils were hand-pollinated 2 days after emasculation (same procedure for selfing and outcrossing plants).
Capsella accessions Cr48.21, Cg88.14 and Cg4a were used for the phenotypic analysis and Cr48.21 and Cg4a for the gene expression and QTL analyses. All accessions originated from wild collections. An F2 hybrid population of C. grandiflora × C. rubella individuals was generated based on an F1 interspecies cross (C. grandiflora × C. rubella; accessions: Cg4a × Cr48.21). F1 hybrid plants were selfed and the resulting F2 seeds were grown. Capsella QTL lines were described in [40]. Hybrid embryos derived from seeds of crosses of C. rubella (Cr48.21) × C. grandiflora (Cg4a) were isolated at 13 days after pollination (DAP). After a short incubation of siliques in 70% ethanol, the embryo was isolated using fine needles and placed on MS medium containing 2% sucrose. Plates were covered with a tissue and incubated in a growth room under conditions as indicated above. Surviving seedlings were transferred to soil.
Seed size and seed weight analysis
Seeds were arranged on white plastic dishes and pictures were taken using a Leica Z16apoA microscope. Images were converted to black and white using the ‘‘threshold” function in ImageJ (http://rsbweb.nih.gov/ij/) and seed size was measured using the ‘‘Analyze Particles” function. For seed weight, 30–150 seeds per replicate (three replicates per cross) were weighed with an Ohaus GA110 balance and the total weight was divided by the number of seeds.
Microscopy
Clearing analysis was performed as previously described [57].
Endosperm nuclei counts were conducted on cleared seeds at defined time points. After one day incubation in chloral hydrate solution (glycerol/chloral hydrate/water in a ratio of 1:8:3) at 4°C pictures in all possible planes of the seed were taken and endosperm nuclei were subsequently counted in all possible seed planes. Endosperm nuclei were counted using Adobe Photoshop CS5. For tissue sections, seeds were fixed and embedded with Technovit 7100 (Heraeus Kulzer, Hanau, Germany) following the procedure described in [15]. Treatment of the embedded seeds was conducted following [30]. Pictures of embedded and cleared samples were taken using a Leica DMI 4000B microscope and Leica DFC360 FX camera. Pictures of mature seeds were taken using a Leica Z16apoA microscope and a Leica DFC425C camera. All pictures were processed using Adobe Photoshop CS5.
RNA sequencing and expression analysis
Two biological replicates with 300–500 seeds per replicate were generated. Seeds were harvested at 6 DAP and stored at -20°C in RNAlater (Sigma-Aldrich, St Louis, USA). RNA extraction was done using RNAqueous Kit with Plant RNA Isolation Aid (Life Technologies, Carlsbad, USA). The RNA-sequencing libraries were prepared using TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, USA) according to the manufacturer's instructions.
For quantitative RT-PCR analysis five siliques at each time point were harvested and frozen in liquid nitrogen (accessions: Cr48.21, Cg4a). Glass beads (1.25–1.55 mm) were added, and the samples were ground in a Silamat S5 (IvoclarVivadent, Ellwangen, Germany). RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Residual DNA was removed using the RNase-free DNase kit (Qiagen) and cDNA was synthesized using the Fermentas First strand cDNA synthesis kit (Fermentas, Burlington, Canada) according to the manufacturer’s instruction.
Quantitative RT-PCR was performed using a Bio-Rad MyiQ single colour Real-Time PCR Detection System (BioRad, Hercules, USA) and Maxima SYBR green qPCR master mix (Fermentas) according to the manufacturer’s instruction. Specificity of the PCR reactions was confirmed by melting curve analysis (55°-95°C). Results were analyzed as described by [58] using PP2A as a reference gene (S6 Table).
High-throughput RNA sequence analysis
Sequencing reads were aligned to the C. rubella genome v1.0 (Phytozome) using Stampy v1.0.23 [59]. Reads mapping to multiple locations were discarded using Samtools [60], by invoking view–q 4 option. The number of reads for each annotated gene were counted using the default option in htseq-count command from the HTSeq Python package [61] (S7 Table). Paired logarithmic fold changes between each replicate were calculated using the R/Bioconductor package DESeq2 [62]. Differentially regulated genes across the two replicates were detected using the rank product method [63] as implemented in the Bioconductor RankProd Package [64]. The test was run with 100 permutations and gene selection was corrected for multiple comparison errors using a pfp (percentage of false prediction) <0.05. Arabidopsis closest homologues of C. rubella genes were determined according to the Phytozome v1.0 annotation of the C. rubella genome. For overlaps between misregulated genes in Arabidopsis and Capsella, only genes having a homologue in the other species were kept. The significance of the overlap between deregulated genes was estimated using a hypergeometric test. Enrichment of GO categories was determined using ATCOECIS [65]. Homologues of A. thaliana type I AGLs were identified in the C. rubella genome according to the Phytozome C. rubella v1.0 genome annotation. A. thaliana type I AGLs were determined as in [66]. Sequencing reads are deposited as fastq files in the Gene Expression Omnibus (GSE67359).
QTL analysis
The QTL analysis was done using Windows QTL Cartographer v2.5 [67]. The crossing type was set to selfing RILs line (Ri1). Genotype data were coded as follows: two (2) for homozygous C. grandiflora alleles, one (1) for heterozygous alleles, and zero (0) for homozygous C. rubella alleles. Marker information and linkage map were obtained from [40]. The number of RILs used in the analysis was 101. Composite Interval Mapping method (CIM) was applied by setting walking speed to 1 cM to recover maximum resolution (the average distance between markers is 6 cM). The background control parameter was set to standard model with backward regression to select any available possible QTLs with standard 5 control markers. Trait values to be tested were entered as binary value (1 for plants with >2% seed abortion and 0 for plants with <2% seed abortion). The 2% threshold was defined based on the log transformed distribution of seed abortion frequencies that showed two distinct distributions bordering at 2%. The LR significant threshold was determined by running 1000 permutation tests with p = 0.05.
Pollen viability assay
Pollen viability among 20 F2 individuals was assessed using Alexander stain [68]. Collected inflorescences were incubated in 10% ethanol. The anthers were dissected on a slide prior to adding a few drops of Alexander stain. The samples were incubated at room temperature for 15 min and then examined under a Zeiss Axioplan microscope. Viable (pink) and aborted (green) pollen grains were counted for each sample (190–320 pollen grains per individual).
Supporting Information
Cleared seeds of the indicated crosses (C. grandiflora × C. grandiflora (Cg), C. grandiflora × C. rubella (Cg × Cr), C. rubella × C. grandiflora (Cr × Cg), C. rubella × C. rubella (Cr)) between 3 to 7 days after pollination (DAP). Scale bars correspond to 100 μM.
(TIF)
Only few genes deregulated in C. rubella × C. grandiflora hybrid seeds compared to both parents overlap with deregulated genes in osd1 × wild type (WT). Likewise, only few genes deregulated in C. grandiflora × C. rubella hybrid seeds compared to both parents overlap with deregulated genes in WT × osd1. The Arabidopsis osd1 mutant produces unreduced gametes, mimicking an interploidy hybridization when crossed with WT. P values reflecting significance of overlap were calculated using a hypergeometric test.
(TIF)
Expression of indicated genes was tested at 4–7 days after pollination (DAP) in whole siliques derived from crosses of Capsella rubella × C. rubella (white bars), C. rubella × C. grandiflora (light grey bars), C. grandiflora × C. grandiflora (black bars), C. grandiflora × C. rubella (dark grey bars). In the cross C. grandiflora × C. rubella expression was undetectable at 6 and 7 DAP. Error bars represent standard deviation.
(TIF)
(A) Genetic model predicting a negative interaction between one maternal C. rubella locus (allele noted R) and three paternal C. grandiflora loci (alleles noted A, B and C) giving rise to seed lethality. The red color symbolizes the negative lethal interaction. Paternal C. rubella a, b, c alleles and maternal C. grandiflora r allele are compatible and result in viable seeds. (B) Viable F1 plants are generated by the cross C. grandiflora ♀ × ♂ C. rubella. Among F2 progeny produced from selfed F1s, those inheriting R (maternal) and ABC (paternal) will not survive. The surviving F2s are predicted to cause seed abortion when backcrossed to C. rubella (C. rubella ♀ × F2 ♂) according to the number of A, B, C alleles they inherited from C. grandiflora. Only the ABC loci combination in a paternal gamete will cause seed abortion by interacting with the R locus in C. rubella maternal plants.
(TIF)
(A) Log2 transformed seed abortion distribution of C. rubella × RILs. (B) Seed abortion of crosses C. grandiflora × C. rubella (Cg ×Cr), C. rubella × C. grandiflora (Cr × Cg) using parental accessions of the RIL population.
(TIF)
(PDF)
(XLSX)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Cecilia Wärdig for technical support, Pernilla Elander for help with crossing experiments, and Michael Lenhard and Adrien Sicard for sharing the RIL lines. Sequencing was performed by the SNP&SEQ Technology Platform, Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council (VRRFI) and the Knut and Alice Wallenberg Foundation.
Data Availability
All fastq files are available from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) Accession number GSE67359.
Funding Statement
This work was supported by a European Research Council Starting Independent Researcher grant (280496) to CK and an Erwin Schrödinger fellowship (J3394) of the Austrian Science Fund to CAR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Guo Y-L, Bechsgaard JS, Slotte T, Neuffer B, Lascoux M, Weigel D, et al. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. PNAS. 2009;106: 5246–5251. 10.1073/pnas.0808012106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Foxe JP, Slotte T, Stahl EA, Neuffer B, Hurka H, Wright SI. Recent speciation associated with the evolution of selfing in Capsella. PNAS. 2009;106: 5241–5245. 10.1073/pnas.0807679106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hurka H, Neuffer B. Evolutionary processes in the genusCapsella (Brassicaceae). Pl Syst Evol. 1997;206: 295–316. 10.1007/BF00987954 [DOI] [Google Scholar]
- 4. Paetsch M, Mayland-Quellhorst S, Neuffer B. Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity. 2006;97: 283–290. 10.1038/sj.hdy.6800854 [DOI] [PubMed] [Google Scholar]
- 5. Brandvain Y, Slotte T, Hazzouri KM, Wright SI, Coop G. Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species Capsella rubella. PLoS Genet. 2013;9: e1003754 10.1371/journal.pgen.1003754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brandvain Y, Haig D. Divergent Mating Systems and Parental Conflict as a Barrier to Hybridization in Flowering Plants. The American Naturalist. 2005;166: 330–338. 10.1086/an.2005.166.issue-3 [DOI] [PubMed] [Google Scholar]
- 7. Haig D, Westoby M. Genomic Imprinting in Endosperm: Its Effect on Seed Development in Crosses between Species, and between Different Ploidies of the Same Species, and Its Implications for the Evolution of Apomixis. Phil Trans R Soc Lond B. 1991;333: 1–13. 10.1098/rstb.1991.0057 [DOI] [Google Scholar]
- 8. Haig D. GENOMIC IMPRINTING AND KINSHIP: How Good is the Evidence? Annual Review of Genetics. 2004;38: 553–585. 10.1146/annurev.genet.37.110801.142741 [DOI] [PubMed] [Google Scholar]
- 9. Gehring M. Genomic imprinting: insights from plants. Annu Rev Genet. 2013;47: 187–208. 10.1146/annurev-genet-110711-155527 [DOI] [PubMed] [Google Scholar]
- 10. Li J, Berger F. Endosperm: food for humankind and fodder for scientific discoveries. New Phytologist. 2012;195: 290–305. 10.1111/j.1469-8137.2012.04182.x [DOI] [PubMed] [Google Scholar]
- 11. Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, et al. Dynamic Analyses of the Expression of the HISTONE::YFP Fusion Protein in Arabidopsis Show That Syncytial Endosperm Is Divided in Mitotic Domains. Plant Cell. 2001;13: 495–509. 10.1105/tpc.13.3.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berger F. Endosperm: the crossroad of seed development. Curr Opin Plant Biol. 2003;6: 42–50. 10.1016/S1369526602000043 [DOI] [PubMed] [Google Scholar]
- 13. Scott RJ, Spielman M, Bailey J, Dickinson HG. Parent-of-origin effects on seed development in Arabidopsis thaliana . Development. 1998;125: 3329–3341. [DOI] [PubMed] [Google Scholar]
- 14. Pignocchi C, Minns GE, Nesi N, Koumproglou R, Kitsios G, Benning C, et al. ENDOSPERM DEFECTIVE1 Is a Novel Microtubule-Associated Protein Essential for Seed Development in Arabidopsis. Plant Cell. 2009;21: 90–105. 10.1105/tpc.108.061812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hehenberger E, Kradolfer D, Köhler C. Endosperm cellularization defines an important developmental transition for embryo. Development. 2012;139: 2031–2039. 10.1242/dev.077057 [DOI] [PubMed] [Google Scholar]
- 16. Watkins AE. Hybrid sterility and incompatibility. Journ of Genetics. 1932;25: 125–162. 10.1007/BF02983249 [DOI] [Google Scholar]
- 17. Cooper DC, Brink RA. The Endosperm as a Barrier to Interspecific Hybridization in Flowering Plants. Science. 1942;95: 75–76. 10.1126/science.95.2455.75 [DOI] [PubMed] [Google Scholar]
- 18. Stebbins GL. The Inviability, Weakness, and Sterility of Interspecific Hybrids In: Demerec M., editor. Advances in Genetics. Academic Press; 1958. pp. 147–215. [DOI] [PubMed] [Google Scholar]
- 19. Valentine DH, Woodell SRJ. Seed Incompatibility in Primula. Nature. 1960;185: 778–779. 10.1038/185778b0 [DOI] [Google Scholar]
- 20. Bushell C, Spielman M, Scott RJ. The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell. 2003;15: 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, Kinoshita T. Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. The Plant Journal. 2011;65: 798–806. 10.1111/j.1365-313X.2010.04466.x [DOI] [PubMed] [Google Scholar]
- 22. Schatlowski N, Köhler C. Tearing down barriers: understanding the molecular mechanisms of interploidy hybridizations. J Exp Bot. 2012;63: 6059–6067. 10.1093/jxb/ers288 [DOI] [PubMed] [Google Scholar]
- 23. Johnston SA, Hanneman RE. Manipulations of Endosperm Balance Number Overcome Crossing Barriers Between Diploid Solanum Species. Science. 1982;217: 446–448. 10.1126/science.217.4558.446 [DOI] [PubMed] [Google Scholar]
- 24. Carputo D, Frusciante L, Peloquin SJ. The Role of 2n Gametes and Endosperm Balance Number in the Origin and Evolution of Polyploids in the Tuber-Bearing Solanums. Genetics. 2003;163: 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Josefsson C, Dilkes B, Comai L. Parent-Dependent Loss of Gene Silencing during Interspecies Hybridization. Curr Biol. 2006;16: 1322–1328. 10.1016/j.cub.2006.05.045 [DOI] [PubMed] [Google Scholar]
- 26. Gutierrez-Marcos JF, Pennington PD, Costa LM, Dickinson HG. Imprinting in the endosperm: a possible role in preventing wide hybridization. Philos Trans R Soc Lond, B, Biol Sci. 2003;358: 1105–1111. 10.1098/rstb.2003.1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Birchler JA, Veitia RA. The Gene Balance Hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 2010;186: 54–62. 10.1111/j.1469-8137.2009.03087.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Birchler JA. Interploidy hybridization barrier of endosperm as a dosage interaction. Front Plant Sci. 2014;5: 281 10.3389/fpls.2014.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kradolfer D, Wolff P, Jiang H, Siretskiy A, Köhler C. An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana . Dev Cell. 2013;26: 525–535. 10.1016/j.devcel.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 30. Kradolfer D, Hennig L, Köhler C. Increased Maternal Genome Dosage Bypasses the Requirement of the FIS Polycomb Repressive Complex 2 in Arabidopsis Seed Development. PLoS Genet. 2013;9: e1003163 10.1371/journal.pgen.1003163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schatlowski N, Wolff P, Santos-González J, Schoft V, Siretskiy A, Scott R, et al. Hypomethylated Pollen Bypasses the Interploidy Hybridization Barrier in Arabidopsis. Plant Cell. 2014; tpc.114.130120. 10.1105/tpc.114.130120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. D’ Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R. Turning Meiosis into Mitosis. PLoS Biol. 2009;7 10.1371/journal.pbio.1000124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murata T, Sano T, Sasabe M, Nonaka S, Higashiyama T, Hasezawa S, et al. Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat Commun. 2013;4 10.1038/ncomms2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiao C, Somerville C, Anderson CT. POLYGALACTURONASE INVOLVED IN EXPANSION1 Functions in Cell Elongation and Flower Development in Arabidopsis. Plant Cell. 2014;26: 1018–1035. 10.1105/tpc.114.123968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L. Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol. 2009;19: 1128–1132. 10.1016/j.cub.2009.05.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Erilova A, Brownfield L, Exner V, Rosa M, Twell D, Mittelsten Scheid O, et al. Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet. 2009;5: e1000663 10.1371/journal.pgen.1000663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tiwari S, Spielman M, Schulz R, Oakey RJ, Kelsey G, Salazar A, et al. Transcriptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biology. 2010;10: 72 10.1186/1471-2229-10-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dobzhansky T. Studies on Hybrid Sterility. II. Localization of Sterility Factors in Drosophila Pseudoobscura Hybrids. Genetics. 1936;21: 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muller HJ. Isolating mechanisms, evolution, and temperature. Biol Symp. 1942;6: 71–125. [Google Scholar]
- 40. Sicard A, Stacey N, Hermann K, Dessoly J, Neuffer B, Bäurle I, et al. Genetics, Evolution, and Adaptive Significance of the Selfing Syndrome in the Genus Capsella. Plant Cell. 2011;23: 3156–3171. 10.1105/tpc.111.088237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Valentine DH, Woodell SRJ. Studies in British Primulas. X. Seed Incompatibility in Intraspecific and Interspecific Crosses at Diploid and Tetraploid Levels. New Phytol. 1963;62: 125–143. [Google Scholar]
- 42. Kato J, Mii M. Production of Interspecific Hybrids in Ornamental Plants In: Loyola-Vargas VM, Ochoa-Alejo N, editors. Plant Cell Culture Protocols. Humana Press; 2012. pp. 233–245. [DOI] [PubMed] [Google Scholar]
- 43. Garcia D, Fitz Gerald JN, Berger F. Maternal Control of Integument Cell Elongation and Zygotic Control of Endosperm Growth Are Coordinated to Determine Seed Size in Arabidopsis. Plant Cell. 2005;17: 52–60. 10.1105/tpc.104.027136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sekine D, Ohnishi T, Furuumi H, Ono A, Yamada T, Kurata N, et al. Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J. 2013;76: 792–799. 10.1111/tpj.12333 [DOI] [PubMed] [Google Scholar]
- 45. Wagner GK, Pesnot T. Glycosyltransferases and their assays. Chembiochem. 2010;11: 1939–1949. 10.1002/cbic.201000201 [DOI] [PubMed] [Google Scholar]
- 46. Burkart-Waco D, Josefsson C, Dilkes B, Kozloff N, Torjek O, Meyer R, et al. Hybrid incompatibility in Arabidopsis is determined by a multiple-locus genetic network. Plant Physiol. 2012;158: 801–812. 10.1104/pp.111.188706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5: e236 10.1371/journal.pbio.0050236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moyle LC, Nakazato T. Hybrid Incompatibility “Snowballs” Between Solanum Species. Science. 2010;329: 1521–1523. 10.1126/science.1193063 [DOI] [PubMed] [Google Scholar]
- 49. Chae E, Bomblies K, Kim S-T, Karelina D, Zaidem M, Ossowski S, et al. Species-wide Genetic Incompatibility Analysis Identifies Immune Genes as Hot Spots of Deleterious Epistasis. Cell. 2014;159: 1341–1351. 10.1016/j.cell.2014.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moyle LC, Graham EB. Genetics of hybrid incompatibility between Lycopersicon esculentum and L. hirsutum. Genetics. 2005;169: 355–373. 10.1534/genetics.104.029546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fishman L, Willis JH. A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evolution. 2006;60: 1372–1381. [DOI] [PubMed] [Google Scholar]
- 52. Sweigart AL, Fishman L, Willis JH. A simple genetic incompatibility causes hybrid male sterility in mimulus. Genetics. 2006;172: 2465–2479. 10.1534/genetics.105.053686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Orr HA, Coyne JA. The genetics of postzygotic isolation in the Drosophila virilis group. Genetics. 1989;121: 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Coyne JA, Orr HA. “Patterns of Speciation in Drosophila” Revisited. Evolution. 1997;51: 295 10.2307/2410984 [DOI] [PubMed] [Google Scholar]
- 55. Sasa MM, Chippindale PT, Johnson NA. Patterns of Postzygotic Isolation in Frogs. Evolution. 1998;52: 1811 10.2307/2411351 [DOI] [PubMed] [Google Scholar]
- 56. Presgraves DC. Patterns of postzygotic isolation in Lepidoptera. Evolution. 2002;56: 1168–1183. [DOI] [PubMed] [Google Scholar]
- 57. Roszak P, Köhler C. Polycomb group proteins are required to couple seed coat initiation to fertilization. PNAS. 2011;108: 20826–20831. 10.1073/pnas.1117111108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simon P. Q-Gene: processing quantitative real-time RT–PCR data. Bioinformatics. 2003;19: 1439–1440. 10.1093/bioinformatics/btg157 [DOI] [PubMed] [Google Scholar]
- 59. Lunter G, Goodson M. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2010; 10.1101/gr.111120.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. bioRxiv. 2014; 002824. 10.1101/002824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. bioRxiv. 2014; 002832. 10.1101/002832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Letters. 2004;573: 83–92. 10.1016/j.febslet.2004.07.055 [DOI] [PubMed] [Google Scholar]
- 64. Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22: 2825–2827. 10.1093/bioinformatics/btl476 [DOI] [PubMed] [Google Scholar]
- 65. Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 2009;150: 535–546. 10.1104/pp.109.136028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bemer M, Heijmans K, Airoldi C, Davies B, Angenent GC. An Atlas of Type I MADS Box Gene Expression during Female Gametophyte and Seed Development in Arabidopsis. Plant Physiol. 2010;154: 287–300. 10.1104/pp.110.160770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang Shengchu, Basten CJ, Zeng Z-B. Windows QTL Cartographer 2.5 In: Department of Statistics, North Carolina State University, Raleigh, NC: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm [Google Scholar]
- 68. Alexander MP. Differential Staining of Aborted and Nonaborted Pollen. Biotech Histochem. 1969;44: 117–122. 10.3109/10520296909063335 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cleared seeds of the indicated crosses (C. grandiflora × C. grandiflora (Cg), C. grandiflora × C. rubella (Cg × Cr), C. rubella × C. grandiflora (Cr × Cg), C. rubella × C. rubella (Cr)) between 3 to 7 days after pollination (DAP). Scale bars correspond to 100 μM.
(TIF)
Only few genes deregulated in C. rubella × C. grandiflora hybrid seeds compared to both parents overlap with deregulated genes in osd1 × wild type (WT). Likewise, only few genes deregulated in C. grandiflora × C. rubella hybrid seeds compared to both parents overlap with deregulated genes in WT × osd1. The Arabidopsis osd1 mutant produces unreduced gametes, mimicking an interploidy hybridization when crossed with WT. P values reflecting significance of overlap were calculated using a hypergeometric test.
(TIF)
Expression of indicated genes was tested at 4–7 days after pollination (DAP) in whole siliques derived from crosses of Capsella rubella × C. rubella (white bars), C. rubella × C. grandiflora (light grey bars), C. grandiflora × C. grandiflora (black bars), C. grandiflora × C. rubella (dark grey bars). In the cross C. grandiflora × C. rubella expression was undetectable at 6 and 7 DAP. Error bars represent standard deviation.
(TIF)
(A) Genetic model predicting a negative interaction between one maternal C. rubella locus (allele noted R) and three paternal C. grandiflora loci (alleles noted A, B and C) giving rise to seed lethality. The red color symbolizes the negative lethal interaction. Paternal C. rubella a, b, c alleles and maternal C. grandiflora r allele are compatible and result in viable seeds. (B) Viable F1 plants are generated by the cross C. grandiflora ♀ × ♂ C. rubella. Among F2 progeny produced from selfed F1s, those inheriting R (maternal) and ABC (paternal) will not survive. The surviving F2s are predicted to cause seed abortion when backcrossed to C. rubella (C. rubella ♀ × F2 ♂) according to the number of A, B, C alleles they inherited from C. grandiflora. Only the ABC loci combination in a paternal gamete will cause seed abortion by interacting with the R locus in C. rubella maternal plants.
(TIF)
(A) Log2 transformed seed abortion distribution of C. rubella × RILs. (B) Seed abortion of crosses C. grandiflora × C. rubella (Cg ×Cr), C. rubella × C. grandiflora (Cr × Cg) using parental accessions of the RIL population.
(TIF)
(PDF)
(XLSX)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All fastq files are available from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) Accession number GSE67359.