Abstract
Background
There is microscopic spatial and temporal heterogeneity of pathologic changes in idiopathic pulmonary fibrosis (IPF) lung tissue, which may relate to heterogeneity in pathophysiological mediators of disease and clinical progression. We assessed relationships between gene expression patterns, pathological features, and systemic biomarkers to identify biomarkers that reflect the aggregate disease burden in IPF patients.
Methods
Gene expression microarrays (N=40 IPF; 8 controls) and immunohistochemical analyses (N=22 IPF; 8 controls) of lung biopsies. Clinical characterization and blood biomarker levels of MMP3 and CXCL13 in a separate cohort of IPF patients (N=80).
Results
2940 genes were significantly differentially expressed between IPF and control samples (|fold change| > 1.5, p < 0.05). Two clusters of co-regulated genes related to bronchiolar epithelium or lymphoid aggregates exhibited substantial heterogeneity within the IPF population. Gene expression in bronchiolar and lymphoid clusters corresponded to the extent of bronchiolization and lymphoid aggregates determined by immunohistochemistry in adjacent tissue sections. Elevated serum levels of MMP3, encoded in the bronchiolar cluster, and CXCL13, encoded in the lymphoid cluster, corresponded to disease severity and shortened survival time (p < 10−7 for MMP3 and p < 10−5 for CXCL13; Cox proportional hazards model).
Conclusions
Microscopic pathological heterogeneity in IPF lung tissue corresponds to specific gene expression patterns related to bronchiolization and lymphoid aggregates. MMP3 and CXCL13 are systemic biomarkers that reflect the aggregate burden of these pathological features across total lung tissue. These biomarkers may have clinical utility as prognostic and/or surrogate biomarkers of disease activity in interventional studies in IPF.
Keywords: MMP3, CXCL13, bronchiolization, lymphoid aggregates
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive, and fatal fibrotic lung disease with no known cause and a median survival of ~3 years (1). The clinical course within the IPF patient population is variable, ranging from a slow, steady loss of lung function over 5 or more years to rapid progression and death within 1–2 years of diagnosis to relatively stable disease punctuated by intermittent precipitous declines in lung function resulting from acute exacerbations (2). The mechanisms underlying this variability in disease progression within the IPF patient population are poorly understood.
The fibrosis observed in IPF is hypothesized to originate from an irregular wound healing response triggered by a loss of integrity in the alveolar epithelium followed by persistent pro-fibrotic signals that fail to resolve (3). The microscopic presentation of IPF in diseased lung tissue is spatially and temporally heterogeneous. Regions of fibroblast accumulation and active matrix remodeling can exist between areas of normal-appearing lung and mature scar tissue (4). A complex mixture of resident and infiltrating cell types can be found in these distinct regions of remodeled lung. It is therefore important to develop a deeper understanding of IPF biology at a local level to better characterize the location and consequences of aberrant signaling processes.
A challenge in designing interventional clinical studies in IPF is the lack of a robust means of identifying and controlling for biological heterogeneity, and of selecting patients at risk for outcomes of interest. Non-invasive biomarkers that reflect the activity of specific pathways and aggregate fibrotic burden across a given patient’s total lung tissue could be valuable tools in identifying stage of disease, and could aid in selecting treatments and monitoring biological responses to therapy. In this study we used gene expression profiling and histology to characterize pathological patterns that display heterogeneous expression levels within lung tissue from a population of IPF patients, and identified systemic biomarkers related to these patterns that correspond to disease severity. The coordinated analyses of gene expression, histology, and peripheral biomarkers provide the opportunity to integrate insight into IPF pathogenesis at multiple levels, with the potential to contribute to the development of novel therapies and diagnostics.
METHODS
Tissues were obtained from clinical samples from IPF patients at the time of biopsy or lung transplantation. Peripheral blood and clinical data were obtained from a separate cohort of IPF patients at the time of initial presentation to the interstitial lung disease clinic. All patients were seen at UCSF and the diagnosis of IPF was established through multidisciplinary review of clinical, radiological, and pathological data according to consensus criteria (5). Non-diseased normal lung tissues were procured from lungs not used by the Northern California Transplant Donor Network. Clinical and demographic information for the IPF cohorts is in Supplementary Table 1 and Table 1. Sample and data collection were approved by the UCSF Committee on Human Research and all patients provided written informed consent.
Table 1. Clinical and demographic features of IPF patients in the prognostic cohort (N=80).
Clinical data are as of the time of initial presentation to the UCSF ILD clinic, when blood samples were collected.
| Variable | mean (SD) unless otherwise specified |
|---|---|
| Sex (M:F) | 63:17 |
| Age at initial visit, median (range) | 70 (50–87) |
| Subsequent lung transplant (Y:N) | 7:63 |
| Ever smoker (Y:N) | 58:16 |
| Current smoker (Y:N) | 1:73 |
| Systemic steroid ± azathioprine (Y: N) | 21:59 |
| FVC % predicted (N=79) | 70.3 (15.8) |
| DLCO % predicted (N=69) | 49.6 (19.0) |
| Dyspnea score* (N=74) | 9.7 (6.2) |
| HRCT fibrosis score** (N=35) | 15.3 (10.9) |
| Serum MMP3 level (ng/ml), median (IQR) (N=78) | 22.0 (15.6–41.9) |
| Serum CXCL13 level, median (IQR) (pg/ml) | 80 (55–136) |
| Survival after initial visit (y), median (IQR)*** | 2.77 (1.43–5.36) |
In cases where data were not available for all 80 patients, number of patients with data are indicated (N)
Dyspnea score is on a scale of 1–20 per Watters et al (37)
HRCT fibrosis score estimates % of lung that is fibrotic per Best et al (38)
Censored as of date of analysis for surviving patients
Statistical inference analyses were performed in R. Relationship of expression signature scores or serum protein concentration to patient diagnosis was tested by the Wilcoxon rank-sum method. Relationship of expression signatures scores to histology scores was tested by a one-term logistic regression model. Correlations between MMP3 or CXCL13 and other variables were performed by the Spearman method. Survival analyses were performed using the Cox proportional hazards model with serum concentration as the only independent variable term and p-values determined by the score test. Transplants were treated as censored events.
Detailed methods for RNA preparation, gene expression, immunohistochemistry, and biomarker analyses are available in the online data repository.
RESULTS
Differential gene expression in IPF lung tissue
We performed genome-wide transcriptomic analysis of lung biopsy tissue from 40 IPF patients and 8 control subjects without IPF (Supplementary Table 1). Eleven IPF samples were from video-assisted thoracoscopic (VATS) biopsies and 29 were from explants taken at the time of lung transplantation. Control tissue was explanted from unused donor lungs. RNA isolated from these samples was analyzed using microarrays. Limited available metadata including sex, tissue source (VATS or explant), and diagnosis were included in a linear model used to identify differentially expressed genes. We identified 2940 probes as differentially expressed (DE): 1531 upregulated and 1409 downregulated in IPF tissues compared to control tissues (fold-change > |1.5|, Benjamini-Hochberg adjusted p-value < 0.05; Supplementary Table 3). Among the significantly upregulated genes in IPF were those encoding multiple matrix metalloproteinases (MMPs), collagens, and cytokines and growth factors. Many DE genes in IPF lung tissue overlapped with those reported in previously published datasets (6–8).
Heterogeneity in clusters of co-regulated genes
Unsupervised 2-way hierarchical clustering of the 2490 DE probes between IPF and control demonstrated two major clusters of samples defined by diagnosis (Figure 1A). Inspection of the dendrogram revealed 2 clusters of co-regulated genes that were upregulated in IPF compared to controls, yet were variable within the IPF population.
Figure 1.
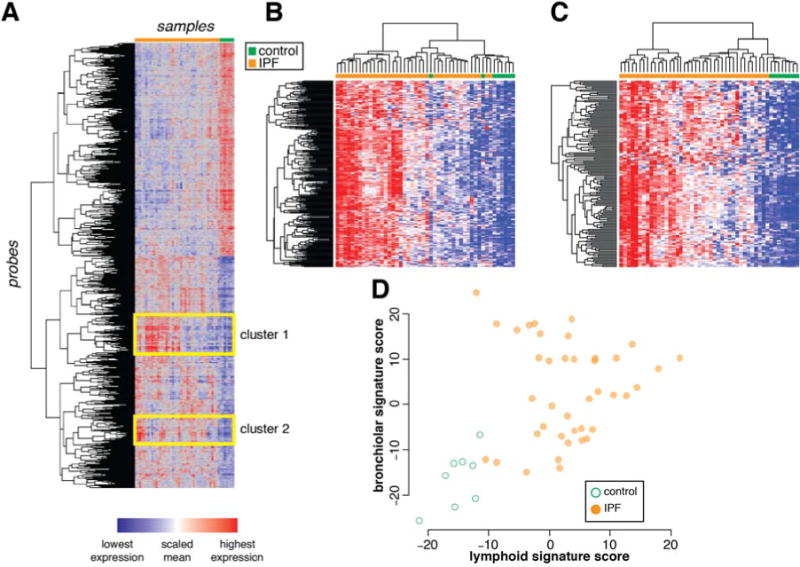
Heterogeneous clusters of co-expressed genes among IPF lung biopsy samples.
A) Unsupervised 2-way clustering of all differentially expressed genes (|fold change| > 1.5; Benjamini-Hochberg adjusted p-value < 0.05) between IPF (N=40) and control (N=8) lung biopsy samples. IPF samples are indicated in orange and control samples are indicated in green. B) Re-clustering of samples on co-regulated genes in cluster 1 (“bronchiolar” cluster). C) Re-clustering of samples on co-regulated genes in cluster 2 (“lymphoid” cluster). D) Normalized mean expression scores for all genes in the bronchiolar and lymphoid clusters.
Cluster 1 comprises genes related to bronchiolar epithelium, including mucins (MUCL1, MUC4, MUC20), proline-rich secreted factors (PRR7, PRR15, SPRR1B, SPRR2D), keratins (KRT5, 6B, 13, 14, 15, 17), serine protease inhibitors (SERPINB3, B4, B5, B13), ion channels and associated factors (TRPV4, CLCA2), and cilium components (BBS5). This is consistent with prior reports of abnormal “bronchiolization” of alveolar spaces in IPF, which may represent epithelialization of honeycombed cystic spaces in regions of dense scarring (9–11), hence we have termed this cluster of co-regulated genes the “bronchiolar signature.” A full list of genes in the bronchiolar signature is in Supplementary Table 4. Re-clustering the samples using only the bronchiolar signature genes emphasizes the range in gene expression levels observed within the IPF population (Figure 1B).
Cluster 2 consists of T and B cell markers (CD19, CD20, CD27, CD28), Fc receptor genes (FCRLA, FCRL2, FCRL5), and chemokines and chemokine receptors (CXCL13, CXCR5, CCR6, CCR7). This pattern is consistent with prior reports showing ectopic lymphoid tissue in regions of established fibrosis in biopsies from IPF (12–14), hence we have termed this cluster of co-regulated genes the “lymphoid signature.” A full list of genes in this cluster is in Supplementary Table 5. Re-clustering the samples using only the lymphoid gene signature genes shows heterogeneity across the IPF samples (Figure 1C).
To determine whether the bronchiolar and lymphoid signatures were related to each other in individual samples, we derived summary gene expression scores for each signature by calculating the normalized mean expression of all the genes in each cluster. While samples from most IPF patients had higher bronchiolar or lymphoid signature scores than controls, there was substantial variability within the IPF population. While nearly all samples with markedly elevated lymphoid signatures also had elevated bronchiolar signatures, the converse was not true (Figure 1D). These results suggest that the signatures may reflect distinct biological processes heterogeneously expressed across IPF patients and/or differential sampling of processes that have heterogeneous spatial distribution within bulk IPF lung tissue.
Localization of signature markers to specific histological features in IPF lung tissue
IPF lung tissue is histologically heterogeneous on a microscopic scale as evidenced by a low magnification H&E image (Figure 2A). Higher magnification of selected regions reveals relatively normal alveolar tissue (Figure 2B) adjacent to “transition zones” of active fibrogenesis with thickened alveolar walls (Figure 2C) and advanced scar tissue with regions of dense fibrosis interspersed with honeycombed cysts lined with columnar epithelium (Figure 2D).
Figure 2.
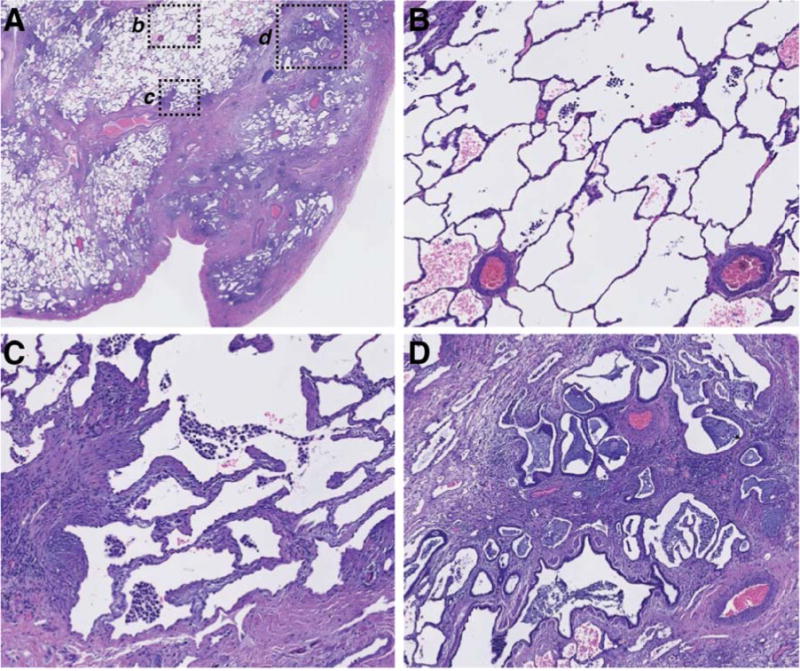
Spatial heterogeneity of pathophysiology in a single representative IPF biopsy.
A) Low-power magnification of an H&E-stained paraffin section of explanted lung tissue from an IPF patient. Pleural surface is at lower right. b, c, and d indicate regions shown at higher magnification in: (B), normal-appearing alveolar structure, (C) transition zone underlying normal-appearing parenchyma with thickened alveolar walls, (D) advanced scar tissue with microscopic honeycombing and bronchiolization.
To determine where selected genes encoded in each of the signatures were spatially distributed in regions of lung tissue, we performed immunohistochemistry (IHC) on biopsy tissue taken from IPF lung explants (N=10). Cystic structures were lined with epithelial cells expressing keratin 5 (KRT5), a gene encoded in the bronchiolar signature (Figure 3A–B). H&E, trichrome, anti-KRT14, and PAS staining of adjacent sections further revealed that these structures were lined with columnar epithelial cells with abundant cytoplasm, near to but distinct from regions of collagen deposition, with coincident expression of mucins (Supplementary Figure 1A–D). Taken together, these patterns are consistent with cystic spaces in IPF lung lined with bronchiolar epithelium.
Figure 3.
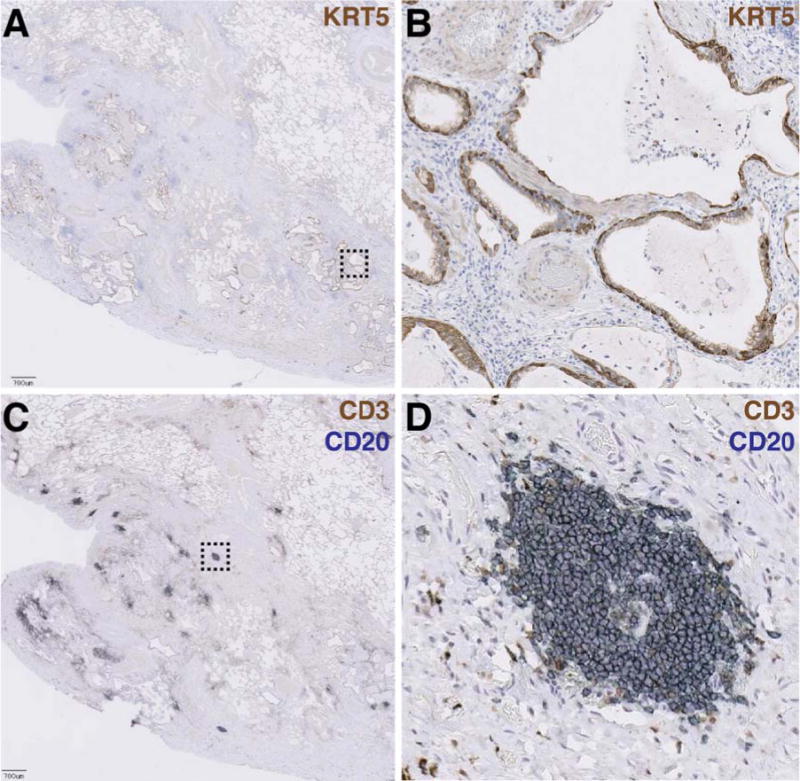
Bronchiolization and lymphoid aggregates occur in spatially distinct regions of advanced scar tissue in IPF biopsies.
A) Low-power magnification of an anti-keratin 5 (KRT5)-stained paraffin section of explanted lung tissue from an IPF patient. Pleural surface is at lower left. Scale bar = 700 μM. Dashed box indicates region shown at higher magnification in panel (B). C) Adjacent section co-stained with anti-CD3 (brown) and anti-CD20 (blue) showing lymphoid aggregates. Dashed box indicates region shown at higher magnification in panel (D).
In spatially discrete areas from bronchiolized regions, we observed numerous aggregates of CD20+ B lymphocytes and CD3+ T lymphocytes (Figure 3C–D). H&E and trichrome staining revealed clusters of cells with darkly staining nuclei, near to but distinct from regions of high collagen deposition; these cells stained positively for CD20 (Supplementary Figure 1E–H). Taken together, these patterns are consistent with lymphoid aggregates.
Gene expression signatures correlate to specific histological features in IPF tissue
To confirm that the gene signatures observed in the microarray experiment corresponded to bronchiolar and lymphoid structures observed histologically in IPF lung tissue, we performed a coordinated analysis of gene expression and histological evaluation on serial tissue sections from lung biopsies, schematically depicted in Figure 4A.
Figure 4.
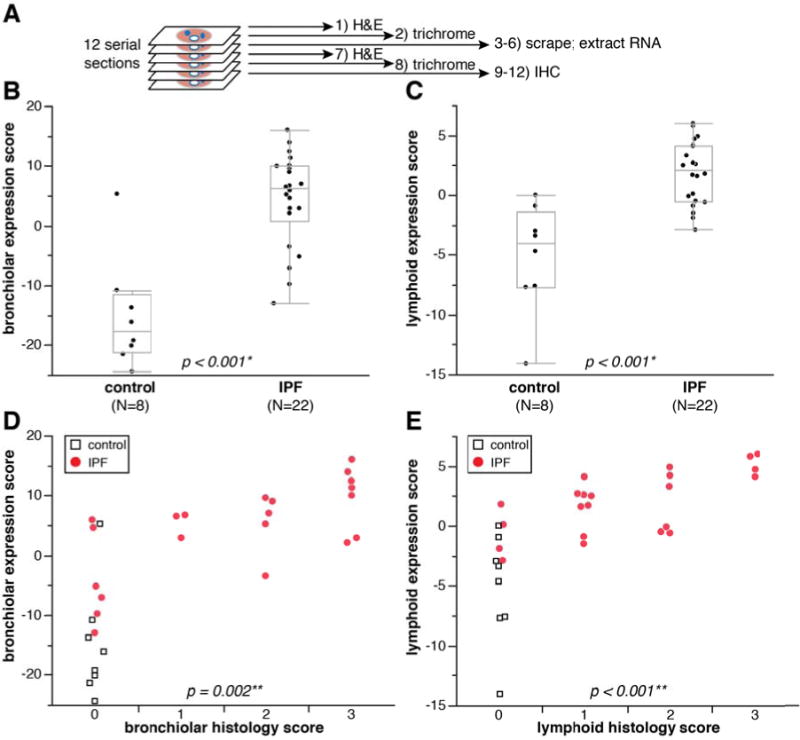
Bronchiolar and lymphoid gene signature expression scores correspond to extent of bronchiolization and lymphoid aggregates in adjacent tissue sections.
A) Schematic depiction of method to compare histopathology and gene expression in adjacent tissue sections. B) qPCR-determined bronchiolar gene signature score in control and IPF tissue scrapings. C) qPCR-determined lymphoid gene signature score in control and IPF tissue scrapings. *p-values for (B) and (C) determined by Wilcoxon rank-sum method. D) qPCR bronchiolar gene signature score as a function of bronchiolar histology score. E) qPCR lymphoid gene signature score as a function of lymphoid histology score. **p-values for (D) and (E) determined by logistic regression (IPF samples only; controls excluded from statistical analysis).
We isolated total RNA from tissue sections and assessed gene expression by qPCR using 24 probes spanning the bronchiolar and lymphoid signatures (Supplementary Table 2) and derived expression indices for each signature from the qPCR data by taking the normalized mean expression of all the genes in each cluster across all samples (centered at 0). We first tested the qPCR platform with the same RNA isolated from bulk lung biopsies used in the microarray analyses of gene expression described above and observed highly concordant signature scores between the microarray and qPCR experiments (Supplementary Figure 2) irrespective of tissue source (VATS biopsy or explant). Gene expression in individual tissue sections was significantly enriched in IPF compared to control lung tissue for both the bronchiolar and lymphoid signature scores (Figure 4B–C). Sections adjacent to those assessed by qPCR were assessed by IHC and scored by a pathologist blinded to the expression scores for the presence of bronchiolization and lymphoid aggregates on a 0–3 scale, with “0” representing the absence of a particular feature and “3” representing a section that displays that feature prominently (examples in Supplementary Figure 3). There were highly significant correlations between histology and gene expression scores for both the bronchiolar and lymphoid signatures (Figure 4D–E). Taken together, these data further substantiate that the gene expression patterns observed by microarray correspond to specific and distinct pathological features of IPF lesions.
Signature genes encoding candidate biomarkers in IPF
As bronchiolization and lymphoid aggregates are heterogeneously distributed within individual IPF biopsies (Figure 3), a small biopsy sample may not accurately represent the aggregate pathological burden across the total lung tissue in a given IPF patient. However, soluble secreted factors produced by those pathological structures may be detectable in peripheral blood and have the potential to reflect the total pathological burden within a patient. Therefore, we identified genes within the bronchiolar and lymphoid signatures that encode soluble proteins known to be detectable in peripheral blood to investigate in peripheral blood as biomarker candidates that may provide insight into clinical presentation and disease progression in IPF. MMP3 is a matrix metalloproteinase encoded in the bronchiolar signature and CXCL13 is a chemokine encoded in the lymphoid signature; both are soluble secreted factors that are detectable in peripheral blood (15, 16). IHC revealed strong staining for MMP3 predominantly in epithelial cells lining bronchiolized regions in dense scar tissue (Figure 5A), with modest immunoreactivity evident in some lymphoid aggregates. CXCL13 protein stained preferentially in the periphery of lymphoid aggregates and was not evident in bronchiolized regions (Figure 5B).
Figure 5.
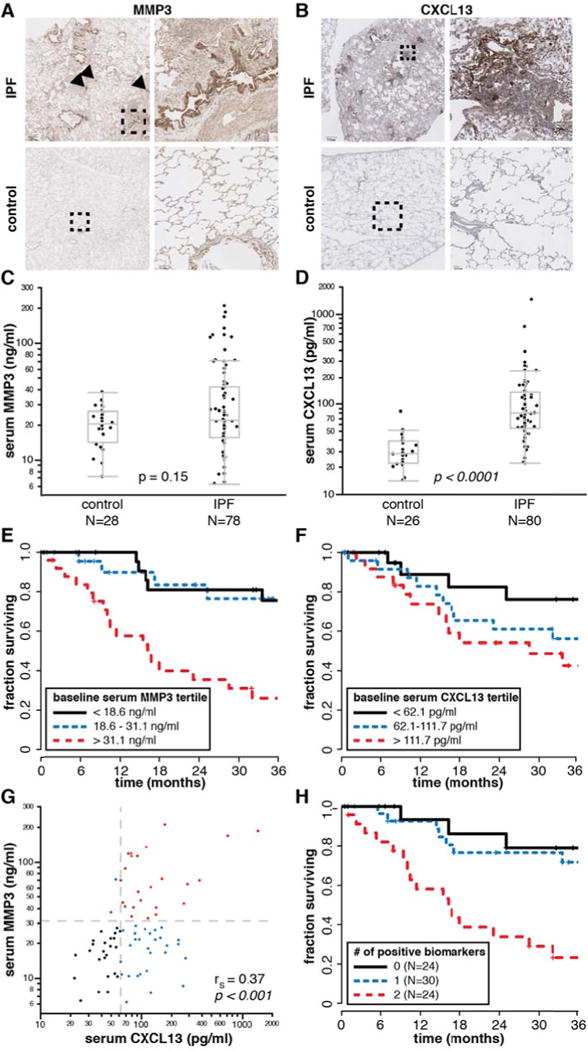
Elevated serum MMP3 and CXCL13 levels are prognostic for decreased survival time in IPF patients.
A) Immunohistochemistry (IHC) for MMP3. Dashed boxes on left panels indicate region shown at higher magnification in right panels. Arrowheads indicate lymphoid aggregates. Scale bar = 700 μM (upper left panel); 90 μM (lower right). B) IHC for CXCL13 as in (A). Scale bar = 700 μM (upper left); 100 μM (lower right). C) Serum MMP3 levels in control and IPF patients. D) Serum CXCL13 levels in control and IPF patients. p-values for (C) and (D) determined by the Wilcoxon rank-sum method. E) Kaplan-Meier (K-M) survival plot for IPF patients by tertiles of baseline serum MMP3 levels. F) K-M survival plot for IPF patients by tertiles of baseline serum CXCL13 levels. G) Correlation between serum MMP3 and CXCL13 levels (IPF patients only). rS, Spearman’s rank-order correlation coefficient. H) K-M survival plot for IPF patients by combined baseline serum MMP3 and CXCL13 levels. A score of 0 corresponds to MMP3 < 31.1 ng/ml AND CXCL13 < 62.1 pg/ml; a score of 1 corresponds to either MMP3 > 31.1 ng/ml OR CXCL13 > 62.1 pg/ml; a score of 2 corresponds to both MMP3 > 31.1 ng/ml AND CXCL13 > 62.1 pg/ml.
We evaluated serum levels of MMP3 and CXCL13 in a separate cohort of 80 IPF patients collected at the time of initial presentation to the ILD clinic (Table 1), and 28 healthy controls (not age or sex-matched; average age 40, range 26 to 64 years old, 56% male). The distribution of serum MMP3 mostly overlapped between controls and IPF patients; however approximately 1/3 of IPF patients had elevated serum MMP3 levels compared to controls (Figure 5C). Serum CXCL13 was significantly elevated in IPF patients compared to controls (Figure 5D).
Among IPF patients, serum MMP3 levels were positively correlated with dyspnea score and negatively correlated with FVC, while serum CXCL13 levels were positively correlated with dyspnea score and negatively correlated with DLCO (Table 2). Both biomarkers were significantly negatively correlated with survival over 3 years subsequent to blood sampling, as assessed using continuous correlations and a Cox proportional hazards model (Table 2). These effects remain significant when the model is adjusted for baseline FVC, DLCO, sex, and age (Supplementary Table 6). We extended the analysis of this relationship via Kaplan-Meier plots of survival by tertiles of serum biomarkers, which suggested that IPF patients in the top tertile of baseline serum MMP3 or the top two tertiles of baseline serum CXCL13 levels (Figure 5E–F) had shortened survival times. Combinations of biomarkers with thresholds of 31 ng/ml MMP3 (top tertile) and 62 pg/ml CXCL13 (top 2 tertiles) demonstrated that patients with elevated levels of both biomarkers had substantially shorter survival times than patients with elevated levels of neither or only one biomarker. However, most of this effect is due to MMP3, as MMP3 and CXCL13 levels were positively correlated in IPF patients; nearly all patients with elevated MMP3 also had elevated CXCL13, but not all patients with elevated CXCL13 had elevated MMP3 (Figure 5G–H). Taken together, these data show that blood biomarkers encoded by genes expressed in pathological structures associated with advanced lesions in IPF lung tissue are correlated with clinical disease severity in IPF patients.
Table 2.
Correlations between blood biomarkers, lung function, and survival among IPF patients
| Variable | by variable | rS | p-value |
|---|---|---|---|
| MMP3 | FVC % predicted | −0.25 | 0.03 |
| DLCO, % predicted | −0.14 | 0.3 | |
| Dyspnea Score | 0.24 | 0.04 | |
| Survival time | −0.33 | 0.004 | |
| CXCL13 | 0.37 | < 0.001 | |
| CXCL13 | FVC % predicted | −0.01 | 0.9 |
| DLCO, % predicted | −0.32 | 0.007 | |
| Dyspnea Score | 0.40 | < 0.001 | |
| Survival time | −0.24 | 0.03 | |
| Survival (Cox proportional hazards model) | Coefficient (CI) | p-value | |
| MMP3 | 1.018 (1.010–1.024) |
5.0 × 10−8 | |
| CXCL13 | 1.003 (1.002–1.005) |
7.3 × 10−6 | |
rS, Spearman’s rank-order correlation coefficient; FVC, forced vital capacity; DLCO, diffusing capacity of carbon monoxide; CI, 95% confidence interval
DISCUSSION
The analysis of gene expression in lung tissue from IPF patients can provide insights into aberrant molecular and cellular processes associated with disease pathology. However, in cross-sectional analyses of bulk biopsy tissue, it is difficult to discern when, where, and how particular sets of differentially expressed genes exert their effect. We have taken complementary approaches, coordinating gene expression with pathological analyses of lung tissue in focal biopsies and extending those findings to peripheral biomarkers and clinical data. Systemic biomarkers that reflect focal pathologic processes in diseased lung have the potential to integrate the cumulative burden of these processes across the entirety of patients’ lung tissue and provide insights into clinically relevant disease features.
We identified 2 clusters of co-regulated genes in IPF lung tissue that display significant heterogeneity across the sample cohort. The genes in these clusters correspond to bronchiolar epithelium and lymphoid aggregates, respectively. Pathological examination of IPF biopsies reveals ciliated columnar epithelial cells lining cystic structures and clusters of lymphocytes in regions of advanced scarring. Elevated expression of genes encoded in the respective signatures was correlated with the relative prevalence of those features in a given tissue section.
Pathological implications of the bronchiolar and lymphoid gene signatures
Bronchiolization of alveolar tissue is a well-described feature of the lung remodeling in IPF lesions (9–11). We observed keratin-expressing columnar cells lining cystic structures in regions of advanced scarring near the pleural surfaces, consistent with the structures being lined with bronchiolar epithelial cells. Whether the columnar epithelial cells lining honeycomb cysts arise from adjacent normal bronchiolar structures or de novo bronchiolization of alveolar tissue remains uncertain, but their presence is a feature of advanced fibrosis and honeycomb cysts found in IPF lung tissue.
MMP3 clusters with genes in the bronchiolar signature (Supplementary Table 4) and exhibits strong immunoreactivity in bronchiolized regions of IPF lung biopsy tissue (Figure 5A). MMP3 can degrade extracellular matrix components, and its activity has been implicated in both wound repair and tumor initiation (17, 18). Elevated levels of MMP3 gene and protein expression have been described in IPF lung tissue, with MMP3 expression localized to alveolar and bronchiolar epithelial cells in IPF but not control lung tissue (19). Elevated MMP3 protein levels have been reported in bronchoalveolar lavage (BAL) fluid from IPF patients (20). Ectopic overexpression of MMP3 in rat lungs promoted pulmonary fibrosis while MMP3-deficient mice were protected from bleomycin-induced pulmonary fibrosis relative to wild-type mice (19). Hence MMP3 may play a role in the chronic pathologic wound repair and tissue remodeling characteristic of advanced IPF lesions.
The bronchiolar signature is similar to a set of genes reported to be upregulated in lung tissue from patients with concomitant pulmonary fibrosis and pulmonary artery hypertension (PAH) (21). Hence it is possible that the decreased tissue compliance and impaired gas exchange associated with advanced scarring, honeycombing, and bronchiolization is associated with increased pulmonary vascular resistance and PAH. PAH in IPF patients is associated with dramatically reduced survival time (22, 23). Unfortunately, we lacked data on whether patients in our cohort had PAH, but we hypothesize that elevated serum MMP3 levels may be indicative of PAH in IPF patients, which should be formally assessed in future studies.
The “lymphoid” signature suggests an inflammatory component associated with chronic lesions in IPF (24), although the exact nature of immune cell aggregates’ role in IPF pathology is unclear. Most T and B cells present in IPF reside in loose focal aggregates as determined by CD3 and CD20 immunolocalization. CXCL13 is a chemokine produced by follicular dendritic cells. It recruits B cells to secondary and tertiary lymphoid structures by binding to its cognate receptor CXCR5, and both CXCL13 and CXCR5 are required for lymphoid follicle formation (25). Lymphoid aggregates have been reported to associate with UIP lesions in IPF biopsies (12) and B cell infiltrates with high CXCL13 expression have also been reported in COPD (26–28). Serum CXCL13 is a biomarker of the severity of joint erosions (29) and of B cell repopulation after rituximab treatment in rheumatoid arthritis (16); a recent report has described increased CXCL13 expression in IPF lungs and plasma CXCL13 as prognostic in an independent cohort of IPF patients (14). In that study, increases in plasma CXCL13 levels over time corresponded to an increased rate of respiratory failure. This trend is consistent with CXCL13 being a biomarker of advanced disease in IPF and suggests that lymphoid aggregates are a late manifestation of the disease.
Whether lymphoid aggregates in IPF are a cause or a consequence of other molecular and cellular processes is presently unclear. Some studies have suggested an autoimmune component to IPF, with increased incidence of autoantibodies against periplakin (30) and Hsp70 (31), and increased levels of BLyS (B lymphocyte stimulator, a B cell survival factor) (32) in IPF patients, each of which was associated with increased disease severity and poor prognosis. Other studies have reported similar prevalence of common autoantibodies in IPF patients and healthy age-matched controls (33). No studies to date have reported whether therapeutic interventions specifically targeting lymphoid biology have any therapeutic benefit in IPF. In our study, lymphoid aggregates were generally found in regions of dense scar tissue, and exhibited immunoreactivity for both CXCL13 and MMP3, which could explain the moderate correlations between bronchiolar and lymphoid signatures and MMP3 and CXCL13 levels in peripheral blood. Bronchiolization and lymphoid neogenesis may be common processes downstream of heavy fibrotic burden within IPF lungs. The prognostic properties of CXCL13 presented here, taken together with prior reports, further substantiate the significance of lymphoid aggregates and adaptive immune responses with respect to disease severity and prognosis in IPF.
Conclusion: potential applications of IPF biomarkers
We have shown that biomarkers encoded in gene expression signatures correlated with distinct pathological structures in IPF lung tissue are detectable at elevated, but variable, levels in the peripheral blood of IPF patients. MMP3 and CXCL13 levels in peripheral blood are associated with features of disease severity, including lung function, diffusing capacity, dyspnea, and survival time. Given the geographic heterogeneity and patchy nature of IPF pathology, systemic biomarkers may provide a more comprehensive picture of the aggregate burden of specific pathological disease features in an individual IPF patient than gene expression levels from or histological examination of a focal biopsy. A potential confounder in this study is that a quarter of the patients in the biomarker cohort were taking systemic immunosuppressants (steroids and/or azathioprine) at the time of sample collection, which could potentially affect systemic biomarker levels and/or survival time. In light of evolving IPF clinical management strategies including recommendations against systemic steroid treatment (34) and new therapies such as pirfenidone and nintedanib (35), future work should be directed at replicating these findings and assessing the dynamics of MMP3 and CXCL13 in additional cohorts of IPF patients over time with respect to treatment and clinical disease progression, and exploring whether these biomarkers can be incorporated along with clinical and physiologic variables into tools that predict rates of disease progression and mortality (36).
Supplementary Material
Key messages.
What is the key question?
How is gene expression in idiopathic pulmonary fibrosis (IPF) lung tissue linked to pathology, systemic biomarkers, and clinical manifestations of disease?
What is the bottom line?
Using transcriptomic and pathological approaches, we identified two distinct features of IPF lesions: bronchiolization and lymphoid aggregates; systemic levels of protein biomarkers encoded in these respective gene signatures (MMP3 and CXCL13) correspond to disease severity and survival in IPF patients.
Why read on?
The unique systematic approach to understanding a complex human disease in this study sheds light on IPF disease progression and is generalizable to many indications.
Acknowledgments
We thank the patients who participated in this study and the providers who referred these patients to the UCSF ILD clinic. Without the generous donation of biological samples from patients, this study could not have been performed.
Funding sources: Genentech, the Nina Ireland Lung Disease program at UCSF, and NIH grant HL 108794.
References
- 1.Adkins JM, Collard HR. Idiopathic pulmonary fibrosis. Seminars in respiratory and critical care medicine. 2012;33:433–9. doi: 10.1055/s-0032-1325154. [DOI] [PubMed] [Google Scholar]
- 2.Ley B, Collard HR. Risk prediction in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;185:6–7. doi: 10.1164/rccm.201111-1960ED. [DOI] [PubMed] [Google Scholar]
- 3.Leslie KO. Idiopathic pulmonary fibrosis may be a disease of recurrent, tractional injury to the periphery of the aging lung: a unifying hypothesis regarding etiology and pathogenesis. Archives of pathology & laboratory medicine. 2012;136:591–600. doi: 10.5858/arpa.2011-0511-OA. [DOI] [PubMed] [Google Scholar]
- 4.Cavazza A, Rossi G, Carbonelli C, et al. The role of histology in idiopathic pulmonary fibrosis: an update. Respiratory medicine. 2010;104(Suppl 1):S11–22. doi: 10.1016/j.rmed.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konishi K, Gibson KF, Lindell KO, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;180:167–75. doi: 10.1164/rccm.200810-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuo F, Kaminski N, Eugui E, et al. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci U S A. 2002;99:6292–7. doi: 10.1073/pnas.092134099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang IV, Coldren CD, Leach SM, et al. Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax. 2013 doi: 10.1136/thoraxjnl-2012-202943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plantier L, Crestani B, Wert SE, et al. Ectopic respiratory epithelial cell differentiation in bronchiolised distal airspaces in idiopathic pulmonary fibrosis. Thorax. 2011;66:651–7. doi: 10.1136/thx.2010.151555. [DOI] [PubMed] [Google Scholar]
- 10.Chilosi M, Poletti V, Murer B, et al. Abnormal re-epithelialization and lung remodeling in idiopathic pulmonary fibrosis: the role of deltaN-p63. Lab Invest. 2002;82:1335–45. doi: 10.1097/01.lab.0000032380.82232.67. [DOI] [PubMed] [Google Scholar]
- 11.Seibold MA, Smith RW, Urbanek C, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PloS one. 2013;8:e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nuovo GJ, Hagood JS, Magro CM, et al. The distribution of immunomodulatory cells in the lungs of patients with idiopathic pulmonary fibrosis. Mod Pathol. 2011 doi: 10.1038/modpathol.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todd NW, Scheraga RG, Galvin JR, et al. Lymphocyte aggregates persist and accumulate in the lungs of patients with idiopathic pulmonary fibrosis. Journal of inflammation research. 2013;6:63–70. doi: 10.2147/JIR.S40673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vuga LJ, Tedrow JR, Pandit KV, et al. C-X-C Motif Chemokine 13 (CXCL13) Is a Prognostic Biomarker of Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2014;189:966–74. doi: 10.1164/rccm.201309-1592OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mamehara A, Sugimoto T, Sugiyama D, et al. Serum matrix metalloproteinase-3 as predictor of joint destruction in rheumatoid arthritis, treated with non-biological disease modifying anti-rheumatic drugs. The Kobe journal of medical sciences. 2010;56:E98–107. [PubMed] [Google Scholar]
- 16.Rosengren S, Wei N, Kalunian KC, et al. CXCL13: a novel biomarker of B-cell return following rituximab treatment and synovitis in patients with rheumatoid arthritis. Rheumatology (Oxford) 2011;50:603–10. doi: 10.1093/rheumatology/keq337. [DOI] [PubMed] [Google Scholar]
- 17.Gill SE, Parks WC. Metalloproteinases and their inhibitors: regulators of wound healing. The international journal of biochemistry & cell biology. 2008;40:1334–47. doi: 10.1016/j.biocel.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sternlicht MD, Lochter A, Sympson CJ, et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–46. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita CM, Dolgonos L, Zemans RL, et al. Matrix metalloproteinase 3 is a mediator of pulmonary fibrosis. The American journal of pathology. 2011;179:1733–45. doi: 10.1016/j.ajpath.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter AG, McKeown S, Rathinam S, et al. Soluble endostatin is a novel inhibitor of epithelial repair in idiopathic pulmonary fibrosis. Thorax. 2009;64:156–61. doi: 10.1136/thx.2008.102814. [DOI] [PubMed] [Google Scholar]
- 21.Mura M, Anraku M, Yun Z, et al. Gene expression profiling in the lungs of patients with Pulmonary Hypertension associated with Pulmonary Fibrosis. Chest. 2011 doi: 10.1378/chest.11-0449. [DOI] [PubMed] [Google Scholar]
- 22.Kimura M, Taniguchi H, Kondoh Y, et al. Pulmonary hypertension as a prognostic indicator at the initial evaluation in idiopathic pulmonary fibrosis. Respiration; international review of thoracic diseases. 2013;85:456–63. doi: 10.1159/000345221. [DOI] [PubMed] [Google Scholar]
- 23.Lettieri CJ, Nathan SD, Barnett SD, et al. Prevalence and outcomes of pulmonary arterial hypertension in advanced idiopathic pulmonary fibrosis. Chest. 2006;129:746–52. doi: 10.1378/chest.129.3.746. [DOI] [PubMed] [Google Scholar]
- 24.Bringardner BD, Baran CP, Eubank TD, et al. The role of inflammation in the pathogenesis of idiopathic pulmonary fibrosis. Antioxidants & redox signaling. 2008;10:287–301. doi: 10.1089/ars.2007.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ansel KM, Ngo VN, Hyman PL, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406:309–14. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 26.Hogg JC, Chu F, Utokaparch S, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. The New England journal of medicine. 2004;350:2645–53. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 27.Litsiou E, Semitekolou M, Galani IE, et al. CXCL13 Production in B Cells via Toll-like Receptor/Lymphotoxin Receptor Signaling Is Involved in Lymphoid Neogenesis in Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2013;187:1194–202. doi: 10.1164/rccm.201208-1543OC. [DOI] [PubMed] [Google Scholar]
- 28.Bracke KR, Verhamme FM, Seys LJ, et al. Role of CXCL13 in Cigarette Smoke-induced Lymphoid Follicle Formation and COPD. Am J Respir Crit Care Med. 2013 doi: 10.1164/rccm.201211-2055OC. [DOI] [PubMed] [Google Scholar]
- 29.Meeuwisse CM, van der Linden MP, Rullmann TA, et al. Identification of CXCL13 as a marker for rheumatoid arthritis outcome using an in silico model of the rheumatic joint. Arthritis and rheumatism. 2011;63:1265–73. doi: 10.1002/art.30273. [DOI] [PubMed] [Google Scholar]
- 30.Taille C, Grootenboer-Mignot S, Boursier C, et al. Identification of periplakin as a new target for autoreactivity in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2011;183:759–66. doi: 10.1164/rccm.201001-0076OC. [DOI] [PubMed] [Google Scholar]
- 31.Kahloon RA, Xue J, Bhargava A, et al. Patients with idiopathic pulmonary fibrosis with antibodies to heat shock protein 70 have poor prognoses. Am J Respir Crit Care Med. 2013;187:768–75. doi: 10.1164/rccm.201203-0506OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue J, Kass DJ, Bon J, et al. Plasma B Lymphocyte Stimulator and B Cell Differentiation in Idiopathic Pulmonary Fibrosis Patients. J Immunol. 2013 doi: 10.4049/jimmunol.1203476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JS, Kim EJ, Lynch KL, et al. Prevalence and clinical significance of circulating autoantibodies in idiopathic pulmonary fibrosis. Respiratory medicine. 2013;107:249–55. doi: 10.1016/j.rmed.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGrath EE, Millar AB. Hot off the breath: triple therapy for idiopathic pulmonary fibrosis–hear the PANTHER roar. Thorax. 2012;67:97–8. doi: 10.1136/thoraxjnl-2011-201398. [DOI] [PubMed] [Google Scholar]
- 35.Hunninghake GM. A new hope for idiopathic pulmonary fibrosis. The New England journal of medicine. 2014;370:2142–3. doi: 10.1056/NEJMe1403448. [DOI] [PubMed] [Google Scholar]
- 36.Ley B, Ryerson CJ, Vittinghoff E, et al. A multidimensional index and staging system for idiopathic pulmonary fibrosis. Annals of internal medicine. 2012;156:684–91. doi: 10.7326/0003-4819-156-10-201205150-00004. [DOI] [PubMed] [Google Scholar]
- 37.Watters LC, King TE, Schwarz MI, et al. A clinical, radiographic, and physiologic scoring system for the longitudinal assessment of patients with idiopathic pulmonary fibrosis. Am Rev Respir Dis. 1986;133:97–103. doi: 10.1164/arrd.1986.133.1.97. [DOI] [PubMed] [Google Scholar]
- 38.Best AC, Meng J, Lynch AM, et al. Idiopathic pulmonary fibrosis: physiologic tests, quantitative CT indexes, and CT visual scores as predictors of mortality. Radiology. 2008;246:935–40. doi: 10.1148/radiol.2463062200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


