SUMMARY
PUF proteins are post-transcriptional regulators that bind to the 3'UTRs of mRNA transcripts. Herein, we show how a yeast PUF protein Puf3p responds to glucose availability to switch the fate of its bound transcripts that encode proteins required for mitochondrial biogenesis. Upon glucose depletion, Puf3p becomes heavily phosphorylated within its N-terminal region of low complexity, associates with polysomes, and promotes translation of its target mRNAs. Such nutrient-responsive phosphorylation toggles the activity of Puf3p to promote either degradation or translation of these mRNAs according to the needs of the cell. Moreover, activation of translation of pre-existing mRNAs might enable rapid adjustment to environmental changes without the need for de novo transcription. Strikingly, a Puf3p phosphomutant no longer promotes translation but also becomes trapped in intracellular foci in an mRNA-dependent manner. Our findings suggest the inability to properly resolve Puf3p-containing RNA-protein granules via a phosphorylation-based mechanism might be toxic to a cell.
INTRODUCTION
Mitochondria play critical roles in cellular energy metabolism, signaling, and survival. The biogenesis of new mitochondria must be properly regulated according to the needs of the cell. In Saccharomyces cerevisiae, mitochondrial biogenesis is repressed in the presence of abundant glucose (Gancedo, 1998). Under such conditions, budding yeast cells prefer a highly glycolytic metabolism. Upon glucose depletion or switch to non-fermentable fuels, yeast cells induce mitochondrial biogenesis to increase their capacity for oxidative catabolism of carbon sources.
However, nuclear-encoded genes required for mitochondrial biogenesis are not transcriptionally silent under glucose-rich conditions. They are transcribed but then rapidly degraded (Scheffler et al., 1998). While the HAP2/3/4/5 DNA-binding complex is involved in the transcriptional activation of genes for respiration (Forsburg and Guarente, 1989), the RNA-binding protein Puf3p, which is not a canonical transcriptional factor, has also been implicated in the regulation of such genes (Gerber et al., 2004). Puf3p is a member of the PUF (PUmilio and FBF) family of RNA-binding proteins (Murata and Wharton, 1995; Zamore et al., 1997; Zhang et al., 1997) and has been shown to specifically bind to the 3' untranslated regions (3'UTRs) of mRNAs encoding mitochondrial proteins (Jackson et al., 2004; Olivas and Parker, 2000; Ulbricht and Olivas, 2008; Zhu et al., 2009). The PUF proteins share eight α-helical repeats that can recognize and bind to specific sequences of mRNA (Edwards et al., 2001; Miller and Olivas, 2011; Wang et al., 2002; Wang et al., 2001). This protein family is conserved from yeast to mammals and has been suggested to function as post-transcriptional regulators. For example, in Caenorhabditis elegans, the PUF protein FBF controls germline stem cells (GSCs) self-renewal by binding to the 3'UTR of genes critical for entry into the meiotic program (Kershner and Kimble, 2010; Merritt and Seydoux, 2010; Suh et al., 2006).
It remains unclear precisely how PUF family proteins regulate their target transcripts. Most studies to date suggest PUF proteins repress translation either through decapping or deadenylation of mRNA (Houshmandi and Olivas, 2005; Kershner and Kimble, 2010; Merritt and Seydoux, 2010; Van Etten et al., 2012). However, other studies suggest their function is to promote mRNA translation perhaps by controlling mRNA localization for the purpose of local translation (Archer et al., 2009; Deng et al., 2008; Gadir et al., 2011; Kaye et al., 2009). Collectively, the current data suggest two opposing functions for PUF family proteins: they paradoxically have been reported to promote both mRNA degradation and translation.
In this study, we decipher how the yeast PUF protein Puf3p functions to regulate its target transcripts that are important for mitochondrial biogenesis. We show how nutrient-responsive phosphorylation of Puf3p within its N-terminal region of low complexity can modulate its function and switch the fate of its target mitochondrial mRNAs from degradation to translation. Furthermore, we show that a phosphomutant of Puf3p becomes trapped within intracellular foci that might be problematic for a cell, suggesting the importance of resolving dynamic RNA-protein granules in a timely manner via phosphorylation. Our results offer a general model by which PUF family proteins function in the post-transcriptional regulation of gene expression.
RESULTS
Demand for mitochondrial biogenesis induces Puf3p phosphorylation
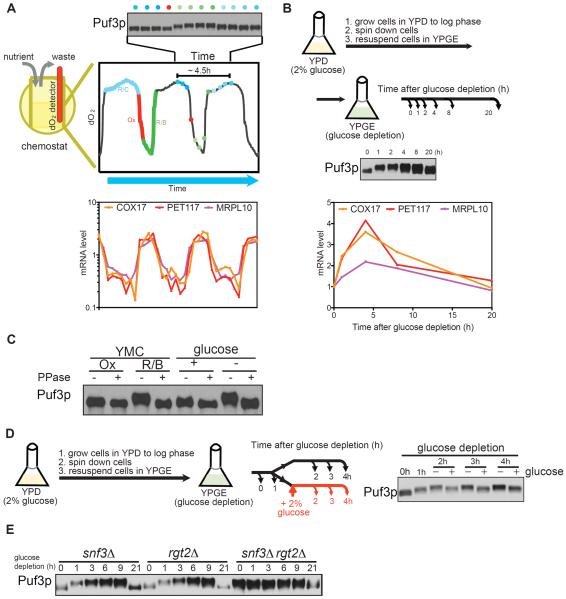
To understand how Puf3p contributes to the regulation of mRNAs required for mitochondrial biogenesis, we first investigated how such gene transcripts are regulated in the context of the yeast metabolic cycle (YMC) (Tu et al., 2005). In the YMC, a highly synchronized cell population transitions between different metabolic phases that are accompanied by the concerted expression of different classes of genes. Transcripts encoding proteins important for mitochondrial biogenesis, including those encoding all mitochondrial ribosomal subunits, coordinately accumulate specifically during the Reductive/Building (R/B) phase when the rate of oxygen consumption begins to decrease. Upon inspection of the cis-regulatory regions of these mitochondrial ribosomal genes, a conserved motif (UGUANAUA) was observed in their 3'UTRs as opposed to the upstream promoter regions (Tu et al., 2005). This motif closely matched a previously determined consensus sequence element for the RNA-binding protein Puf3p, and had suggested a key role for Puf3p in the regulation of these transcripts (Olivas and Parker, 2000; Tu et al., 2005).
We harvested cycling cells that expressed a C-terminal FLAG-tagged version of Puf3p at various time points across the YMC. Following protein extraction, SDS-PAGE and Western blot analysis with FLAG-antibody, we observed a slower-migrating form of Puf3p specifically during the R/B phase precisely when levels of mitochondrial transcripts increase (Figure 1A). We then examined if this slower-migrating form of Puf3p might also be observed in batch cultures upon shifting the cells from glucose medium (YPD) to “glucose depletion” medium (YPGE, which contains glycerol and ethanol), as a means of inducing mitochondrial biogenesis for the utilization of non-fermentable carbon sources (Figure 1B). We observed the rapid appearance of a slower-migrating form of Puf3p that coincided with the increase in mitochondrial transcripts upon switch from glucose to glucose depletion medium. As the slower-migrating form of Puf3p is suggestive of phosphorylation, we then treated cell extracts with phosphatase and observed that the slower-migrating forms of Puf3p collapsed to a single band (Figure 1C). We therefore concluded that Puf3p is subject to phosphorylation modifications that correlate with an increase in transcript levels of nuclear-encoded mitochondrial genes, both in the YMC and in batch culture (Figure 1C). Such transcripts harboring a Puf3p binding sequence in their 3'UTR have been shown to have a shorter half-life in the presence of Puf3p and in glucose medium (Foat et al., 2005; Miller et al., 2014), which is consistent with the lower steady-state levels of such mRNAs observed here in glucose medium.
Figure 1. Glucose depletion triggers the phosphorylation of Puf3p.
(A) Cells expressing PUF3-FLAG were collected at the indicated 12 evenly-spaced time points across one yeast metabolic cycle (YMC). Puf3p protein amounts were assessed by Western blot analysis. The YMC mRNA expression data for 4 genes involved in mitochondrial biogenesis are shown (Tu et al., 2005). Note that a slower-migrating form of Puf3p emerges in the R/B phase, in tune with the observed increase in mitochondrial transcripts.
(B) Cells expressing PUF3-FLAG were grown in batch culture, shifted from glucose (YPD) to glucose depletion (YPGE) medium, and collected at the indicated time points for analysis of Puf3p by Western blot. The abundance of the indicated transcripts was measured by qRT-PCR. Note that a slower-migrating form of Puf3p emerges following glucose depletion.
(C) Puf3p is a phosphoprotein. Puf3p-FLAG was immunoprecipitated from the indicated samples and treated with phosphatase, and then resolved by SDS-PAGE followed by Western blot. Ox and R/B denote samples collected from those phases of the YMC; +glucose and −glucose denote samples collected in batch culture before glucose depletion, or after 4 h glucose depletion.
(D) Glucose inhibits phosphorylation of Puf3p. Cells were grown in YPD, switched to glucose depletion medium (YPGE), and then the culture was split into two after 1 h. One culture was supplemented with 2% glucose, while the other was kept in YPGE. Samples were collected at the indicated time points and Puf3p phosphorylation was assessed by Western blot.
(E) Puf3p is constitutively phosphorylated in the absence of glucose sensors. Puf3p phosphorylation was assessed in the indicated mutant strains at the indicated time points following switch to glucose depletion medium.
Phosphorylation of Puf3p is regulated specifically by glucose
Having observed that Puf3p is subject to phosphorylation modifications, we then aimed to understand how its phosphorylation is regulated. We switched cells to glucose depletion medium to induce Puf3p phosphorylation, and then tested whether the addition of glucose itself or other downstream metabolites could inhibit the phosphorylation of Puf3p. Notably, glucose itself, but not simpler carbon sources such as glycerol, ethanol or acetate, could inhibit the subsequent phosphorylation of Puf3p (Figures 1C and S1). Budding yeast have two regulators of glucose transport, Rgt2p and Snf3p, that function as glucose sensors on the cell surface (Ozcan et al., 1996). We observed phosphorylated Puf3p even in the presence of glucose, in a rgt2Δsnf3Δ mutant lacking both glucose sensors (Figure 1E). Thus, yeast cells appear to sense primarily glucose itself to regulate the phosphorylation status of Puf3p.
We next tested several candidate nutrient-responsive kinases and phosphatases for possible roles in the regulation of Puf3p phosphorylation. Interestingly, the inhibition of either PKA or Sch9p partially inhibited the phosphorylation of Puf3p upon glucose depletion (Figure S1). Both PKA and Sch9p are thought to be most active under glucose-replete conditions (Rolland et al., 2002). Our results suggest that these kinases might have alternate substrates (Roosen et al., 2005), such as Puf3p, when glucose becomes depleted. Moreover, the PP2A-related phosphatase Sit4p contributes to the dephosphorylation of Puf3p in glucose, as sit4Δ mutant exhibited phosphorylated Puf3p even in glucose medium (Figure S1). Thus, while the phosphorylation status of Puf3p is influenced by multiple nutrient-responsive kinases and phosphatases, the phosphorylation of Puf3p is driven primarily by the lack of glucose.
Phosphorylation of Puf3p is important for mitochondrial biogenesis
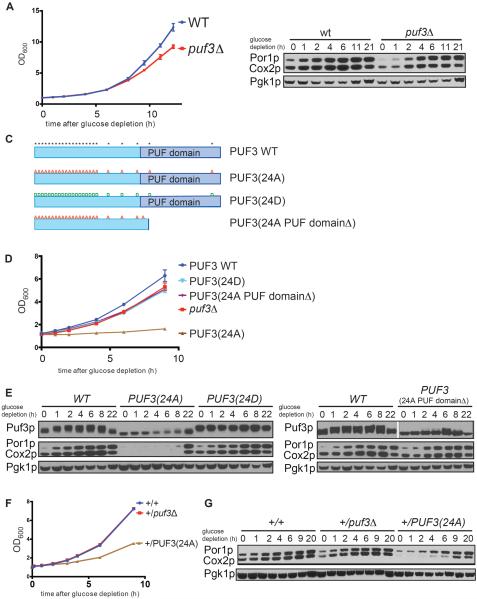
We hypothesized that the phosphorylation of Puf3p, triggered by glucose deprivation, could be a key clue to understanding how the function of this PUF protein is modulated in a manner dependent on the nutrient environment. In wild type (WT) cells, upon glucose depletion the increase in Puf3p phosphorylation and mitochondrial transcript abundance is accompanied by an substantial increase in mitochondrial protein abundance as assessed by the mitochondrial markers Por1p and Cox2p (Figure 2B). Although puf3Δ cells reportedly exhibit a mitochondrial distribution phenotype (Garcia-Rodriguez et al., 2007), Puf3p has been shown to be dispensable for growth in the presence of non-fermentable carbon sources (Eliyahu et al., 2010; Gerber et al., 2004). We compared the growth rates of WT and puf3Δ cells, and confirmed that the absence of Puf3p caused only a small growth defect and delay in mitochondrial biogenesis upon switch to glucose depletion medium (Figures 2A and 2B). It has been proposed that the function of Puf3p may be partially redundant with the mitochondrial outer membrane translocase complex (Eliyahu et al., 2010). Tom20p, a component of this complex, recruits Puf3p target mRNAs to the mitochondrial surface for translation and a synthetic growth defect was observed in puf3Δtom20Δ double mutants in non-fermentable carbon sources.
Figure 2. Puf3p phosphorylation is required for proper mitochondrial biogenesis.
(A) Growth curves of wild type and puf3Δ strains following switch to glucose depletion medium (YPGE).
(B) The abundance of mitochondrial proteins was assessed in WT and puf3Δ strains at the indicated time points following switch to glucose depletion medium (YPGE). Western blot analysis of Por1p (mitochondrial porin) and Cox2p (subunit II of cytochrome c oxidase) indicate abundance of mitochondrial proteins; Pgk1p (phosphoglycerate kinase) serves as a loading control.
(C) Schematic representation of the domain structure of Puf3p. “*” indicates identified phosphorylation sites. “A or D” denote serine or threonine residues that were mutated to alanine or aspartate. The PUF domain mediates mRNA-binding.
(D) Growth curves of WT and the indicated Puf3p mutants in glucose depletion medium.
(E) Mitochondrial biogenesis is severely compromised in the Puf3p(24A) mutant. Possible phosphorylation of Puf3p, Puf3p(24A), Puf3p(24D), and Puf3p(24A PUF domainΔ) at the indicated times following glucose depletion was assessed by Western blot using FLAG-tagged versions of these proteins. The 24A PUF domainΔ protein runs at a smaller size but the gel was cropped to show the relevant band. The accumulation of mitochondrial proteins was assessed in the same samples by Western blot. Por1p and Cox2p are mitochondrial markers, Pgk1p is loading control.
(F) The Puf3p(24A) mutant acts as a dominant negative. Growth curves of the indicated diploid strains: +/+ (WT), +/puf3Δ and +/PUF3(24A) following switch to glucose depletion medium.
(G) Mitochondrial biogenesis following glucose depletion in the indicated strains was assayed by Western blot.
Nonetheless, we assessed whether Puf3p phosphorylation is functionally important for the translation of transcripts encoding mitochondrial proteins. To this end, we identified 24 potential phosphorylation sites within Puf3p by mass spectrometry analysis with ~95% sequence coverage (Experimental Procedures). Most of these modified serine and threonine residues were located in the N-terminal region that has no obvious homology to any known proteins, while a few were located near or within the PUF domain that mediates mRNA-binding (Figure 2C). We initially mutated a subset of these serine and threonine residues to alanine and did not observe substantial decreases in Puf3p phosphorylation or growth phenotypes (data not shown). We then constructed mutants in which all possible phosphorylated serine or threonine residues in Puf3p were mutated to alanine (24A) or aspartate (24D) to mimic either non-phosphorylated or phosphorylated Puf3p (Figure 2C). Strikingly, unlike puf3Δ, the Puf3p(24A) mutant could barely grow at all after switch to glucose depletion medium (Figures 2D and S2). We confirmed that phosphorylation was largely abolished in the Puf3p(24A) mutant following glucose depletion as assessed by SDS-PAGE (Figure 2E). Mitochondrial protein accumulation was severely decreased in the Puf3p(24A) mutant compared to WT, and also substantially lower compared to the puf3Δ mutant which lacks Puf3p altogether (Figure 2E). This substantial defect in mitochondrial biogenesis in the Puf3p(24A) mutant likely explains why it grows very poorly in response to glucose depletion.
We then tested whether these severe phenotypes of Puf3p(24A) were dependent on mRNA binding. The 24 S/T -> A mutations combined did not compromise the ability of immunoprecipitated full-length protein to bind a target mRNA in a gel-shift assay (Figure S3). Moreover, Puf3p was able to bind a target mRNA comparably in both glucose and glucose depletion conditions as assayed by RIP, suggesting that its phosphorylation does not significantly impact RNA binding (Figure S3). A previous study suggested that mutation of one of these sites (S866A) near the end of the PUF domain reduces its binding affinity for RNA (Zhu et al., 2009). However, a single S866A mutant in the context of the full-length protein was still able to bind RNA and grew at identical rates compared to WT following glucose depletion (Figure S3). Moreover, a 23A mutant in which residue 866 was restored to serine, and a 21A mutant in which the three sites most proximal to the PUF domain (515, 563, 866) were restored to serine, both still exhibited the same severe growth defect as the 24A mutant (Figure S3). However, deletion of the PUF domain (24A PUF domainΔ) abolished RNA binding, and relieved the severe phenotype of Puf3p(24A) (Figures 2D, S2, and S3). Puf3p(24A PUF domainΔ) behaved similarly to puf3Δ, exhibiting only a small defect in growth. Therefore, the severe phenotypes of the Puf3p(24A) mutant are dependent on the ability of this protein to bind mRNA. In contrast to Puf3p(24A), the Puf3p(24D) mutant exhibited only a small growth defect following switch to glucose depletion medium, again similar to puf3Δ (Figure 2D). The extent of mitochondrial biogenesis in Puf3p(24D) was not enhanced compared to WT (Figure 2E), suggesting that Puf3p(24D) may not function as an appropriate mimic of the Puf3p phosphorylated state.
These prominent phenotypes of Puf3p(24A) compared to puf3Δ suggest that the Puf3p(24A) mutant protein might be acting in a dominant manner. Indeed, we observed that a diploid strain expressing one copy of PUF3(24A) exhibited a notable growth defect in glucose depletion medium compared to a diploid expressing only one copy of PUF3 (Figures 2F and S2). Accordingly, mitochondrial biogenesis was also compromised in the diploid expressing one copy of PUF3(24A) (Figure 2G). Thus, the function of the Puf3p(24A) protein is impaired in a manner that dominantly compromises mitochondrial biogenesis and growth in glucose depletion medium.
Puf3p promotes translation of its bound mRNAs upon glucose depletion
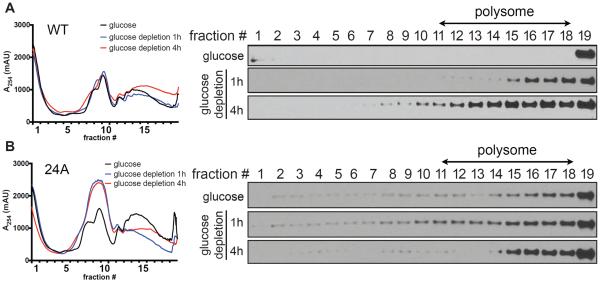
To directly test whether the phosphorylation of Puf3p functions to promote the translation of its target mitochondrial transcripts, we performed polysome profiling to assess whether Puf3p might be associated with actively translating ribosomes upon glucose depletion. Puf3p sedimented in the bottom fraction and was not distributed to polysome fractions in glucose medium (Figure 3A). Following glucose depletion, Puf3p then became strongly associated with the actively translating polysome fractions over time. In contrast, Puf3p(24A) was constitutively distributed to both translationally active and inactive fractions in both glucose and glucose depletion medium (Figure 3B), suggesting that the ability of Puf3p to dynamically associate with translating polysomes is abolished in this phosphomutant. The Puf3p(24A) mutant did not appear to affect global translation in glucose medium, as the polysome profile and rRNA distribution were similar to WT (Figures 3 and S4). However, Puf3p(24A) caused a notable increase in the inactive monosome fraction following glucose depletion, which is suggestive of defects in polysome formation. These data suggest that upon glucose depletion, the phosphorylation of Puf3p might promote the translation of its target mitochondrial transcripts.
Figure 3. Puf3p associates with polysomes following glucose depletion.
(A) Puf3p WT polysome profiles in glucose or following glucose depletion for 1 h or 4 h.
(B) Puf3p(24A) polysome profiles in glucose or following glucose depletion for 1 h or 4 h. Polysomes were fractionated by sucrose gradients (7–47%, w/v). Samples were subjected to continuous A254 measurements and separated into 19 fractions. Proteins from each fraction were precipitated by TCA and analyzed by Western blot. Note that Puf3p associates with polysomes only following glucose depletion, whereas Puf3p(24A) is mislocalized to all fractions regardless of glucose availability.
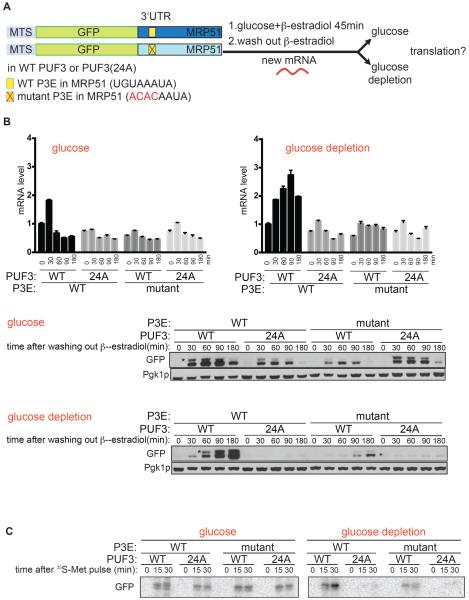
We then developed an assay to determine the extent of translation of an mRNA following the shutoff of its transcription, thereby enabling us to assess the fate of pre-existing mitochondrial mRNAs upon glucose depletion (Figure 4A). We constructed reporter genes consisting of a mitochondrial targeting signal, the GFP coding sequence, and the 3'UTR of a mitochondrial ribosomal gene (MRP51) which contains a single Puf3p binding element (P3E) (Figures 4A and S3). Reporter genes were integrated into a strain expressing a chimeric GAL4DBD-ER-VP16 transcription factor that enables inducible expression by addition of β-estradiol (McIsaac et al., 2011). Thus, following a pulse of expression of the reporter mRNA, the extent of its translation into GFP protein can be assessed by Western blot analysis (Figure 4A). Using this method, we could then monitor the subsequent translation of mRNAs with or without P3E in their 3'UTRs, and in cells expressing either WT Puf3p or Puf3p(24A).
Figure 4. Puf3p promotes the translation of its target mRNAs following glucose depletion.
(A) Schematic representation of RNA-pulse experiment to assay mRNA translation. The indicated reporter constructs with different 3'UTRs were placed under the control of the PGAL1 promoter and integrated into strains expressing GAL4DBD-ER-VP16, along with either Puf3p or Puf3p(24A). After cells were grown in YPD (glucose) to log phase, β-estradiol was added to induce a pulse of transcription of the reporter mRNA. After 45 min, cells were washed twice to remove β-estradiol and then split into YPD (glucose) or YPGE (glucose depletion) medium.
(B) Extent of translation of mRNA reporters assessed by Western blot. Cells were harvested at 0, 30, 60, 90, and 180 min for analysis of reporter mRNA levels by qRT-PCR and protein levels by Western blot. Top panel: reporter mRNA levels assayed by qRT-PCR. Bottom panel: reporter translation assayed by Western blot using anti-GFP antibody, “*” denotes the slightly larger, unprocessed MTS-GFP. Pgk1p denotes loading control. The increase in mRNA level observed in PUF3 WT / 3'UTR WT after medium switch is due to an unavoidable washing step to remove β-estradiol. Note that Puf3p is not absolutely required for translation of its target mRNAs (Figure 2B).
(C) 35S-Met Pulse assay. After cells were grown in SCD-Met (glucose) to log phase, β-estradiol was added to induce a pulse of transcription of the reporter mRNA. After 30 min, cells were washed twice to remove β-estradiol and then split into SCD-Met (glucose) or SCGE-Met (glucose depletion) medium, and then grown for 15min before adding 35S-Met (100mCi/ml). Cells were harvested 15 or 30 min after the pulse. Newly translated GFP was immunoprecipitated by GFP antibody and analyzed by phosphorimaging.
We observed that the translation of the mRNA reporter containing a MRP51 3'UTR increased following switch to glucose depletion medium in cells expressing WT Puf3p, as can be seen by the increases in both GFP as well as the slightly larger, unprocessed MTS-GFP (Figure 4B). However, if cells were left in glucose medium, translation of these reporter mRNAs decreased over time. In contrast, cells expressing Puf3p(24A) showed minimal translation of reporter mRNA into GFP protein upon glucose depletion, consistent with the severe defects in mitochondrial biogenesis in this mutant (Figure 4B). Importantly, the enhanced translation of these reporter mRNAs upon glucose depletion was dependent on an intact P3E in their 3'UTRs. Mutation of four nucleotides within the P3E largely eliminated translation of the reporter mRNA (Figure 4B).
In parallel, we performed a pulse-labeling experiment with 35S-methionine followed by immunoprecipitation to assess translation of the GFP reporter mRNA. In agreement with the Western blot assay, upon glucose depletion, only mRNA containing an intact P3E was actively translated over time in cells expressing normal Puf3p (Figure 4C). Moreover, translation of the reporter mRNA, and translation in general, were substantially reduced in cells expressing Puf3p(24A) following glucose depletion (Figures 4C and S4). Collectively, these data show that the phosphorylation of Puf3p functions to promote the translation of preexisting target mRNAs that contain a P3E in a timely manner. As such, Puf3p responds to a glucose depletion cue to switch the fate of its target mRNAs, enabling them to be translated into protein.
A Puf3p phosphomutant forms punctate foci following glucose depletion
The dominant negative phenotype of the Puf3p(24A) phosphomutant prompted us to further examine the basis of its strong inhibitory effects on mitochondrial biogenesis and cell growth. We constructed C-terminal GFP-tagged versions of Puf3p and Puf3p(24A) expressed at endogenous levels from the native chromosomal locus to assess the subcellular localization of these proteins before or after glucose depletion. We observed that both the WT and phosphomutant protein were localized throughout the cytosol in glucose medium (SCD/YPD) (Figures 5 and S5 and Supplemental Video). However, following switch to glucose depletion medium (SCGE/YPGE), remarkably Puf3p(24A)-GFP, but not Puf3p-GFP, became localized to punctate foci (Figures 5 and S5 and Supplemental Video). These foci emerged specifically upon glucose depletion, and disappeared following glucose repletion. Moreover, the Puf3p(24A)-containing foci exhibited partial but not complete co-localization with the decapping enzyme Dcp2p, a marker of p-bodies, following glucose depletion (Figure S5). Unlike previous reports, we observed that Dcp2p did not localize to foci but was instead uniform throughout the cytosol in the presence of continuous glucose (Figure S5) (Sheth and Parker, 2003; Teixeira et al., 2005).
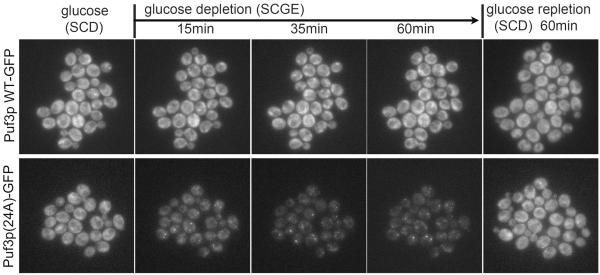
Figure 5. Puf3p(24A) phosphomutant forms punctate foci following glucose depletion.
Live cell imaging of cells expressing Puf3p-GFP or Puf3p(24A)-GFP using the CellASIC microfluidics platform, before and after switch to glucose depletion medium for the indicated times. Puf3p-GFP was uniformly distributed in the cytosol and not specifically localized to mitochondria regardless of glucose availability. Strikingly, Puf3p(24A) forms foci only after switch to glucose depletion medium. Approximately 40% of cells expressing Puf3p(24A)-GFP exhibited foci following glucose depletion, in contrast to 0% of cells expressing WT Puf3p-GFP. See also Figure S5 and Supplementary Videos. These foci exhibited partial co-localization with the p-bodies marker Dcp2p (Figure S5). Both Puf3p(24A) and Dcp2p formed foci only after switch to glucose depletion medium (Figure S5).
Interestingly, the ability of the Puf3p(24A) protein to form foci was dependent on the PUF domains that bind mRNA (Figure S6), suggesting that any bound mRNAs become trapped within these foci as well. Since the Puf3p(24A) mutant acts as a dominant negative, these foci might contribute to the toxicity of this phosphomutant. We speculate that these Puf3p(24A)-containing foci might retain mitochondrial mRNAs too strongly, or compromise critical components of the translational machinery (Figure S4). Intriguingly, these localization data suggest that phosphorylation of Puf3p not only promotes translation of its bound mRNAs, but also functions to resolve its association with RNA-protein granules that influence mRNA fate. Failure to properly resolve or “unfurl” these granules may be problematic for the cell (Figures 2D and 5).
Notably, the phosphorylation of Puf3p occurs primarily within its N-terminal region that is predicted to be disordered and is not conserved among PUF proteins (Olivas and Parker, 2000). To further demonstrate the importance of phosphorylation within this region, we applied Staggered Extension Process (StEP) (Zhao and Zha, 2006) to generate a library of Puf3p phosphomutants with random numbers and positions of S/T to A mutations within this N-terminal region (Figure 6A). We observed a strong correlation between the number of S/T to A mutations and the severity of the growth defect in glucose depletion medium (Figure 6B). This allelic series suggests the accumulation of phosphorylation promotes the “unfurling” of Puf3p from RNA-protein granules and translation of its bound mRNAs. Interestingly this N-terminal region is still dynamically phosphorylated in response to glucose depletion in the absence of the PUF domain (Figure S6). Moreover, a deletion of this N-terminal region abolishes the ability of Puf3p to properly regulate mitochondrial biogenesis and causes the protein to be localized to the mitochondria independent of glucose (Figure S6). Taken together, these data strongly suggest the N-terminal low complexity region of Puf3p confers upon it the ability to dynamically interact with other factors involved in mRNA degradation or translation, and that phosphorylation of this region regulates the fate of its bound transcripts and association with RNA-protein granules.
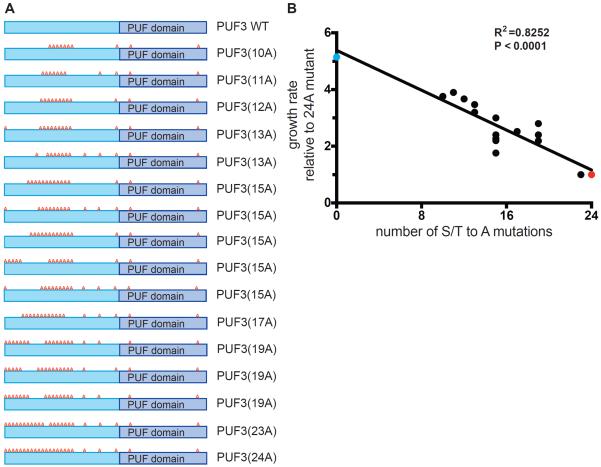
Figure 6. Correlation between number of phosphosite mutations within the N-terminal region of Puf3p and severity of the growth defect following glucose depletion.
(A) A series of 15 phosphomutants containing anywhere from 10 to 23 S/T -> A mutations (10A to 23A) within the N-terminal low complexity region were generated as indicated.
(B) Growth rates of the various phosphomutants (black dots) in YPGE were plotted against the number of phosphosite mutations. Growth rates were normalized against PUF3(24A). Growth rates of PUF3 WT (blue dot) and PUF3(24A) (red dot) are also shown.
DISCUSSION
A general model to explain the function and regulation of the PUF family of RNA-binding proteins
Previous studies of PUF proteins have suggested they promote degradation of bound mRNAs (Goldstrohm et al., 2007; Kershner and Kimble, 2010; Olivas and Parker, 2000), promote their translation (Archer et al., 2009; Kaye et al., 2009), or exert dual, opposing roles on their target transcripts (Suh et al., 2009). For yeast Puf3p, it has been proposed to promote degradation of its target mRNAs in glucose, but not under non-fermentable carbon sources (Miller and Olivas, 2011; Miller et al., 2014). However this model does not explain the delayed growth of puf3Δ cells upon glucose depletion, despite having comparable levels of mRNAs compared to WT (Figures 2 and S7). Here, we show that Puf3p can in fact promote either degradation or translation of its bound transcripts, but in a manner specifically dependent on the nutrient environment. Thus, a consensus model emerges whereupon following glucose depletion, pre-existing mitochondrial transcripts are stabilized, and then activated for translation by Puf3p (Figure 7). The ability of Puf3p to switch the fate of these mRNAs is regulated by its phosphorylation, which occurs in response to glucose depletion. In the wild, yeast cells must be readily able to adapt to fluctuating nutrient environments which may include varying amounts of glucose. As such, the post-transcriptional regulation of mRNA fate by Puf3p could facilitate the prompt acclimation to changes in carbon source availability. Activating the translation of a pre-existing pool of mRNAs by phosphorylated Puf3p could bypass the need for transcriptional induction of such genes and provide a fitness advantage (New et al., 2014). Such a mechanism would enable mRNA transcripts to be rapidly mobilized for translation “on cue”, without the need for de novo transcription. As such, we propose that PUF family proteins in general might function as environmental sensors to toggle the fate of bound mRNAs between degradation and translation. Consistent with this idea, other PUF proteins have been shown to act downstream of nutrient or growth factor signaling, in some cases to regulate cell cycle progression (Archer et al., 2009; Kedde et al., 2010; Souza et al., 1999). In addition, phosphorylation may emerge as a general mechanism to regulate the function of PUF proteins (Deng et al., 2008; Kedde et al., 2010). Future studies will reveal whether other PUF proteins in yeast and other species might function to regulate their target mRNAs with a similar logic.
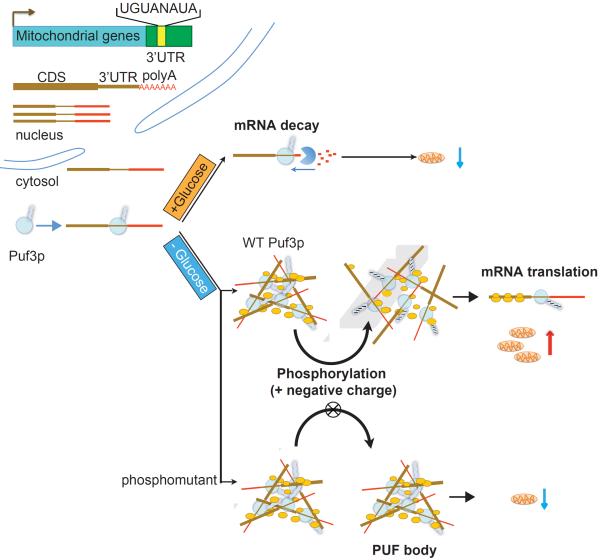
Figure 7. Model depicting the role of Puf3p in regulating the fate of its bound mRNAs.
Many mRNA transcripts important for mitochondrial biogenesis harbor a Puf3p binding element in their 3'UTRs. In the presence of glucose, these mRNAs are transcribed, but then degraded in a Puf3p-dependent manner. Upon glucose depletion, Puf3p becomes phosphorylated which then enables it to associate with polysomes and promote the translation of these mRNAs. A mutant of Puf3p that prevents its phosphorylation becomes trapped within PUF-bodies that might be toxic to the cell. As such, phosphorylation of Puf3p is not only required to promote translation of its bound mRNAs, but is also important for proper and timely resolution of its association with RNA-protein granule components.
General implications for mitochondrial biogenesis
A computational analysis of mRNA transcripts encoding mitochondrial proteins in the context of the YMC provided insights into the spatiotemporal dynamics of mitochondrial biogenesis (Lelandais et al., 2009). Interestingly, transcripts encoding mitochondrial assembly factors and translational machinery tended to increase in abundance earlier than transcripts encoding other mitochondrial components, such as those encoding respiratory chain components. Notably, many of these mitochondrial gene transcripts in the “early” module contain Puf3p binding elements in their 3'UTRs and their asymmetric mRNA localization is dependent on Puf3p (Saint-Georges et al., 2008). For example, more than 70% of genes encoding subunits of the mitochondrial ribosomes are Puf3p targets (Gerber et al., 2004). Such mRNAs with Puf3p-binding elements may therefore function in the earlier stages of mitochondrial biogenesis, necessitating their timelier activation. These considerations suggest an exquisite temporal organization of mitochondrial biogenesis that may be set in motion post-transcriptionally by Puf3p. Activating the translation of Puf3p targets may “kick-start” the earlier steps of mitochondrial biogenesis in order to facilitate the subsequent steps of this elaborate process. As one example, COX2 is a mitochondrially-encoded gene whose translation relies on the translation of nuclear-encoded mitochondrial ribosomal transcripts, most of which are Puf3p targets. Thus, levels of Cox2p protein can serve as a proxy for the proper translation of Puf3p targets (Figure 2). Mitochondrial mRNAs which do not contain Puf3p elements may be regulated more at the transcriptional level, for example by the HAP2/3/4/5 transcription factor complex (Buschlen et al., 2003). Thus, in response to a glucose depletion cue, pre-existing Puf3p target mRNAs are poised to be activated for translation to initiate the early steps of mitochondrial biogenesis so that subsequent steps of this process can ensue.
The inability to properly resolve RNA-protein granules might be toxic to cells
The profound phenotype of the Puf3p phosphomutants (24A, as well as 23A and 21A) may provide insights into the dynamic nature of RNA-protein granules in the cell. Since this mutant dominantly inhibits both mitochondrial biogenesis and growth in glucose depletion medium, we hypothesize the punctate foci, or “PUF-bodies”, formed by the mutant protein might be toxic to the cell. PUF-bodies exhibit partial co-localization with the p-bodies marker Dcp2p (Figure S5), but no obvious co-localization with stress granule markers (data not shown). Under a variety of nutrient conditions, thus far we have observed an increased propensity to form PUF-bodies only in those cells expressing Puf3p(24A), specifically upon glucose depletion. Moreover, both the ability of Puf3p(24A) to form foci and inhibit growth are dependent on its ability to bind mRNA (Figures 2D, S3 and S6), suggesting that these PUF-bodies also trap mRNA transcripts.
We interpret these observations as follows: upon glucose depletion, phosphorylation of Puf3p brings the protein and its bound transcripts to an assembly that promotes mRNA translation, perhaps in the vicinity of mitochondria (Garcia-Rodriguez et al., 2007; Saint-Georges et al., 2008). Failure to properly phosphorylate within its N-terminal region not only compromises mRNA translation, but also substantially increases its residence time at a particular RNA-protein assembly, leading to the apparent formation of PUF-bodies (Figure 7). Notably, this N-terminal region lacking any defined domains is critical for regulating its subcellular localization and ability to associate with RNA-protein granules (Figure S6). The number of available phosphorylation sites in this region correlates well with the growth rate in non-fermentable, glucose depletion conditions (Figure 6). Therefore, phosphorylation of this low-complexity region serves as a mechanism to help resolve its association with granule components that could include mRNA decay or translation factors, perhaps via the accumulation of negative charge that functions to unfurl the protein from interacting partners (Kwon et al., 2013; Kwon et al., 2014). PUF-bodies are not simply dead-end structures or aggregates, as glucose repletion immediately eliminates PUF-bodies and cells can begin to grow again (Figure 5). An alternative explanation for the toxicity of the Puf3p(24A) phosphomutant is that it could exhibit enhanced mRNA decay. However, we observed little difference in the levels of several endogenous mRNAs containing a Puf3-binding element in their 3'UTRs between WT and PUF3(24A) strains, even in glucose (Figure S7). These data suggest the dominant negative phenotype of the phosphomutant in the absence of glucose is not simply due to enhanced mRNA degradation.
Intriguingly, loss of canonical components of P-bodies or stress granules often has little to no phenotypic consequence, raising the question of the general importance of such RNA-protein granules. Here, we have identified a mutant form of a granule-associated RNA-binding protein that exhibits a profound dominant negative phenotype, in a manner specifically dependent on mRNA-binding. As the Puf3p(24A) phosphomutant causes inactive monosome accumulation and inhibits global translational capacity upon glucose depletion (Figures 3B and S4), we speculate that mutants which compromise the timely resolution of RNA-protein granules may be toxic due to sequestration of key mRNA transcripts, or critical components of the translational machinery. The toxic phenotype of the Puf3p(24A) mutant should facilitate the subsequent identification of in vivo modifiers of PUF-bodies that mediate the formation or dissolution of RNA-protein granules in cells. As aggregates of a variety of proteins containing RNA-binding domains have been strongly linked to various forms of neurodegeneration (Kwiatkowski et al., 2009; Ramaswami et al., 2013; Ross and Poirier, 2004; Vance et al., 2009), future studies of the regulation of mRNA fate by PUF proteins may improve our understanding of the etiology of such diseases.
EXPERIMENTAL PROCEDURES
Yeast strains and media
Strains used in this study are listed in Table S1. Gene deletions were carried out using standard PCR-based strategies to amplify resistance cassettes with appropriate flanking sequences, and replacing the target gene by homologous recombination (Longtine et al., 1998). C-terminal tags were similarly made using PCR to amplify resistance cassettes with flanking sequences. Media used in this study: YPD (1% yeast extract, 2% peptone and 2% glucose); YPGE (1% yeast extract, 2% peptone, 3% glycerol and 2% ethanol); SCD (yeast nitrogen base 6.7 g/L, 1X complete supplement mixture, 2% glucose); SCGE (yeast nitrogen base 6.7 g/L, 1X complete supplement mixture, 3% glycerol and 2% ethanol).
Yeast metabolic cycle
The YMC was established as previously described (Tu, 2010). Briefly, 12.5 mL of an overnight culture of PUF3-FLAG cells was inoculated into a 1 L fermentor vessel. In continuous mode, consecutive samples of ~100 OD cells were quickly spun down, flash-frozen in liquid nitrogen, and stored at −80°C until further processing.
Glucose depletion in batch culture
Cells from overnight cultures were inoculated into fresh YPD to 0.2 OD/mL and grown at least two generations to log phase. Cells were then spun down, washed, and re-suspended in same volume of YPGE. Samples were collected at indicated times.
Quantitative RT-PCR
Approximately 1 OD of cells were collected and total RNA was extracted with MasterPure Yeast RNA Purification Kit following the manufacturer's protocol. qRT-PCR was done with SYBR green method. mRNA levels were normalized against ACT1 or PGK1 mRNA. Primers are listed in Supplemental Table S2.
Mapping phosphorylation sites
PUF3-FLAG cells were inoculated to 0.2 OD/mL in fresh YPD. After reaching 1 OD/mL, cells were spun down, washed with YPGE, and resuspended in the same volume of YPGE. The cells were then grown for 4 h to induce maximal Puf3p phosphorylation, then ~100 OD of cells were harvested and resuspended in 0.5 mL lysis buffer (100 mM Tris-Cl pH 7.5, 50 mM NaCl, 0.1% Tween-20, glycerol 10%, 0.1% β-mercaptoethanol, 1X EDTA-free protease inhibitor cocktail (Roche), 1 mM EDTA 1mM EGTA, 1 mM PMSF, 5 μM Pepstatin A, 10 μM Leupeptin, 0.2 mM sodium orthovanadate, 10 mM β-glycerolphosphate, 60 mM NaF, and 10 mM NaN3) and then mixed with 0.5 mL glass beads. The cells were lysed by bead beating 6 × 20 s. After spinning at 16000×g 1 min at 4°C to collect the supernatant, 0.5 mL lysis buffer was added to the tube, bead-beated 2 × 20 s, and the supernatant was collected after centrifugation at 16000×g 1 min at 4°C. The supernatant was then incubated with anti-FLAG M2 antibody coated Dynabead (Invitrogen) at 4°C for 1 h. After extensive washing, Puf3p-FLAG was eluted with 3X FLAG peptide (1 mg/mL) (Sigma) and resolved by SDS-PAGE. The gel was silver stained, then subjected to in-gel digestion by trypsin, chymotrypsin and elastase, followed by LC-MS/MS analysis (MSBioworks, USA). Data are included as Supplemental Table S2. In total, 24 phosphorylation sites were identified: Ser21, Ser55, Ser56, Thr59, Ser65, Ser76, Ser77, Ser86, Thr89, Ser117, Ser166, Ser178, Ser203, Ser205, Ser210, Thr213, Thr216, Thr252, Thr255, Ser371, Thr427, Ser515, Ser563, Ser866.
N-terminal phosphorylation site mutations were made by gene synthesis (Genewiz, USA). Other phosphorylation site mutations (Ser371, Thr427, Ser515, Ser563 and Ser866) were made by fusion PCR with mutation sites introduced in the primer sequence. These two fragments were fused by another fusion PCR and then cloned into pFA6a-3xFLAG-NatMX6 vector. Then the sequence including PUF3 coding sequence, C-terminal 3xFLAG and NatMX6 genes were amplified by PCR and transformed into puf3∷KanMX6. The entire sequence was introduced to the endogenous PUF3 locus by recombination and verified by DNA sequencing. PUF3(24A PUF domainΔ) was made by deleting the sequence of PUF domain (538–844) in PUF3(24A) backbone.
Phosphatase treatment
Puf3p-FLAG was first immunoprecipitated by Anti-FLAG and eluted with 3X FLAG peptide, following the same procedure as mapping phosphorylation sites. The samples were treated with 400 units of Lambda phosphatase (NEB) at 30°C for 30 min.
Western blot
Yeast total extracts were prepared following trichloroacetic acid precipitation method (Knop et al., 1999). 5 OD of cells were resuspended with 1150 μL alkaline lysis buffer (0.24 N NaOH, 1% β-mercaptoethanol, 1X EDTA-free protease inhibitor cocktail (Roche), 1 mM EDTA, 1 mM PMSF, 5 μM Pepstatin A, 10 μM Leupeptin, 0.2 mM sodium orthovanadate, 10 mM β-glycerolphosphate, 10 mM NaF and 10 mM NaN3). After incubation on ice for 15 min, 150 μL of 55% TCA was added to precipitate protein for 10 min and followed by 16000×g centrifugation for 10 min at 4°C. The pellet was resuspended in 250 μL HU buffer (8 M urea, 5% SDS, 200 mM Tris-HCl pH 6.8, 1 mM EDTA, 5% β-mercaptoethanol and 1% Bromophenol blue) and incubated in 65°C for 10 min, followed by 16000×g centrifugation for 5 min at RT. The supernatant was subjected to SDS-PAGE for protein analysis.
Antibodies: mouse anti-FLAG M2 antibody (Sigma), mouse anti-Por1 monoclonal antibody (Invitrogen), mouse anti-Cox2 monoclonal antibody (Invitrogen), mouse anti-Pgk1 monoclonal antibody (Invitrogen), mouse anti-GFP monoclonal antibody (Roche, clone 7.1 and 13.1), and mouse anti-HA monoclonal antibody (Roche, clone 12CA5).
Polysome analysis
~100 OD of cells were mixed with 100 μg/mL cycloheximide and agitated at 30°C for 15 min. The cells were centrifuged and resuspended in 0.5 mL lysis buffer (10 mM Tris-Cl pH 7.5, 100 mM NaCl, 30 mM MgCl2, cycloheximide 200 μg/mL, Heparin 200 μg/mL, 0.1% β-mercaptoethanol, 1X EDTA-free protease inhibitor cocktail (Roche), 1 mM EDTA, 1 mM PMSF, 5 μM pepstatin A, 10 μM leupeptin, 0.2 mM Na-orthovanadate, 10 mM β-glycerolphosphate, 10 mM NaF, 10m M NaN3 and 23 U/mL RNase inhibitor) and then mixed with 0.5 mL glass beads. The cells were lysed by bead beating 6 × 20 s. After 16000×g 1min at 4°C to collect the supernatant, 0.5 mL lysis buffer was added to the tube, bead-beated for 20 s and the supernatant was collected after centrifugation at 16000×g 1min at 4°C. After another 16000×g centrifugation for 5min at 4°C, 200 μL supernatant was loaded on top of pre-cooled sucrose gradients (7–47 %). Polysomes were fractionated by centrifugation after 35000 rpm for 3 h at 4°C with SW41 Ti rotor. The gradient was continuously collected from the top by densi-flow (Buchler), and the collection line was connected to UV detector to continuously monitor the 254nm absorbance. 19 fractions (0.6 mL/fraction) were collected by fraction collector (AKTA Frac-900). Protein was precipitated with 6% TCA and blotted with anti-FLAG antibody. RNA was purified with Masterpure Yeast RNA Purification kit (Epicentre) according to the protocol and run in 1.5% agarose gel to visualize rRNA.
Live cell imaging
Cells were grown in SCD to log phase before being imaged using the Cellasic microfluidics platform (Y04C-02). Cells were exposed to continuous flow (2psi) of either SCD or SCGE. Images were taken with Deltavision microscope with 100X oil immersion objective and processed by Fiji Is Just ImageJ (FIJI).
β-Estradiol inducible reporter system
For GFP Western blot analysis, the PATC1-GEV and PGAL1-yEGFP-3UTR sequences were cloned into HO-Kan-HO plasmid. The whole sequence was amplified by PCR and then introduced into the HO locus using homologous recombination. For reporter mRNA induction, cells were grown in YPD to log phase and 100 nM of β-estradiol was added for 45 min to induce transcription. Cells were then spun down and washed twice with YPD or YPGE to remove residual β-estradiol. Then the cells were grown in either YPD or YPGE for 0, 30, 60, 90, and 180 min to assess the amount of yEGFP mRNA and protein. To quantitate reporter mRNA, total RNA was purified with Masterpure Yeast RNA Purification kit (Epicentre) according to the protocol. cDNA was made with Superscript III reverse transcriptase (Invitrogen). Gene expression level was analyzed by qRT-PCR with SYBR Green (Invitrogen).
For the 35S-Met pulse assay, a similar procedure was used with a few modifications. Cells were grown in SCD-Met to log phase and 100 nM of β-estradiol was added for 30 min to induce transcription. Cells were then spun down and washed twice with SCD-Met or SCGE-Met to remove residual β-estradiol. Then the cells were grown in either SCD-Met or SCGE-Met for 15 min before adding 35S-Met to 100mCi/ml. Samples were taken in 15 min, 30 min after adding 35S-Met. GFP was immunoprecipitated with GFP antibody (Roche) and the samples were subjected to SDS-PAGE and analyzed by phosphorimaging.
Generation of a library of Puf3p phosphomutants and growth rate measurements
1:9 molar ratio of WT and PUF3(24A) templates were mixed in the PCR reaction to perform the staggered extension process (StEP) technique. PCR was performed with CloneAmp HiFi PCR premix and the PCR cycling condition was 94°C 30 s, 48°C 10 s, 94°C 30 s, and 68°C 10 s for 40 cycles. The mixed product was then amplified to transform puf3Δ cells. After transformation, cells were plated on YPGE plates. 14 growth-competent colonies and 1 slow-growing colony, as a negative control, were picked and their PUF3 allele was sequenced to identify the number of S/T to A mutations. Their growth rates in YPGE compared to PUF3 WT and PUF3(24A) were measured for 9 h. All growth rates were normalized to PUF3(24A).
Ribonucleoprotein immunoprecipitation (RIP)
Approximately 100 ODs of PUF3-FLAG cells were collected in YPD and after 4 h in YPGE. Cells were harvested and resuspended in 0.5 mL lysis buffer (100 mM Tris-Cl pH 7.5, 50 mM NaCl, 0.1% Tween-20, 10% glycerol, 0.1% β-mercaptoethanol, 1X EDTA-free protease inhibitor cocktail (Roche), 1 mM EDTA 1 mM EGTA, 1 mM PMSF, 5 μM Pepstatin A, 10 μM Leupeptin, 0.2 mM sodium orthovanadate, 10 mM β-glycerolphosphate, 60 mM NaF, 10 mM NaN3, and 10U Riboguard (Epicentre) per milliliter) and lysed by bead beating. Cell lysate was incubated with non-coated (negative control) or anti-FLAG M2 antibody coated Dynabead (Invitrogen) at 4°C for 1 h. Beads were then washed with 1 ml lysis buffer for three times. 1/8 amount of beads were treated with sample buffer to elute bound Puf3p and subjected to SDS-PAGE and Western blot for protein quantification. 7/8 of beads were subjected to RNA purification with MasterPure Yeast RNA Purification Kit following the manufacturer's protocol. qRT-PCR was done with SYBR green method. The sample with non-coated Dynabead was used as a negative control to quantify the nonspecific RNA binding. The fold of RNA enrichment was the RNA amount bound by Puf3p divided by the background and normalized with Puf3p level. COX17 is a Puf3p target RNA and COX12 is non-Puf3p target. Primers are listed in Supplemental Table S2
Supplementary Material
Highlights.
The PUF protein Puf3 is phosphorylated upon glucose depletion
Phosphorylation of Puf3 switches fate of mRNAs from degradation to translation
The low complexity region of Puf3 is highly phosphorylated and regulates localization
A phosphomutant of Puf3 becomes retained in punctate foci and may be toxic
ACKNOWLEDGMENTS
We thank members of the Tu Lab for helpful discussions, Randal Halfmann for providing plasmids and helpful discussions, E. O'Shea for providing the PKA analog-sensitive strain, K. Shokat for the 1NM-PP1 inhibitor, and M. Springer for CellAsic plates. This work was supported by a HHMI International Student Fellowship (C-D.L.), NIH grant R01GM094314 (B.P.T.), and a Packard Fellowship (B.P.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTARY INFORMATION Supplementary Information includes 7 figures, 3 tables, and 2 movies.
AUTHOR CONTRIBUTIONS C-D.L. and B.P.T. conceived the project, designed the experiments, and wrote the manuscript. C-D.L. performed all the experiments.
REFERENCES
- Archer SK, Luu VD, de Queiro RA, Brems S, Clayton C. Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS pathogens. 2009;5:e1000565. doi: 10.1371/journal.ppat.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschlen S, Amillet JM, Guiard B, Fournier A, Marcireau C, Bolotin-Fukuhara M. The S. Cerevisiae HAP complex, a key regulator of mitochondrial function, coordinates nuclear and mitochondrial gene expression. Comparative and functional genomics. 2003;4:37–46. doi: 10.1002/cfg.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes & development. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- Eliyahu E, Pnueli L, Melamed D, Scherrer T, Gerber AP, Pines O, Rapaport D, Arava Y. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Molecular and cellular biology. 2010;30:284–294. doi: 10.1128/MCB.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17675–17680. doi: 10.1073/pnas.0503803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes & development. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, Gerst JE. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA (New York, N.Y.) 2011;17:1551–1565. doi: 10.1261/rna.2621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. Yeast carbon catabolite repression. Microbiology and molecular biology reviews : MMBR. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. The Journal of cell biology. 2007;176:197–207. doi: 10.1083/jcb.200606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS biology. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. The Journal of biological chemistry. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- Houshmandi SS, Olivas WM. Yeast Puf3 mutants reveal the complexity of Puf-RNA binding and identify a loop required for regulation of mRNA decay. RNA (New York, N.Y.) 2005;11:1655–1666. doi: 10.1261/rna.2168505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JS, Jr., Houshmandi SS, Lopez Leban F, Olivas WM. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA (New York, N.Y.) 2004;10:1625–1636. doi: 10.1261/rna.7270204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JA, Rose NC, Goldsworthy B, Goga A, L'Etoile ND. A 3'UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron. 2009;61:57–70. doi: 10.1016/j.neuron.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3' UTR controls miR-221 and miR-222 accessibility. Nature cell biology. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3936–3941. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science (New York, N.Y.) 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science (New York, N.Y.) 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelandais G, Saint-Georges Y, Geneix C, Al-Shikhley L, Dujardin G, Jacq C. Spatio-temporal dynamics of yeast mitochondrial biogenesis: transcriptional and post-transcriptional mRNA oscillatory modules. PLoS computational biology. 2009;5:e1000409. doi: 10.1371/journal.pcbi.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast (Chichester, England) 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- McIsaac RS, Silverman SJ, McClean MN, Gibney PA, Macinskas J, Hickman MJ, Petti AA, Botstein D. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Molecular biology of the cell. 2011;22:4447–4459. doi: 10.1091/mbc.E11-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Seydoux G. The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development (Cambridge, England) 2010;137:1787–1798. doi: 10.1242/dev.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley interdisciplinary reviews. RNA. 2011;2:471–492. doi: 10.1002/wrna.69. [DOI] [PubMed] [Google Scholar]
- Miller MA, Russo J, Fischer AD, Lopez Leban FA, Olivas WM. Carbon source-dependent alteration of Puf3p activity mediates rapid changes in the stabilities of mRNAs involved in mitochondrial function. Nucleic acids research. 2014;42:3954–3970. doi: 10.1093/nar/gkt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- New AM, Cerulus B, Govers SK, Perez-Samper G, Zhu B, Boogmans S, Xavier JB, Verstrepen KJ. Different levels of catabolite repression optimize growth in stable and variable environments. PLoS biology. 2014;12:e1001764. doi: 10.1371/journal.pbio.1001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. The EMBO journal. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM. Glucose-sensing and -signalling mechanisms in yeast. FEMS yeast research. 2002;2:183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x. [DOI] [PubMed] [Google Scholar]
- Roosen J, Engelen K, Marchal K, Mathys J, Griffioen G, Cameroni E, Thevelein JM, De Virgilio C, De Moor B, Winderickx J. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Molecular microbiology. 2005;55:862–880. doi: 10.1111/j.1365-2958.2004.04429.x. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nature medicine. 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PloS one. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler IE, de la Cruz BJ, Prieto S. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. The international journal of biochemistry & cell biology. 1998;30:1175–1193. doi: 10.1016/s1357-2725(98)00086-7. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza GM, da Silva AM, Kuspa A. Starvation promotes Dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development (Cambridge, England) 1999;126:3263–3274. doi: 10.1242/dev.126.14.3263. [DOI] [PubMed] [Google Scholar]
- Suh N, Crittenden SL, Goldstrohm A, Hook B, Thompson B, Wickens M, Kimble J. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA (New York, N.Y.) 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP. Ultradian metabolic cycles in yeast. Methods in enzymology. 2010;470:857–866. doi: 10.1016/S0076-6879(10)70035-5. [DOI] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science (New York, N.Y.) 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Ulbricht RJ, Olivas WM. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA (New York, N.Y.) 2008;14:246–262. doi: 10.1261/rna.847408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, Goldstrohm AC. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. The Journal of biological chemistry. 2012;287:36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science (New York, N.Y.) 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Molecular cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA (New York, N.Y.) 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- Zhao H, Zha W. In vitro 'sexual' evolution through the PCR-based staggered extension process (StEP) Nature protocols. 2006;1:1865–1871. doi: 10.1038/nprot.2006.309. [DOI] [PubMed] [Google Scholar]
- Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TM. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20192–20197. doi: 10.1073/pnas.0812079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.